Abstract
The vegetative cells of the filamentous cyanobacterium Nostoc punctiforme can differentiate into three mutually exclusive cell types: nitrogen-fixing heterocysts, spore-like akinetes, and motile hormogomium filaments. A DNA microarray consisting of 6,893 N. punctiforme genes was used to identify the global transcription patterns at single time points in the three developmental states, compared to those in ammonium-grown time zero cultures. Analysis of ammonium-grown cultures yielded a transcriptome of 2,935 genes, which is nearly twice the size of a soluble proteome. The NH4+-grown transcriptome was enriched in genes encoding core metabolic functions. A steady-state N2-grown (heterocyst-containing) culture showed differential transcription of 495 genes, 373 of which were up-regulated. The majority of the up-regulated genes were predicted from studies of heterocyst differentiation and N2 fixation; other genes are candidates for more detailed genetic analysis. Three days into the developmental process, akinetes showed a similar number of differentially expressed genes (497 genes), which were equally up- and down-regulated. The down-regulated genes were enriched in core metabolic functions, consistent with entry into a nongrowth state. There were relatively few adaptive genes up-regulated in 3-day akinetes, and there was little overlap with putative heterocyst developmental genes. There were 1,827 differentially transcribed genes in 24-h hormogonia, which was nearly fivefold greater than the number in akinete-forming or N2-fixing cultures. The majority of the up-regulated adaptive genes were genes encoding proteins for signal transduction and transcriptional regulation, which is characteristic of a motile filament that is poised to sense and respond to the environment. The greatest fraction of the 883 down-regulated genes was involved in core metabolism, also consistent with entry into a nongrowth state. The differentiation of heterocysts (steady state, N2 grown), akinetes, and hormogonia appears to involve the up-regulation of genes distinct for each state.
The vegetative cells of the filamentous, oxygenic, phototrophic cyanobacterium Nostoc punctiforme have the potential for four developmental alternatives (32). In the presence of replete nutrients, including a source of combined nitrogen such as ammonium, light, and CO2 or in the dark with glucose or fructose as a source of reduced carbon, a permissive vegetative cell cycle consisting of growth and division is maintained. Nutrient depletion or change elicits three mutually exclusive differentiation responses. The differentiated cells are morphologically and physiologically distinct from vegetative cells. The question to be asked is, what is the extent of differential gene expression in the developmental states of N. punctiforme?
Depletion of combined nitrogen induces the transcription of genes encoding the nitrogenase enzyme complex. Nitrogenase expression is preceded by the differentiation of heterocysts, which are specialized microoxic cells in which oxygen-sensitive nitrogen fixation takes place. The microoxic environment of heterocysts is achieved by inactivation of the photosystem II oxygen-evolving reaction, an increased rate of aerobic respiration, and the synthesis of a bilayer envelope that retards diffusion of gases (44). Heterocysts are terminally differentiated; they constitute 10% or less of the cells in a filament and are singly spaced in a nonrandom pattern (33). They are the source of organic nitrogen for vegetative cells and a sink for photosynthate from vegetative cells. Heterocyst differentiation allows continued vegetative cell growth in a habitat depleted of combined nitrogen. The formation and maintenance of heterocysts are variously estimated to involve the differential expression of 140 (43) to 600 (12) to more than 1,000 (28) genes, and this is the most highly studied cyanobacterial differentiation event (for reviews, see references 1, 17, 22, 33, 43, 44, and 47).
When limited in cellular energy, typically in response to phosphate deprivation, vegetative cells differentiate into spore-like cells termed akinetes. Although akinetes retain some metabolic activity (40), they do not grow. Akinetes are resistant to cold and desiccation, thereby allowing perennation or longer periods of survival. This is a transient differentiation process; when environmental conditions are appropriate for growth, akinetes germinate, and the emerging filaments continue a vegetative cell cycle. Akinete differentiation is confined to the heterocyst-forming cyanobacteria (36). Based on this taxonomic clustering and some biochemical and genetic similarities, there is speculation that akinetes served as the evolutionary progenitors of heterocysts (44). Although a priori there is no obvious reason that heterocyst differentiation and akinete differentiation should be mutually exclusive, continued heterocyst differentiation is generally not observed when environmental conditions favor akinete differentiation. In heterocyst-containing filaments of N. punctiforme, akinetes first appear midway in the interval between two heterocysts, with subsequent differentiation events radiating in both directions away from the first akinete and towards the adjacent heterocysts (32). However, in a glucose-6-phosphate dehydrogenase (zwf) mutant of N. punctiforme that is unable to fix nitrogen or grow in the dark, vegetative cells synchronously differentiate into akinetes when they are placed in the dark (4). Five genes that are differentially transcribed during akinete differentiation have recently been identified (6, 48).
Vegetative filaments of N. punctiforme are nonmotile. Gliding and buoyant mobility can be induced following the transient differentiation of vegetative filaments into small-celled filaments called hormogonia. No single unique environmental factor induces hormogonium differentiation; rather, changes in a multiplicity of factors, including depleted and replete nutrients, light quality, the presence of symbiotic plant partners, or dilution into fresh medium, stimulate hormogonium differentiation to various degrees (33, 39). Once induced, N. punctiforme filaments proceed through a non-N2-fixing hormogonium differentiation cycle that lasts for 72 to 96 h (9). Upon entry into the cycle, there is a cessation of net increases in biomass, DNA, protein, and pigments (9, 10, 20), while cell division continues, resulting in an increase in the number of cells, all of which are smaller and shaped differently than the parental vegetative cells (9, 10, 32, 36, 39). Since filamentous cyanobacteria contain multiple copies of the chromosome (19), there is a high probability that hormogonium cells contain at least one genome copy. In N. punctiforme, gliding motility is maximal between 18 and 72 h after induction (9). Once the filaments become sessile, biomass increase resumes and the filaments exit the hormogonium cycle to reenter the vegetative cell cycle in the presence or absence of combined nitrogen. There have been few molecular studies of events in the hormogonium cycle. It is known that the transcription of genes encoding gas vesicle proteins, pili, (39), an alternative sigma subunit of RNA polymerase, and a Clp protease (8) is up-regulated during entry into the cycle, while synthesis of phycobiliproteins is down-regulated (39).
The complete 9.059-Mbp genome sequence of N. punctiforme strain ATCC 29133 has been determined. A preliminary report has been published (34), and the sequence can be accessed at the Integrated Microbial Genomes site (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). The complete genome sequence allows entry into systems-level approaches to identification of gene expression profiles. We report here the construction of a DNA microarray of the N. punctiforme genome. This array was used to identify genes that are transcribed differentially during heterocyst, akinete, and hormogonium differentiation or function by reference to transcription during growth of vegetative cells on NH4+.
MATERIALS AND METHODS
Cultures and culture conditions.
N. punctiforme strain ATCC 29133 (= PCC 73102) and its derivatives were grown with shaking at 100 to 120 rpm and at 23 to 25°C under ca. 19 to 46 μmol photons/m2/s of cool white fluorescent light in the standard minimal salts medium of Allan and Arnon, diluted fourfold, as described previously (14). Since heterocyst differentiation results in perpetuation of a modified vegetative cell cycle, comparisons were made between steady-state NH4+- and N2-grown cultures. A spontaneously derived strain of N. punctiforme, UCD 102, which does not form hormogonia, was used for these and subsequent N2 growth experiments so that heterocyst differentiation was not complicated by formation of hormogonia. Duplicate 50-ml cultures from the same N2-grown inoculum (200 μl) were incubated for 7 days under the same environmental conditions, with one culture supplemented with 2.5 mM NH4Cl and 5.0 mM morpholinepropanesulfonic acid (MOPS) and the other culture without these compounds (N2 steady state). Cultures grown with NH4+ were devoid of heterocysts, as determined by microscopy, and were used as the reference. Under these low-cell-density culture conditions, the heterocyst frequency of the N2 steady-state culture was, on average, 8.7%. The cultures were harvested at densities of 2 to 3 μg chlorophyll a per ml by centrifugation at 1,000 × g for 5 min and stored at −80°C for subsequent RNA extraction.
The gliding motility of N. punctiforme hormogonia is pronounced 24 h after induction, and this time point was chosen for these experiments, using the symbiotically competent strain UCD 153. Cultures were grown in 500 ml of minimal medium supplemented with 2.5 mM NH4Cl and 5.0 mM MOPS for 7 days, reaching densities of 2 to 3 μg chlorophyll a/ml; the cultures were then synchronized for differentiation of hormogonia by centrifugation as described above and suspended in fresh medium without supplementation with combined nitrogen. Time zero (T0) samples were collected at this time. After 24 h under standard incubation conditions, cultures were examined by microscopy for verification of hormogonium formation, and samples showing essentially 100% differentiation were collected by centrifugation and stored as described above.
Akinete differentiation occurs over a period of up to 6 days (4). A gene with enhanced expression in akinetes, avaK, shows strong transcription 3 days following induction (4), as well as does the onset of resistance to desiccation, cold, or lysozyme treatments (5). Therefore, cells in the process of differentiation into akinetes were induced by incubation in the dark for 3 days of a 50-ml culture of the zwf::PpsbA-npt insertion mutant CSUN 466 (a reconstruction of UCD 466) (18) previously grown for 5 days in minimal medium supplemented with 2.5 mM NH4Cl and 5.0 mM MOPS plus 50 mM fructose. Under these conditions, strain CSUN 466 shows synchronized differentiation into akinetes, and approximately 50% of the cells are resistant to cold and desiccation at day 3 (4, 5). Cells were collected at T0 and at 3 days by centrifugation and stored as described above.
RNA isolation, cDNA synthesis, and dye labeling.
Frozen samples were subjected to breakage using a mini bead beater (Biospec) in the presence of phenol, 0.5-mm zirconium/silicate beads, and Celite as described previously (38). RNA in the aqueous phase was precipitated using 4 M lithium chloride, 20 mM Tris (pH 7.4), 10 mM EDTA (pH 8). The samples were then further purified using QIAGEN RNeasy columns by following the manufacturer's specifications, including the on-column DNase digestion to eliminate DNA contamination. mRNA was enriched from 10 μg of total RNA using a cyanobacterial capture module in a MicrobExpress rRNA subtraction kit (Ambion); the average yield was 1.05 μg of RNA. Fifteen micrograms of total RNA was used in each cDNA synthesis reaction in the presence of 4 μg of a random 18-mer, 10 μg of random hexamers, and a deoxynucleoside triphosphate mixture containing aminoallyl-modified nucleotides or 1 μg of purified mRNA and 10 μg of random hexamers; otherwise, the manufacturer's instructions (Invitrogen) were used. Alexa 647 and Alexa 555 dyes (Invitrogen) were coupled to samples prior to hybridizations. The fluorescently labeled probes were purified using QIAGEN PCR “Qiaquick” columns and were quantified according to the manufacturer's instructions.
Array construction and hybridization.
The N. punctiforme array consists of internal gene fragments ranging in size from 53 to 725 bp, with the great majority between 200 and 700 bp. Unique primer pairs for each gene in the array were designed and synthesized under a contract with Operon. PCRs were performed under standard conditions using genomic DNA as the template. The products were then purified using QIAGEN columns in a 96-well format (Qiaquick 96), and the concentrations were adjusted to between 0.1 and 0.3 μM. There are 6,893 individual gene fragments in the array; in addition, five genes are spotted in duplicate as controls (rbcS [NpF4197], rbcL [NpF4195], rpoC [NpF4987], glnA [NpR5387], and hemL [NpR4096]), four concentrations of sheared genomic DNA, and salmon sperm DNA as a negative control. Absent in the array are 351 of the 416 transposase genes with multiple members in various families, three psbA and one psbD family member genes, and 75 hypothetical genes less than 200 bp long. DNA fragments were spotted in duplicate onto aminosilane A+-coated slides (Schott Nexterion) by the ArrayCore Facility at UC Davis (array.ucdavis.edu). All hybridizations were done with an automated Tecan HS4800 hybridization station at 50°C. Slides were scanned using a GenePix 4000A scanner (Axon Instruments).
Data analysis.
Raw data in a .gpr file format were generated using the GenePix Pro 3.0 software. Three biological replicates were used in the analysis for each of the differentiation states, and each of the replicates was analyzed with dye swaps. Thus, data from six hybridizations, with T0 as the reference, for each experiment (three biological replicates plus the reciprocal dye swaps) for a total of 12 technical replicates (since each array was printed in duplicate on the slide) were used in each analysis. To determine which genes showed statistically significant differential expression in the three developmental states relative to T0 expression, the free source software package LIMMA GUI (Linear model for microarray analysis graphical user interface) (37), written in the R-programming language (www.R-project.org) and available through Bioconductor (www.Bioconductor.org), was employed. The data were first normalized within arrays using Print-Tip Loess and across arrays in the analysis using Aquantile methods. The Linear model fit was computed using the duplicates on the arrays and the least-squares method, and all P values were adjusted using the Benjamini method (reference 37 and references therein) for controlling for the false discovery rate. Statistically significantly differentially expressed genes were defined for N. punctiforme arrays as the genes with B values of >0; this corresponded to adjusted P values of <1.49 × 10−2 for the N2 steady state, 4.0 × 10−3 for hormogonia, and 1.02 × 10−2 for akinetes. The raw data, as .gpr files, are available at http://microbiology.ucdavis.edu/faculty/jcMeeks/arrays/.
Functional gene assignments.
Open reading frames were initially assigned to a functional category based on Clusters of Orthologous Groups (COG) identities from the genome computational annotation. The COG identities were manually verified or corrected by computational sequence comparisons to protein databases; the proteins with Interpro (TIGRfam, Pfam, and SwissPro) identities were accepted as accurate. Gene products were assigned to subcategories based on their physiological roles from metabolic tables and pathways (EcoCyc) or, in some instances, genetic phenotypes.
As a means to organize large data sets in a physiological framework, the NH4+-grown and differentially transcribed genes were grouped into the following five functional categories based on their gene products (2): (i) core metabolism, which includes proteins required for vegetative cell growth and involved in the synthesis of precursors and monomers, ion incorporation, cofactors, polymers, assembly of cell structures, and cell division and genome segregation; (ii) transport metabolism, which involves import and export protein complexes; (iii) adaptive metabolism, including alternative C, N, P, and S macronutrient sources, signal transduction, transcriptional regulation, stress, secondary products, and developmental processes; (iv) selfish metabolism, including phage and transposition functions; and (v) unassigned metabolism, including proteins with known domains or motifs but unknown physiological functions, conserved hypothetical and hypothetical proteins, and pseudogenes (Table 1) (a detailed breakdown is provided in File S1 in the supplemental material). Some category assignments, such as proteases and DNA/RNA metabolic enzymes, are subjective and were defined based on the presence or absence in an NH4+-grown soluble proteome of N. punctiforme (2) and differential expression profiles in this study. This is a dynamic database, in which category assignments can change as experimental identification of the functional role of a gene product occurs.
TABLE 1.
Functional distribution of genes expressed in the NH4+-grown transcriptome and the differentially expressed transcriptomes of N2-grown cultures, 3-day developing akinetes, and 24-h hormogonia
| Functional group | No. of genes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genome | NH4+ transcriptome | N2-grown cultures, up-regulated | N2-grown cultures, down-regulated | Akinetes, up-regulated | Akinetes, down-regulated | Hormogonia, up-regulated | Hormogonia, down-regulated | |
| Core metabolism | 1,590 | 1,081 | 90 | 29 | 67 | 154 | 179 | 430 |
| Precursor | 101 | 73 | 15 | 4 | 4 | 19 | 13 | 39 |
| Cofactor | 356 | 103 | 6 | 7 | 8 | 8 | 17 | 39 |
| Energy | 137 | 101 | 17 | 1 | 2 | 40 | 8 | 58 |
| Monomers | 256 | 171 | 15 | 1 | 14 | 26 | 23 | 79 |
| Polymers | 529 | 398 | 19 | 5 | 26 | 51 | 51 | 148 |
| Cell envelope | 272 | 145 | 9 | 11 | 9 | 5 | 51 | 35 |
| Cell division | 29 | 16 | 0 | 0 | 0 | 0 | 1 | 4 |
| Reactive oxygen species/metabolic protection | 68 | 46 | 5 | 0 | 2 | 2 | 8 | 19 |
| Storage | 41 | 28 | 4 | 0 | 1 | 3 | 7 | 9 |
| Transport metabolism | 399 | 174 | 19 | 12 | 8 | 12 | 62 | 50 |
| ABC systems | 206 | 85 | 10 | 8 | 3 | 3 | 23 | 21 |
| Permease | 15 | 9 | 1 | 0 | 0 | 0 | 6 | 3 |
| Porter | 68 | 30 | 2 | 3 | 1 | 4 | 8 | 15 |
| Channel | 26 | 12 | 1 | 0 | 0 | 4 | 2 | 4 |
| Ion ATPase | 23 | 8 | 1 | 0 | 1 | 0 | 8 | 3 |
| Secretion/export | 58 | 28 | 4 | 1 | 3 | 0 | 13 | 4 |
| Regulation | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| Adaptive metabolism | 992 | 317 | 104 | 14 | 29 | 13 | 204 | 63 |
| Carbon | 24 | 12 | 0 | 0 | 1 | 0 | 2 | 6 |
| Nitrogen | 59 | 13 | 32 | 0 | 1 | 0 | 18 | 2 |
| Phosphate/sulfur | 10 | 3 | 0 | 0 | 1 | 0 | 5 | 0 |
| Development | 63 | 14 | 32 | 0 | 3 | 1 | 17 | 2 |
| Circadian | 5 | 2 | 0 | 0 | 0 | 0 | 1 | 0 |
| Secondary | 113 | 16 | 5 | 0 | 2 | 5 | 5 | 10 |
| Conjugation | 8 | 2 | 0 | 1 | 0 | 0 | 2 | 1 |
| Signal transduction | 362 | 162 | 16 | 7 | 12 | 2 | 85 | 29 |
| Stress | 31 | 25 | 8 | 1 | 5 | 2 | 21 | 3 |
| Motility | 16 | 8 | 0 | 4 | 0 | 0 | 10 | 0 |
| Taxis | 33 | 14 | 1 | 0 | 0 | 0 | 18 | 0 |
| Transcriptional regulation | 119 | 47 | 10 | 1 | 4 | 2 | 20 | 10 |
| Energy, monomer, polymer | 144 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Selfish metabolism | 457 | 15 | 1 | 5 | 1 | 0 | 3 | 6 |
| Phage | 23 | 9 | 0 | 5 | 1 | 0 | 2 | 6 |
| Transposase | 416 | 6 | 1 | 0 | 0 | 0 | 1 | 0 |
| Unassigned metabolism | 3,898 | 1,348 | 159 | 62 | 150 | 65 | 496 | 334 |
| Conserved hypothetical | 1,945 | 712 | 80 | 41 | 88 | 38 | 265 | 178 |
| Hypothetical | 570 | 103 | 15 | 4 | 9 | 7 | 67 | 24 |
| Unassigned | 1,103 | 515 | 62 | 16 | 53 | 18 | 155 | 128 |
| Pseudogenes | 280 | 18 | 2 | 1 | 1 | 1 | 9 | 4 |
| Total | 7,336 | 2,935 | 373 | 122 | 255 | 242 | 944 | 883 |
RESULTS AND DISCUSSION
Overall extent of differential gene expression.
Hybridization results, including signal strength, background, and optimal incubation temperature, were similar whether cDNA was synthesized from total RNA or from enriched mRNA (data not shown). Therefore, all subsequent experiments were conducted with total RNA.
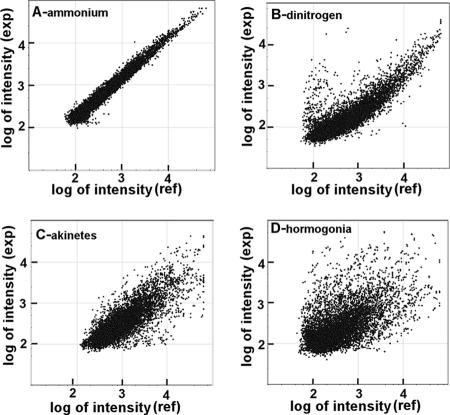
The results of a single biological and two technical replicates for NH4+-grown cells and each developmental state are shown in the scatter plots in Fig. 1. When the sets of differently labeled cDNAs from NH4+-grown cultures were self-hybridized, few gene spots showed more than a 1.5-fold departure from a linear function (Fig. 1A), and plots of replicate self-hybridizations showed different low-level outliers. These results verified a lack of dye bias in labeling of the cDNAs and established that in replicate experiments departures from linearity are experimentally significant and not technical artifacts.
FIG. 1.
Scatter plots of a single biological and two technical replicates of two-color hybridization intensities. The y axes represent experimental (exp) intensities, and the x axes represent the reference (ref) (T0 or steady-state NH4+-grown) intensities.
Ehira and Ohmori (13) and Ehira et al. (12) reported 396 (see below) and 600 differentially expressed genes in Anabaena sp. strain PCC 7120, respectively, 24 h following deprivation of combined nitrogen. The number of statistically significant N. punctiforme N2 steady-state differentially expressed genes (495 genes) is between the two Anabaena sp. strain PCC 7120 values, and 75% (373) of these genes were up-regulated (Fig. 1B). Developing akinetes displayed a comparable number, 497 differentially expressed genes, but these genes were about equally up-regulated (255 genes) and down-regulated (242 genes) (Fig. 1C). Actively motile 24-h hormogonia exhibited the greatest extent of differential gene expression, which involved 1,827 genes; 52 and 48% of these genes were up- and down-regulated, respectively (Fig. 1D).
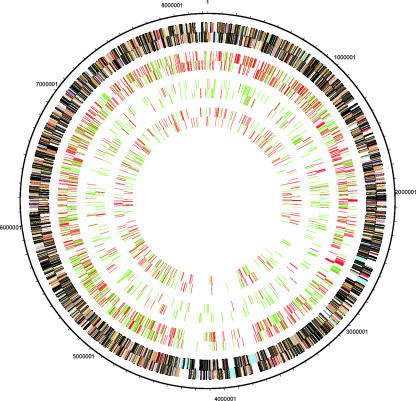
There are no distinct and extended genetic islands of developmentally up- or down-regulated genes in any of the three differentiation states; the genes appear to be randomly distributed in the N. punctiforme chromosome (Fig. 2). The only appearance of island distribution in Fig. 2 is the nondifferentially transcribed blocks of genes at approximately 6:00 o'clock that putatively encode proteins for the synthesis of cyclic peptide secondary metabolites.
FIG. 2.
Circular map of the chromosome. The outside circle depicts the annotated genome with color-coded COG categories of genes. The next circle shows differentially expressed genes in 24-h hormogonia, with up-regulated genes indicated by red and down-regulated genes indicated by green; this is followed by circles showing differentially expressed genes in steady-state N2-grown cultures and then 3-day akinetes.
Functional categories of ammonium-grown cultures.
Gene expression levels from the NH4+-grown self-hybridization shown in Fig. 1A that were three times the mean red channel background signal (75 fluorescent units) were arbitrarily accepted as evidence for expression (Table 1). This analysis yielded expression signals from 2,935 genes, nearly twice the number of genes for the soluble NH4+-grown proteome (1,572 genes) (2). As one approach to validate the minimum threshold, the list was compared to the composite differentially transcribed genes in all developmental states. If a gene is statistically down-regulated, it should have been expressed at some level in the NH4+-grown culture. Down-regulated genes were 14% of the differentially transcribed genes with NH4+ expression levels that were less than three times the background, 40% of the genes with expression levels that were between three and four times the background, and 59% of the genes with expression levels that were more than five times the background. These values imply a conservative threshold for the lower level of expression.
In general, the fraction of expressed genes clustered similar to the genome categories (Table 1), although the fraction of genes encoding core metabolic functions was proportionally higher, while the fractions of genes encoding adaptive and selfish metabolism were proportionally lower. This result is reasonable since the array contains few gene fragments encoding putative transposases, and many genes encoding adaptive functions are expected to be more highly expressed under adaptive conditions. The increased proportion of expression of genes involved in core metabolism is similar to the proportion observed in the soluble proteome (2) and would be physiologically expected in a photoautotrophic NH4+-grown transcriptome, as these conditions are the most permissive culture conditions for N. punctiforme. The core metabolic category contains more than 60 gene families with two to eight members, 9 of which are involved in precursor carbon metabolism. One hundred fifteen genes were expressed from the five N. punctiforme plasmids (691 total open reading frames). This set of genes is a reasonable NH4+-grown transcriptome of N. punctiforme, but we do not suggest that it is definitive. It provides an internal control for genes that are differentially transcribed in cellular differentiation.
Functional categories of dinitrogen-grown cultures.
The most striking result obtained with N2-grown cultures is that the majority of the statistically significantly regulated genes were up-regulated (373 genes). Of these genes, 43% were genes involved in unassigned metabolism, 28% were genes involved in adaptive metabolism, and 24% were genes involved in core metabolism (Table 1). The unassigned metabolic genes in all differentiation states are clearly a reservoir of undescribed developmental, stress, and metabolic potential in N. punctiforme. The majority of the N2-grown up-regulated adaptive and core genes can be predicted from the results of many studies of heterocyst differentiation and function (17, 22, 31, 33, 43, 44, 47). The presence of such known genes (e.g., het, dev, pat, hgl, hep, nif, hup, cyo, zwf, and gnd [see below]) provides an internal control to assess genome coverage.
The adaptive metabolic category includes known nitrogenase (nif), hydrogenase (hup), and nitrate assimilation structural genes and genes encoding enzymes for synthesis of the unique polysaccharide (hep) and glycolipid (hgl) heterocyst envelope. It is worth noting that steady-state expression of nitrate assimilation genes was relatively high in the absence of nitrate or nitrite. In Anabaena sp. strain PCC 7120, these genes are derepressed upon combined-nitrogen starvation but are repressed again as cultures grow with N2 as the nitrogen source (21). Six genes encoding regulatory proteins involved in heterocyst differentiation and nitrogen assimilation are in the adaptive category; hetR (NpR1722), hetF (NpF3553), nrrA (NpR3907), and patB (NpR0334) are in the development subgroup, while ntcA (NpF5511) and ntcB (NpF1534) plus two genes encoding alternative sigma subunits of RNA polymerase (NpF0307 and NpF4153, neither of which has yet been shown to be essential for differentiation) are in the transcriptional regulation group. Genes encoding other identified heterocyst regulatory proteins, such as devH (NpR6193), hepK (NpR1012), patA (NpF5682), patN (NpF6624), and patU (NpR1831), were not present in the differentially expressed N2 transcriptome, although devH, patA, patN, and patU are in the NH4+-grown transcriptome and DevH, HepK, and PatN are in the NH4+-grown proteome (2). Expression of five genes encoding serine/threonine protein kinases (PK) (NpF1910, NpF5105, NpF6098, NpF6345, and NpR5250), two PK phosphatases (NpF1519 and NpR4927), three histidine kinases (HK) (NpF0957, NpR1236, and NpR4835), and four response regulators (RR) (NpF1800, NpF5044, NpF5659, and NpR1903) was enhanced in the N2 transcriptome, implying that these genes have roles in heterocyst differentiation or steady-state N2 growth. These signal transduction genes are obvious targets for genetic analyses, especially analyses to identify the cognate HKs of NrrA (13) and PatA (27).
The differentially up-regulated genes in the core metabolic category include two putative operons encoding terminal cytochrome c oxidases (NpF0336 [cyoA], NpF0337 [cyoB], and NpF0338 [cyoC]; NpR3535 [cyoC], NpR3536 [cyoB], and NpR3537 [cyoA]), none of which is present in the proteome or expressed in the NH4+ transcriptome, and genes encoding 15 carbon catabolism enzymes that are likely to be involved in reductant generation in heterocysts. Enhanced expression of two cytochrome oxidase complexes (41) and of oxidative pentose phosphate proteins (42) is known to occur in heterocysts. Thirteen genes encoding amino acid metabolic enzymes, including predicted heterocyst enhanced glutamine synthetase (NpR5387) (35), were up-regulated, while the level of the gene encoding vegetative cell-localized glutamate synthase (NpR3877) (29) was not different from the NH4+-grown level. A glutamate dehydrogenase gene (NpF3641) was expressed in the NH4+-grown transcriptome, but it was not differentially transcribed in steady-state N2-grown cultures. The sequential activities of glutamine synthetase and glutamate synthase constitute the dominant route of NH4+ assimilation in cyanobacteria, and glutamate dehydrogenase does not have a significant role (15). The remaining genes do not appear to encode enzymes specific for any complete amino acid biosynthetic pathways. The expression of five genes for synthesis of proteins for metabolism of reactive oxygen species was enhanced in the N2-grown differential transcriptome; three of these genes (NpF2660 [ggt], NpF6212 [dps], and NpR0901 [gor], but not NpR5469 [dps] and NpR6212 [katE]), were also present in the NH4+-grown proteome. The NH4+-grown proteome contains another 25 proteins for metabolism of reactive oxygen (2). Whether the up-regulated gene products are localized in the heterocyst is unknown, but metabolism of reactive oxygen is clearly an important process in N. punctiforme. Nineteen genes encoding import and export transport functions are up-regulated. Their substrates are unknown, but several of the gene products are annotated as Fe and Mo ion transporters, consistent with the demands of nitrogenase. Neither of the two genes encoding an NH4+ transport channel (NpF4661 and NpR3288) was significantly up- or down-regulated compared with the NH4+-grown transcriptome level.
The fact that 122 genes were down-regulated in N2-grown cultures implies that at least some of the gene products may enhance growth in the presence of NH4+. A change from nitrogen-replete (NH4+) to nitrogen-limited (N2) growth is predicted to involve down-regulation of proteins for low-affinity NH4+ assimilation. This was not the result observed, as exemplified by low-affinity glutamate dehydrogenase (see above). The majority of the down-regulated genes are in the unassigned (51%) and core (24%) categories, and most of the core genes encode enzymes of cell envelope and cofactor metabolism. The functionally identified core genes do not exhibit an obvious physiological pattern so that one could predict the basis of their differential expression. Genes encoding three HKs (NpF3675, NpF3677, and NpR3054), two PKs (NpF01289 and NpF1616), and two RRs (NpF3676 and NpR2902) were significantly down-regulated. The NpF3675, NpF3676, and NpF3677 genes may be transcribed as an operon, and the NpF3677 gene product is a hybrid HK with an N-terminal RR receiver domain. Since the collocated RR (NpF3676) has no DNA binding output domain, there may be additional components of this putative phosphorelay signal transduction pathway, or the regulator may modulate activity of another protein.
A recent analysis (46; see also http://www.people.vcu.edu/∼elhaij/microarray/) of the transcriptome data generated by Ehira and Ohmori (13) using Anabaena sp. strain PCC 7120 yielded 396 differentially transcribed genes 24 h following deprivation of combined nitrogen; 189 genes were up-regulated, and 207 genes were down-regulated. There are 144 and 177 N. punctiforme orthologs in the up- and down-regulated clusters of Anabaena sp. strain PCC 7120 differentially transcribed genes, respectively. However, when the Anabaena sp. strain PCC 7120 24-h differential expression database was compared to the N. punctiforme differential steady-state N2-grown organism database, only 42 of the up-regulated and 6 of the down-regulated genes were in both databases (see File S2 in the supplemental material). Most of the common up-regulated genes fell into three categories: 11 genes for heterocyst development (including nrrA but not hetR, hetF, patB, or ntcA); 12 genes for nitrogenase catalytic proteins, assembly, or function; and 12 genes encoding either unassigned or conserved hypothetical proteins. Three of the down-regulated genes also encode conserved hypothetical proteins, plus a cofactor (NpF3770), a cell wall catabolic enzyme (NpF1408), and the ATP binding protein of a transporter (NpR2354). We note that many genes predicted to be transcriptionally enhanced in developing or functional steady-state heterocysts are present in the N. punctiforme steady-state N2-grown database but are not present in the Anabaena sp. strain PCC 7120 24-h database. These genes include the above-mentioned developmental genes, as well as the metabolic genes glnA, zwf, opcA, pgl, and idh, the hydrogen recycling genes hupS and hupL, and the second cyo operon. Moreover, the Anabaena sp. strain PCC 7120 24-h database contains a marked number of up-regulated genes encoding ribosomal proteins and down-regulated genes encoding phycobiliproteins and photosynthetic reaction center and electron transport proteins that are not seen as differentially transcribed genes in N2-grown N. punctiforme. A difference in the two databases could result from the fact that the N. punctiforme data are data from a steady-state culture, while the data for Anabaena sp. strain PCC 7120 were obtained 24 h following nitrogen deprivation. This rationale implies that the Anabaena sp. strain PCC 7120 cultures had not reached steady-state N2 growth conditions by 24 h.
Functional categories of differentiating akinetes.
Unlike N2-grown cultures, 3-day akinete-forming cultures had relatively equal numbers of up- and down-regulated genes (Table 1). The majority of the 255 genes that are up-regulated as akinetes differentiate are in the unassigned category (59%). Akinete differentiation appears to involve relatively few differentially expressed adaptive proteins (11%), and the low number of these proteins may facilitate analysis of their developmental role. Transcription of avaK (NpF5452), encoding an akinete marker protein (6, 48), was enhanced, as was transcription of patA and hetF, whose products are assigned to heterocyst differentiation. The transcription of 12 signal transduction proteins was up-regulated, but only one of these proteins (encoded by NpF6364; a hybrid HK with an RR receiver domain in the N-terminal region) was from a not detectably expressed level in the NH4+-grown state. Four putative transcriptional regulatory proteins were up-regulated: two DNA binding proteins (encoded by NpR3597 and NpR1565, with a Crp family binding domain and a PAS sensor domain) and two sigma subunits (encoded by NpF4153 and NpR4091, the latter of which appears to be specifically expressed in developing akinetes).
The up-regulated core metabolic genes fall into all functional subgroups. The genes involved in cell envelope and cofactor metabolism are the functional counterparts of down-regulated genes in N2-grown cultures, but they are not homologous. Genes encoding polymer synthetic or modification activity do not appear to be essential for growth and survival and, in most cases, appear to be functionally duplicated by other genes. There was not a dramatic increase in the transcription of genes encoding proteins for carbon, nitrogen, or phosphate storage, although akinetes contain glycogen and cyanophycin granules that are visible by light microscopy (1). Presumably, the constitutive level of expression of these genes is sufficient to ensure accumulation of the polymers. None of the four genes determined by differential display to be up-regulated during akinete development (NpF0062, NpF5999, NpF6000, and NpR4070) (6) was present in the akinete transcriptome or in the NH4+-grown transcriptome or proteome. NpR4070 was up-regulated in steady-state N2-grown cultures at a significant but low level. The reasons for this discrepancy are not known, but the discrepancy may be a consequence of the particular time point chosen for analysis; the differential display results were confirmed by highly sensitive real-time quantitative PCR and reporter gene localization (6).
Also in contrast to N2-grown cultures, the majority (64%) of the 242 genes down-regulated in akinete differentiation are involved in core metabolism. These genes include genes encoding proteins essential for energy, precursor, monomer, and polymer metabolism. The down-regulation of such genes is consistent with a cell entering a metabolically quiescent state. Nevertheless, few genes for constitutively transcribed transport proteins were down-regulated during akinete differentiation, implying that there was maintenance of an external assimilatory capacity. Nearly one-half of the down-regulated adaptive genes encode proteins for production of secondary metabolites. The most conspicuously down-regulated adaptive gene is the gene encoding HetR. HetR has been suggested to also regulate akinete differentiation in Nostoc ellipsosporum (26). However, a hetR mutant of N. punctiforme differentiates akinete-like cells that lack granulation (45). It is possible that HetR is involved in very early steps of akinete differentiation and that the gene is down-regulated by the 3-day time point. Time course transcription profiles of akinete differentiation are currently being analyzed and should resolve this possibility for N. punctiforme (M. Summers, E. Campbell, and J. Meeks, unpublished data).
Functional categories of differentiating hormogonia.
This is the first systems-level assessment of differential gene expression at any stage of hormogonium development. The most striking feature of the data is the number of differentially transcribed genes in hormogonia 24 h after induction, which is nearly five times higher than the number of differentially transcribed genes in akinete-forming or N2-fixing cultures (Table 1). This was an unpredicted result, especially the up-regulation of 944 genes in cells that have entered a nongrowth state. The impression that one gets from the hormogonium database is that the hormogonium is a clearly metabolically active, but nongrowing, motile filament that is highly tuned to sense the environment and respond to it.
Whereas most up-regulated genes were again in the unassigned category (52%), the second largest cluster was the adaptive metabolism cluster, which contained 204 genes. Slightly more than 50% of the adaptive genes encode proteins for signal transduction (85 genes) and transcriptional regulation (20 genes). On average, 85% of these genes are up-regulated uniquely in the hormogonium state. The products of the genes encoding transcriptional regulators likely account for part of the extensive differential gene expression in hormogonia at 24 h. The N. punctiforme genome contains genes encoding 156 HK, 103 RR, 33 PK, and 119 transcriptional regulators (31, 34). Some of the sensory proteins are organized with a multiple-domain hybrid kinase and/or regulator architecture. In the cluster of up-regulated hormogonium sensory proteins there are four hybrid HK-PKs and 24 HK-RRs, and in 13 of the HK-RRs the receiver domain is in the N terminus of the protein, where it likely functions as a sensor domain. An alternative sigma subunit (sigH [NpR1771]), which is induced during coculture of N. punctiforme with a symbiotic plant partner and whose mutation results in an altered infection frequency (8), was not up-regulated in the combined-nitrogen starvation-induced 24-h hormogonia. This result implies that hormogonia may be subtly different depending on the environmental signal for their induction.
In addition, 18 genes encoding putative chemotaxis proteins from five separate genomic loci, five gas vesicle proteins, and five pilus formation proteins were up-regulated. Up-regulation of the gas vesicle and pilus genes confirms results for hormogonia of Calothrix sp. strain PCC 7601 (10). Cyanobacterial gliding motility has been reported to be dependent on type IV pilus retraction (7) and/or polysaccharide secretion (23). There are several gene clusters encoding proteins of polysaccharide synthesis assigned to core metabolism (cell envelope synthesis) and development (heterocyst envelope synthesis); eight genes in the latter group were also up-regulated in hormogonia. A cluster of genes that encode polysaccharide biosynthetic and type II secretion proteins was up-regulated uniquely in hormogonia (NpF0066 to NpF0073; NpF0073 [pulG] was also up-regulated in akinetes). If polysaccharide extrusion is a mechanism of gliding motility in hormogonia, there appears to be sufficient biosynthetic capacity to support it, although there is currently no experimental evidence for either the motility mechanisms or specific polysaccharide synthetic genes.
Since hormogonia in this study were induced by transfer from NH4+ into combined-nitrogen-free medium, we anticipated some overlap in the hormogonium differentiation and N2-grown databases. Seventeen developmental and 18 alternative nitrogen (N) source genes were up-regulated in 24-h hormogonia. The developmental genes include the akinete marker avaK and the heterocyst-specific genes devH, hetF, and patU. Hormogonia are the infective units of Nostoc sp.-plant symbioses (30), and an N. punctiforme hetF mutant infects a symbiotic plant partner (45), which implies that HetF, at least, is not essential for hormogonium differentiation. The alternative N source genes include the genes encoding nitrate (NpR1526) and nitrite (NpF1528) reductases and a nitrate/nitrite permease (NpR1527), as well as 11 nitrogenase-associated proteins, including the catalytic subunit proteins NifH (NpR0415), NifD (NpR0414), and NifK (NpR0390). As in essentially all heterocyst-forming cyanobacteria, the N. punctiforme nifD gene is interrupted by an insertion element (31, 34). Northern hybridization established that the nifD element was excised in 24-h hormogonia and that nifK can also be transcribed in the absence of the excision rearrangement (data not shown). There is evidence, however, that hormogonia do not fix N2 in normal atmospheric conditions (9), implying that an active protein complex is not assembled. We suggest that these alternative N source genes and some of the developmental genes are expressed in response to the NH4+ starvation method of hormogonium induction. Heterocysts were not present in the 24-h hormogonia, and their differentiation does not occur until 48 to 56 h later. Moreover, HetR, which is essential for heterocyst differentiation, was strongly down-regulated during hormogonium development (see below), indicating that a complete developmental pathway was not operational in 24-h hormogonia.
A substantial number of core metabolic genes (179 genes) were up-regulated in 24-h hormogonia. Most of these genes are involved in cell envelope, polymer, monomer, cofactor, and precursor metabolism. The up-regulated cell envelope proteins include the cell shape-determining proteins MreC (NpR1840) and MreD (NpR1839); MreB (NpR1841) is constitutively expressed in the NH4+-grown transcriptome and proteome. The presence of these proteins may account for the change in shape of hormogonium cells. The polymer proteins are enriched in DNA metabolism and protein processing. The 16 adaptive and core DNA metabolic genes include the genes encoding five helicases, four nucleases, three proteins for modification, two proteins for replication (polA [NpF1050] and priA [NpR0306]), and two proteins for replication repair (sbcC [NpF4484] and ssb [NpF5942]). The presumptive presence of these proteins indicates that there is some DNA metabolism in hormogonia, perhaps chromosome stabilization, although there is no net accumulation of DNA. There are 13 genes encoding protein processing factors and 12 proteases in the adaptive and core categories. The chaperones include two different DnaKs (NpF0122 and NpF5907), two DnaJs (NpF0150 and NpR3872), and two IbpAs (Hsp20) (NpF0198 and NpF0784). A protease whose structural gene was significantly up-regulated is NblA (NpF0291), which degrades phycobilisomes and accounts for the decrease in phycobiliprotein-induced fluorescence in hormogonia. The hormogonium cultures were incubated in the absence of an environmental source of combined nitrogen. Therefore, the products of phycobiliprotein degradation most likely supply amino nitrogen for synthesis of adaptive proteins, aided by the action of the products of the up-regulated amino acid biosynthetic genes. Thus, substantial protein metabolism by reorganization and processing may occur in hormogonia.
Nearly 50% of the 883 genes down-regulated in 24-h hormogonia encode proteins of core metabolism, and most of the remainder cluster in the unassigned metabolism category (38%). Except for the cell division genes, every subgroup of core metabolic genes was substantially down-regulated, and the down-regulation was much more extensive than that of akinetes 3 days into the differentiation process. These results are consistent with a stringent nongrowth state of hormogonia. Notably down-regulated were two genes encoding the chaperone GroEL (NpF1230 and NpR0829) and one gene encoding GroES (NpR0830). Many genes for energy metabolism (e.g., ATP synthase) and photosynthetic and respiratory electron transport were down-regulated, although their transcripts remained detectable. A specific Fe-S protein (putative PsaC; encoded by NpF5213) has been shown to be down-regulated in hormogonia using a reporter gene (5). While nongrowing, hormogonia maintain a measurable rate of photosynthetic CO2 assimilation (9). The photosynthetic rate may be sufficient to provide photosynthate for polysaccharide synthesis and extrusion or energy for pilus assembly and retraction for hormogonium gliding.
Almost as many genes encoding transport proteins were down-regulated as were up-regulated. This observation may imply that hormogonia acquire different classes of compounds from the environment than NH4+-grown filaments acquire. There is a smaller, but still numerically substantial, fraction of adaptive genes that were down-regulated in hormogonia. Approximately 35% as many genes encoding signal transduction proteins were down-regulated as were up-regulated. This also suggests that there are major specific changes in the signal transduction pathways as hormogonia differentiate from NH4+-grown filaments. The notable down-regulated genes include patA and hetR involved in heterocyst differentiation and glnB (NpF4466) encoding PII, which has an intracellular nitrogen- and carbon-sensing role in unicellular cyanobacteria (16). The down-regulation of 10 transcriptional regulators, coupled with the up-regulation of 20 similar proteins, indicates that there is a shift in the cellular composition of transcription factors concurrent with a change in physiology. Included in the down-regulated adaptive category are four genes (NpF2182, NpF2183, NpF2186, and NpF2187) located in a cluster of genes encoding proteins for the synthesis of a cyclic peptide (nostopeptide) (nosB, -C, -E, and -F). The differential expression of these genes in hormogonia and other genes in akinetes and N2-grown cultures may lead to an understanding of the physiological role(s) of such secondary metabolites.
The differential expression in hormogonia of seven genes (up-regulated genes NpF0152 [tonB], NpF2507 [pilT], NpF3829 [atoC], NpF4811 [fliA], NpF5963 [che, encoding methyl-accepting chemotaxis protein], and NpR0414 [nifD] and down-regulated gene NpR0830 [groES]) was verified by Northern hybridization (data not shown).
Overlap of genes differentially expressed in the developmental states.
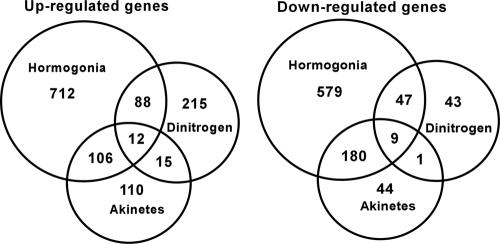
The extent of overlap of up- and down-regulated genes between developmental states is depicted in the Venn diagrams in Fig. 3. This depiction does not include the genes that were up-regulated in one developmental state in any of the two- or three-way comparisons and down-regulated in another developmental state and the converse. A detailed list of the up- and down-regulated genes shared by all three developmental states and by the akinete-forming state and the N2 steady state is presented in Table 2.
FIG. 3.
Numerical relationships between up- and down-regulated genes in the N2-grown, hormogonium, and akinete developmental states.
TABLE 2.
Genes up- and down-regulated in common in the three developmental states (N2-grown cultures, 3-day developing akinetes, and 24-h hormogonia) and in developing akinetes and steady-state N2-grown cultures
| Developmental states | Regulation | Gene(s)a |
|---|---|---|
| N2-grown cultures, akinetes, and hormogonia | Up | NpF0152 (membrane; U-con hyp), NpF0291 (nblA; A-alt N), NpF1845 (amiC; C-cell env), NpF3553 (heft; A-dev het), NpF3573 (aroC; C-mono AA), NpF4024 (tal; C-precursor), NpF5710 (nadD; C-cofactor), NpR1565 (crp; A-trans reg), NpR2514 (rbn; C-polymer tRNA), NpR3990 (ald; C-mono AA), NpR4415 (membrane; U-con hyp) NpR5108 (aarF; U-unassign) |
| Down | NpF1541 (rpe; C-precursor), NpF3687 (cynT; C-presursor), NpF3770 (ispA; C-cofactor), NpF6252 (nfnB; U-unassign), NpR1544 (DUF608; U-unassign), NpR2771 (pys; C-cofactor), NpR3741 (tkt; C-precursor), NpR5546 (nuoC; C-energy), NpR5598 (signal sequence; U-con hyp) | |
| N2-grown cultures and akinetes | Up | NpF0817 (signal sequence; U-con hyp), NpF2746 (filamentous cyanobacteria; U-con hyp), NpF2782 (tcm; A-secondary), NpF4153 (rpoE; A-trans reg), NpF4976 (mmsB; U-unassign), NpF5429 (hemY; C-porphyrin), NpF5770 (DUF1400; U-unassign), NpR0727 (degQ; A-stress), NpR1041 (signal sequence; U-con hyp), NpR2814 (ubiE; C-cofactor), NpR2885 (tolC; T-export), NpR4700 (null; U-hyp), NpR4927 (pp; A-PK Pase), NpR4977 (membrane; U-con hyp), NpR5530 (ctaA; C-energy) |
| Down | NpR2397 (signal sequence; U-con hyp) |
Where possible, the gene designation is given; for unassigned proteins, either the COG gene call or a potential compartmental property (predicted membrane or signal sequence) or taxonomic distribution of the protein is indicated. Abbreviations: U-con hyp, unassigned metabolism group and conserved hypothetical subgroup; A-alt N, adaptive metabolism group and alternate N source subgroup; C-cell env, core metabolism group and cell envelope subgroup; A-dev het, adaptive metabolism group and development subgroup; C-mono AA, core metabolism group and monomer amino acid subgroup; C-precursor, core metabolism group and precursor subgroup; C-cofactor, core metabolism group and cofactor subgroup; A-trans reg, adaptive metabolism group and transcriptional regulation subgroup; C-polymer tRNA, core metabolism group and polymer tRNA subgroup; U-unassign, unassigned metabolism group and unassigned subgroup; C-energy, core metabolism group and energy subgroup; A-secondary, adaptive metabolism group and secondary subgroup; C-porphyrin, core metabolism group and porphyrin subgroup; A-stress, adaptive metabolism group and stress subgroup; T-export, transport metabolism group and secretion/export subgroup; U-hyp, unassigned group and hypothetical subgroup; A-PK Pase, adaptive metabolism group and PK phosphatase subgroup.
Very few differentially transcribed genes were shared by the three states or by the akinete-forming and N2-grown states. Hormogonium genes are more in common with both 3-day differentiating akinetes and N2-grown cultures than the latter two are with each other. This sharing is especially true for the common core metabolic genes that are down-regulated as akinetes and hormogonia enter their nongrowth states. The majority of the 12 up-regulated genes that are in common group in the core metabolism cluster, and the gene products are involved in various functions. One shared up-regulated gene encodes AmiC, which breaks peptidoglycan linker bonds and could be involved in rearrangement of the vegetative cell wall, leading to the different shapes of the three types of differentiated cells. The three adaptive genes include nblA for degradation of phycobiliproteins, hetF encoding the putative caspase-hemoglobinase fold protease (3), and a gene encoding a Crp family transcriptional regulator. The increased transcription of nblA is anomalous with the color and intense fluorescence emission of akinetes that indicate the presence of substantial phycobiliproteins. Most of the nine common down-regulated genes encode core functions, dominated by cofactor and precursor metabolism. Comparisons between 3-day akinete-forming and steady-state N2-grown cultures yielded 15 up-regulated genes and one down-regulated gene in common. One-half of these 16 genes encode unassigned proteins. The up-regulated adaptive gene products include a sigma subunit, a stress-induced protease, a PK phosphatase, and a polyketide cyclase, which are not reflective of a common physiological function.
Conclusions.
The collective data indicate that the three alternative developmental states of N. punctiforme do have some genes in common, but their differentiation largely involves up-regulation of genes distinct for each state. The most remarkable result is the extent of differential gene expression by 24-h hormogonia. These filaments were once characterized as growth precursor cells (9, 11). Our data support such a conclusion; hormogonia have a distinctly down-regulated capacity for core metabolic functions, similar to the swarmer cells of Caulobacter crescentus (24, 25). Nevertheless, hormogonia are transcriptionally very active, especially in genes encoding adaptive metabolic environmental sense and response functions. Hormogonia appear to be poised to move in specific directions, perhaps depending on the light quality and quantity or the presence of a chemoattractant, to a microhabitat which is somehow perceived as permissive for subsequent growth and reproduction.
Including the hetR results, there is little evidence in the differential expression profiles of N2-grown cultures and the profiles for 3 days into the differentiation of akinetes to support an evolutionary sequence of akinete to heterocyst in the ancestors of N. punctiforme by, for example, gene duplication. It is possible that such evidence will emerge from comparative time course experiments during differentiation, but it is also possible that too much evolutionary divergence has occurred in developmental pathways of the two cell types.
Supplementary Material
Acknowledgments
This work was supported by grant EF0317104 from the NSF to J.C.M. and by grant GM048680 from the NIH to M.L.S.
We thank Ian Campbell and Michael Goodson for assistance in database and statistical analyses, Jeff Elhai for sharing data, and Michael Schultz for help in the development of hybridization procedures. We also thank Dawei Lin and Brad Sickler of the UC Davis Genome Center and Bioinformatics Program for collaboration in database analysis and visualization tools.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, D. G., and P. S. Duggan. 1999. Heterocyst and akinete differentiation in cyanobacteria. New Phytol. 144:1-33. [Google Scholar]
- 2.Anderson, D. C., E. L. Campbell, and J. C. Meeks. 2006. 3D LC/MS/MS analysis of the soluble proteome of the cyanobacterium Nostoc punctiforme ATCC 29133. J. Proteome Res. 5:3096-3104. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of eukaryotic separins. Proteins Struct. Funct. Genet. 46:355-367. [DOI] [PubMed] [Google Scholar]
- 4.Argueta, C., and M. L. Summers. 2005. Characterization of a model system for the study of Nostoc punctiforme akinetes. Arch. Microbiol. 183:338-346. [DOI] [PubMed] [Google Scholar]
- 5.Argueta, C., K. Yuksek, and M. L. Summers. 2004. Construction and use of GFP reporter vectors for analysis of cell-type-specific gene expression in Nostoc punctiforme. J. Microbiol. Methods 59:181-188. [DOI] [PubMed] [Google Scholar]
- 6.Argueta, C., K. Yuksek, R. Patel, and M. L. Summers. 2006. Identification of Nostoc punctiforme akinete-expressed genes using differential display. Mol. Microbiol. 61:748-757. [DOI] [PubMed] [Google Scholar]
- 7.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941-951. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, E. L., B. Brahamsha, and J. C. Meeks. 1998. Mutation of an alternative sigma factor in the cyanobacterium Nostoc punctiforme results in increased infection of its symbiotic plant partner, Anthoceros punctatus. J. Bacteriol. 180:4938-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, E. L., and J. C. Meeks. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 55:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damerval, T., G. Guglielmi, J. Houmard, and N. Tandeau de Marsac. 1991. Hormogonium differentiation in the cyanobacterium Calothrix: a photoregulated developmental process. Plant Cell 3:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dow, C. S., R. Whittenbury, and N. G. Carr. 1983. The “shut down” or “growth precursor” cell—an adaptation for survival in a potentially hostile environment, p. 187-247. In J. H. Slater, R. Whittenbury, and J. W. T. Wimpenny (ed.), Microbes in their natural environments. Cambridge University Press, Cambridge, United Kingdom.
- 12.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome wide expression of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 13.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 14.Enderlin, C. S., and J. C. Meeks. 1983. Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158:157-165. [DOI] [PubMed] [Google Scholar]
- 15.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA.
- 16.Forchhammer, K. 2004. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28:319-333, [DOI] [PubMed] [Google Scholar]
- 17.Golden, J. W., and H. S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557-563. [DOI] [PubMed] [Google Scholar]
- 18.Hagen, K. D., and J. C. Meeks. 2001. The unique cyanobacterial protein OpcA is an allosteric effector of glucose-6-phosphate dehydrogenase in Nostoc punctiforme ATCC 29133. J. Biol. Chem. 276:11477-11486. [DOI] [PubMed] [Google Scholar]
- 19.Herdmann, M., M. Janvier, R. Rippka, and R. Y. Stanier. 1979. Genome size in cyanobacteria. J. Gen. Microbiol. 111:73-85. [Google Scholar]
- 20.Herdmann, M., and R. Rippka. 1988. Cellular differentiation: hormogonia and baeocytes. Methods Enzymol. 167:232-242. [Google Scholar]
- 21.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 23.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 8:1161-1168. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs-Wagner, C. 2004. Regulatory proteins with a sense of direction: cell cycle signaling network in Caulobacter. Mol. Microbiol. 51:7-13. [DOI] [PubMed] [Google Scholar]
- 25.Laub, M., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 26.Leganés, F., F. Fernandéz-Piñas, and C. P. Wolk. 1994. Two mutations that block heterocyst differentiation have different effects on akinete differentiation in Nostoc ellipsosporum. Mol. Microbiol. 12:679-684. [DOI] [PubMed] [Google Scholar]
- 27.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynn, M. E., J. A. Bantle, and J. D. Ownby. 1986. Estimation of gene expression in heterocysts of Anabaena variabilis by using DNA-RNA hybridization. J. Bacteriol. 167:940-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Figueroa, E., F. Navaro, and F. J. Florencio. 2000. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of the glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 476:282-286. [DOI] [PubMed] [Google Scholar]
- 30.Meeks, J. C. 1998. Symbiosis between nitrogen-fixing cyanobacteria and plants. BioScience 48:266-276. [Google Scholar]
- 31.Meeks, J. C. 2005. The genome of the filamentous cyanobacterium Nostoc punctiforme. What can we learn from it about free-living and symbiotic nitrogen fixation?, p. 27-70. In R. Palacios and W. E. Newton (ed.), Nitrogen fixation: 1888-2001, vol. VI. Genomes and genomics of nitrogen-fixing organisms. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 32.Meeks, J. C., E. L. Campbell, M. L. Summers, and F. C. Wong. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178:395-403. [DOI] [PubMed] [Google Scholar]
- 33.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 65:94-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth. Res. 70:85-106. [DOI] [PubMed] [Google Scholar]
- 35.Renström-Kellner, E., A. N. Rai, and B. Bergmann. 1990. Correlation between nitrogenase and glutamine synthetase expression in the cyanobacterium Anabaena cylindrica. Physiol. Plant. 80:12-19. [Google Scholar]
- 36.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdmann, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbial. 111:1-61. [Google Scholar]
- 37.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carry, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 38.Summers, M. L., and J. C. Meeks. 1996. Transcriptional regulation of zwf, encoding glucose-6-phosphate dehydrogenase, from the cyanobacterium Nostoc punctiforme ATCC 29133. Mol. Microbiol. 22:473-480. [DOI] [PubMed] [Google Scholar]
- 39.Tandeau de Marsac, N. 1994. Differentiation of hormogonia and relationships with other biological processes, p. 825-842. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA.
- 40.Thiel, T., and C. P. Wolk. 1983. Metabolic activities of spores of Nostoc spongiaeforme. J. Bacteriol. 156:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 47:1239-1249. [DOI] [PubMed] [Google Scholar]
- 42.Winkenbach, F., and C. P. Wolk. 1973. Activities of enzymes of the oxidative and the reductive pentose phosphate pathways in heterocysts of a blue-green alga. Plant Physiol. 52:480-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, DC.
- 44.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA.
- 45.Wong, F. C., and J. C. Meeks. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte hornwort Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148:315-323. [DOI] [PubMed] [Google Scholar]
- 46.Xu, X., J. Elhai, and C. P. Wolk. 2007. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383-422. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Horizon Scientific Press, Norwich, United Kingdom.
- 47.Zhang, C.-C., S. Laurent, S. Sakr, L. Peng, and S. Bédu. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59:367-375. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, R., and C. P. Wolk. 2002. Identification of an akinete marker gene in Anabaena variabilis. J. Bacteriol. 184:2529-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.