Abstract
The recent discovery that the protein DksA acts as a coregulator of genes controlled by ppGpp led us to investigate the similarities and differences between the relaxed phenotype of a ppGpp-deficient mutant and the phenotype of a strain lacking DksA. We demonstrate that the absence of DksA and ppGpp has similar effects on many of the observed phenotypes but that DksA and ppGpp also have independent and sometimes opposing roles in the cell. Specifically, we show that overexpression of DksA can compensate for the loss of ppGpp with respect to transcription of the promoters PuspA, PlivJ, and PrrnBP1 as well as amino acid auxotrophy, cell-cell aggregation, motility, filamentation, and stationary phase morphology, suggesting that DksA can function without ppGpp in regulating gene expression. In addition, ppGpp and DksA have opposing effects on adhesion. In the course of our analysis, we also discovered new features of the relaxed mutant, namely, defects in cell-cell aggregation and motility.
The ability of the cell to adapt to nutrient limitation depends on its capacity to change its gene expression immediately when nutrients are depleted. One important mechanism for regulating this change in gene expression is the stringent response, mediated by the alarmone ppGpp (6, 17). Two different ppGpp synthetases exist in Escherichia coli: the ribosome-associated RelA synthesizing ppGpp in response to amino acid starvation and the cytoplasmic SpoT synthesizing ppGpp in response to carbon starvation as well as most other starvations (6). SpoT is also responsible for ppGpp hydrolysis. During the stringent response the production of the protein-synthesizing system is repressed while some genes involved in stress resistance and survival are induced. Cells unable to produce ppGpp (deleted of relA and spoT, called ppGpp0) are largely decontrolled upon entry to stationary phase and behave as if they were still growing exponentially (18). It has been suggested that this shift from growth mode to maintenance mode by ppGpp results in part from the regulation of the activity of alternative sigma factors (12, 17).
The array of phenotypes of a cell deficient in making ppGpp, the so-called relaxed phenotype, has been expanded, and it is obvious not only that gene regulation by ppGpp is important in response to starvation but that ppGpp also has a role in regulating traits involved in virulence (8) and other phenotypes both during growth and in stationary phase.
DksA was first described as a multicopy suppressor of the temperature-sensitive growth and filamentation of a dnaK deletion mutant in E. coli (13). The protein DksA has since then been shown to influence the regulatory activities of ppGpp even though DksA in itself is not a typical regulatory protein. It is still under debate how DksA alters the activity of ppGpp (or vice versa). The levels of DksA in E. coli have been shown to be similar at all growth rates and in all growth phases tested (5, 22). DksA is structurally similar to the transcription factors GreA and GreB, although they share no sequence homology, and has been modeled into the same binding site in the secondary channel of RNA polymerase (RNAP) (24). In vitro experiments show that DksA copurifies with RNAP (24) and thus may exert its action as a transcriptional regulator through direct interaction with the polymerase. In vitro transcription experiments show that DksA can potentiate the ppGpp effect on both stringently induced (23) and repressed promoters (22, 26), and it has been suggested that DksA exerts its effects by stabilizing the interaction between RNAP and ppGpp (24). Nevertheless, in vivo experiments show that DksA can inhibit the stringently repressed promoter of rrnBP1 independently of ppGpp (26). It was recently discovered that GreB has similar effects as DksA on rrn promoters, suggesting that the negative effect of DksA on rrn promoter activity is mediated through interactions in the secondary channel (27). GreB did not, on the other hand, affect the positively regulated promoters, suggesting that the negative and positive effects of DksA work by different mechanisms. GreA has been shown to activate rrnBP1 expression and functionally compete with DksA (26), also indicating that binding in the secondary channel is important for the effects of DksA on rrn. In Pseudomonas aeruginosa, Perron and coworkers (25) showed that the DksA protein interacts with both RNAP and DNA at the rrn promoter; hence, they provided an alternative model of how DksA together with ppGpp alters promoter expression, i.e., by modulating the interaction between RNAP and DNA (25). They suggest a model where DksA increases RNAP binding to rrn promoters and traps RNAP in an inactive state at these promoters. Perron et al. (24) also show that DksA negatively regulates rrn both in exponential and in stationary phase.
The complexity of the stringent response makes it difficult to distinguish between direct and indirect effects of ppGpp. Also, the different stress responses are linked to each other in multiple ways. To elucidate the role of DksA in cellular physiology and in relation to the effects of ppGpp, we compared the phenotypes of the ppGpp0 and ΔdksA mutant strains. We also looked at the possibility that one regulatory molecule compensates for the lack of the other. We examined the regulation of a set of known ppGpp-regulated promoters: the rrn promoter controlling the expression of rRNA which is negatively regulated by ppGpp and two positively regulated promoters, the uspA promoter responsible for expression of the universal stress protein A, and the livJ promoter controlling expression of a protein facilitating transport of branched-chain amino acids in the periplasm. Additionally, other phenotypes of the ppGpp0 mutant were examined also in the ΔdksA mutant. The data show that ppGpp and DksA do not have completely overlapping roles in the cell and that DksA can function without ppGpp.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains and plasmids used in this work are listed in Table 1. Cultures were grown in liquid LB or M9 defined medium (28) aerobically in Erlenmeyer flasks in a rotary shaker at 37°C. The M9 medium was supplemented with a limiting concentration of glucose (0.08%), all the amino acids in excess, and thiamine (10 mM) (30). Minimal medium plates contained M9 salts, glucose (0.4%), and thiamine (10 mM). Carbenicillin (50 μg/ml) was added in all experiments with strains containing plasmids. Kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml) were used to select for transductants.
TABLE 1.
E. coli strains and plasmids
| E. coli strain or plasmid | Relevant genotype | Reference |
|---|---|---|
| Derived strains | ||
| BG143 | MG1655 λ Φ(PlivJ-lacZ)/pBR322 | This work |
| BG144 | MC4100 λ Φ(PuspA-lacZ) ΔdksA::kan/pBR322 | This work |
| BG145 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ)a ΔdksA::kan/pBR322 | This work |
| BG146 | MG1655 λ Φ(PlivJ-lacZ) ΔdksA::kan/pBR322 | This work |
| BG147 | MG1655 λ Φ(PlivJ-lacZ) ΔdksA::kan/pJK537 | This work |
| BG152 | RLG4422 ΔrelA::kan ΔspoT::cam | This work |
| BG154 | MG1655 λ Φ(PlivJ-lacZ) ΔrelA::kan ΔspoT::cam/pJK537 | This work |
| BG155 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ) ΔrelA::kan ΔspoT::cam/pJK537 | This work |
| KK371 | MG1655 ΔrelA ΔspoT (markerless) | K. Kvint |
| LM569 | MG1655/pJK537 | This work |
| LM571 | MC4100 λ Φ(PuspA-lacZ)/pJK537 | This work |
| LM574 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ)/pJK537 | This work |
| LM576 | MG1655 λ Φ(PlivJ-lacZ)/pJK537 | This work |
| LM608 | RLG4422 ΔdksA::kan | This work |
| LM624 | MG1655 ΔdksA::kan/pJK537 | This work |
| LM657 | MG1655 ΔrelA::kan ΔspoT::cam/pJK537 | This work |
| LM659 | MC4100 λ Φ(PuspA-lacZ)/pBR322 | This work |
| LM661 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ) ΔrelA::kan ΔspoT::cam/pBR322 | This work |
| LM663 | MG1655 λ Φ(PlivJ-lacZ) ΔrelA::kan ΔspoT::cam/pBR322 | This work |
| LM667 | MG1655 ΔrelA::kan ΔspoT::cam/pBR322 | This work |
| LM669 | MG1655 Δlac λ Φ(PuspA-lacZ)/p JK537 | This work |
| LM671 | MG1655 Δlac λ Φ(PuspA-lacZ)/pBR322 | This work |
| LM683 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ)/pBR322 | This work |
| LM694 | MG1655 ΔdksA::kan/pBR322 | This work |
| LM699 | MG1655 Δlac λΦ(PuspA-lacZ) ΔrelA::kan ΔspoT::cam/pBR322 | This work |
| LM700 | MG1655 Δlac λΦ(PuspA-lacZ) ΔrelA::kan ΔspoT::cam/p JK537 | This work |
| LM701 | MG1655 Δlac λΦ(PuspA-lacZ) ΔdksA::kan/pBR322 | This work |
| LM702 | MG1655 Δlac λΦ(PuspA-lacZ) ΔdksA::kan/pJK537 | This work |
| PJ1 | MG1655/pBR322 | This work |
| PJ11 | MC4100 λ Φ(PuspA-lacZ) ΔdksA::kan/pJK537 | This work |
| PJ12 | MC4100 λ Φ(PrrnBP1-51-+50-lacZ) ΔdksA::kan/pJK537 | This work |
| PJ13 | MC4100 λ Φ(PuspA-lacZ) ΔrelA::kan ΔspoT::cam/pBR322 | This work |
| PJ14 | MC4100 λ Φ(PuspA-lacZ) ΔrelA::kan ΔspoT::cam/pJK537 | This work |
| RLG4422 | MG1655 λ Φ(PlivJ-lacZ) | R. L. Gourse |
| Parental strains | ||
| AF634 | MC4100 λ Φ(PuspA-lacZ) | A. Farewell |
| CF9239 | ΔdksA::kan | M. Cashel |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | M. Giskov |
| MG1655 | Lab stock | |
| RLG2259 | λ Φ(PrrnBP1-51-+50-lacZ) | R. L. Gourse |
| Plasmids | ||
| pBR322 | Laboratory stock | |
| pJK537 | dksA in pBR322 Apr | 13 |
rrnBP1−51-+50 contains the promoter sequence between −51 and +50 relative to the transcriptional strut site (+1).
General methods.
The ΔdksA, ΔrelA, and ΔspoT alleles were introduced into the different strains by standard P1 transduction. Transformation was performed by standard methods.
Measurements of cellular components.
Relative β-galactosidase levels were determined as described previously (19), with modifications (2). The β-galactosidase activity of a culture is expressed by the following calculation: (1,000 × OD420)/(OD420 × reaction time × sample volume), where the OD420 is the optical density at 420 nm, time is measured in minutes, and volume is measured in milliliters. Overexpression of DksA from the plasmid pJK537 was confirmed using Western blotting. Gel electrophoresis was carried out using 12% sodium dodecyl sulfate-polyacrylamide gels (NuPage). Immunoblotting was performed according to standard procedures using chicken polyclonal antibodies specific for DksA (kind gift from M. Cashel) or rabbit polyclonal antibodies specific for the α-domain of Ag43 (kind gift from P. Owen) as primary antibody. For detection, we used IRDye 800CW-labeled goat anti-chicken immunoglobulin G antibodies (Rockland) and IRDye 800CW-labeled goat anti-rabbit immunoglobulin G antibodies (Westburg). Blots were exposed and quantified using LI-COR's Odyssey Infrared Imaging System and software. All experiments were repeated several times in order to ensure the reproducibility of the results.
Autoaggregation assay.
Overnight cultures of the E. coli strains were adjusted to the same OD600, and 4 ml of each was placed in a sterile 15-ml tube. At the beginning of each experiment, all cultures were vigorously shaken. Cell settlings were visually observed. Samples were also taken for microscopic observation.
Yeast cell agglutination assay.
Saccharomyces cerevisiae (BY4741) and E. coli cells were pelleted after overnight culture, washed, and resuspended in phosphate-buffered saline at a OD600 values of 5 and 3, respectively. Equal volumes of E. coli and yeast cell suspensions were mixed on a glass slide. Aggregation was monitored visually, and the titer was recorded as the highest dilution of bacteria giving a positive agglutination reaction. A value of 1 corresponds to a nondiluted bacterial suspension (see Table 2). To establish that the agglutination observed was due to the adhesion of bacterial fimbriae to mannose residues of the yeast cell surface, we also performed the experiments in the presence of d-mannose (25 mg ml−1), and under this condition, fimbriae-dependent agglutination was abolished. All assays were performed at least three times and gave similar results.
TABLE 2.
Result of the yeast cell agglutination
| Straina | Agglutination titer of strainb
|
||
|---|---|---|---|
| Wt | ΔdksA | ppGpp0 | |
| Control plasmid (pBR322) | 1/4 | 1/16 | − |
| DksA overproduction (pJK537) | 1 | 1/4 | − |
The strains used are PJ1, LM569, LM667, LM657, LM694, and LM624 (Table 1).
The agglutination titer refers to the highest dilution of bacteria giving a positive yeast agglutination reaction. A value of 1 corresponds to a nondiluted bacterial suspension, and 1/16 corresponds to very high agglutination that could be detected even with very diluted bacteria. The minus sign indicates that no agglutination was detected.
Motility assay.
Bacterial cells were picked from colonies grown on LB agar plates and inoculated onto low-agar (0.3%) LB plates. The plates were incubated at room temperature for 24 to 48 h, and motility was assessed qualitatively by examining the circular halo formed by the motile bacterial cells. Photographs were captured using a GelDoc 2000 instrument (Bio-Rad).
Cell morphology observed by microscopy.
The cells were grown overnight on LB plates, stained with crystal violet according to Xiao et al. (31), and viewed with a Leica DMRXA microscope (Deerfield, IL). Photographs were captured using a Hamamatsu ORCA cooled charge-coupled-device camera (Bridgewater, NJ). For the stationary-phase morphology the strains were grown to stationary phase in liquid LB medium and then stained and viewed as above.
Scanning electron microscopy.
Strains were grown overnight in LB medium at 37°C without shaking. From these cultures, 30 μl was taken, and cells were allowed to sediment and adhere to Au-sputtered Thermanox plates for 15 min. The plates were then transferred to 2.5% glutaraldehyde for 15 min, rinsed with Na cacodylate buffer, and postfixed in 1% OsO4 (15 min). Specimens were then dehydrated in a graded series of ethanol and dried with hexamethyldisilazane two times for 5 min each time. All specimens were then sputter-coated with palladium and investigated with a Zeiss DSM 982 Gemini electron microscope.
RESULTS AND DISCUSSION
DksA can compensate for the lack of ppGpp in regulating uspA, livJ, and rrnBP1.
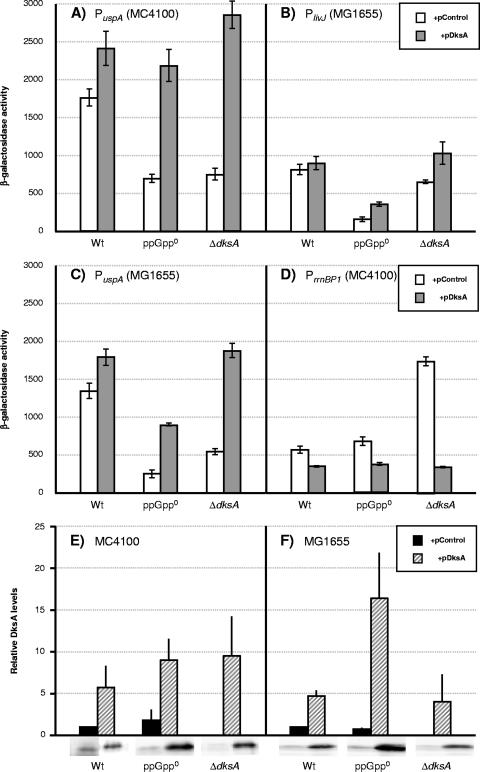
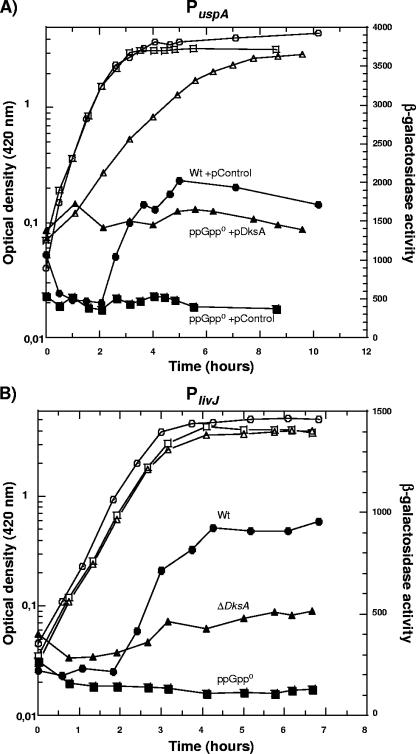
We studied the regulation of genes that are subjected to positive and negative control by ppGpp in cells lacking or overproducing DksA. The stringently induced promoter PlivJ has been shown before to be under positive regulation of DksA (23), and its expression is lower in stationary phase in the ΔdksA strain than in wild type (Fig. 1B and 2B). There is still a partial induction of expression from the livJ promoter upon entry to stationary phase, most likely due to the action of ppGpp, although the induction ratio is not as high as in wild type (Fig. 2B). Here, we show that the expression from the stringently induced promoter PuspA (15) is also lower in the ΔdksA strain in two different backgrounds and, therefore, under positive regulation of DksA (Fig. 1A and C). Lower expression compared to the wild type was also seen in the ppGpp0 strain (Fig. 1A and B), as has been shown previously (4, 15). The expression of the following set of stringently induced promoters is also lower in the ΔdksA strain (data not shown): Phis (4); PthrABC (4); PuspC, PuspD, and PuspE (9); and PuspF (Laurence Nachin, personal communication). Overexpression of DksA (from a multicopy plasmid with dksA under its own promoter, pJK537) (13) complements the expression of PuspA and PlivJ in the ΔdksA strain (Fig. 1A and B). In addition, overexpression of DksA can compensate for the loss of ppGpp and increase expression from both PuspA and PlivJ in the ppGpp0 background (Fig. 1A, B, C, and E). DksA is overproduced approximately fivefold in both ppGpp0 and wild type, and the overexpression level in the ΔdksA mutant is similar (Fig. 1E). Although DksA is overproduced to the same extent, it gives a growth defect only when ppGpp is absent (Fig. 2A and data not shown). Compensation for the loss of ppGpp by DksA overproduction has previously been shown for the expression of RpoS (5). Thus, DksA can function independently of ppGpp. For PuspA expression this was possible both in the MC4100 (Fig. 1A) and MG1655 backgrounds (Fig. 1C). The effect on PuspA expression was not dependent on the growth defect seen in Fig. 2A since similar results were seen in the MG1655 background (Fig. 1C), where growth was only slightly affected (data not shown). It is also interesting that the growth defect is not proportional to the degree of DksA overexpression since DksA is overproduced approximately 20-fold in the MG1655 background (Fig. 1F). The growth defect in the MC4100 background could, in fact, be a consequence of larger effects on stringently controlled genes in this background. DksA overexpression not only suppresses the reduced stationary-phase expression of uspA in a ppGpp0 strain but also leads to an elevated expression in exponential phase, which supports the notion that DksA can work independently from ppGpp (Fig. 2A). Overexpression of DksA in the wild type did not significantly affect expression of uspA in the exponential phase (data not shown).
FIG. 1.
Overproduction of DksA can compensate for the lack of ppGpp in regulating uspA, livJ and rrnBP1. (A to D) Gene expression measured as β-galactosidase activity in overnight cultures of reporter strains. Cultures were grown in M9 defined medium including glucose, amino acids, and thiamine supplemented with carbenicillin. (A) PuspA-lacZ (MC4100 background). The strains used are LM659, LM571, PJ13, PJ14, BG144, and PJ11. (B) PlivJ-lacZ. The strains used are BG143, LM576, LM663, BG154, BG146, and BG147. (C) PuspA-lacZ (MG1655 background). The strains used are LM671, LM669, LM699, LM700, LM701 and LM702. (D) PrrnBP1-lacZ. The strains used are LM683, LM574, LM661, BG155, BG145, and PJ12. (E) Western blot analysis of the levels of DksA in an MG1655 background. The strains used are LM659, LM571, PJ13, PJ14, BG144, and PJ11. (F) Western blot analysis of the levels of DksA in an MG1655 background. The strains used are PJ1, LM569, LM667, LM657, LM694 and LM624. Wt, wild type.
FIG. 2.
(A) Overproduction of DksA can compensate for the lack of ppGpp in regulating uspA. Growth curve (open symbols) and gene expression (closed symbols) measured as β-galactosidase activity from PuspA-lacZ (MC4100 background) in M9 defined medium including glucose, amino acids, and thiamine supplemented with carbenicillin. The strains used are LM659 (circles), PJ13 (squares), and PJ14 (triangles). (B) Deletion of DksA causes a reduction in expression of livJ but not to the same extent as a deficiency in ppGpp production. Growth curve and gene expression measured as β-galactosidase activity from PlivJ-lacZ in M9 defined medium including glucose, amino acids, and thiamine supplemented with antibiotics when appropriate. The strains used are RLG4422 (circles), BG152 (squares), and LM608 (triangles). Relevant genotypes of the strains used are listed in Table 1. Wt, wild type.
The stringently repressed promoter PrrnBP1 displays higher levels of expression in the ΔdksA strain (Fig. 1D), which is consistent with previous results (22). While we do not see an effect of the ppGpp0 mutation in the late-stationary-phase measurements (Fig. 1D), we do see it in transition and early stationary phase in this strain (data not shown). For the ΔdksA mutant, on the other hand, the higher expression is obvious also in late stationary phase. Overexpression of DksA from a multicopy plasmid can complement the ΔdksA mutant as well as suppress the elevated expression in the ppGpp0 mutant (Fig. 1D). In addition, overexpression of DksA marginally attenuated the expression from PrrnBP1 in the wild-type background (Fig. 1D).
DksA overproduction suppresses amino acid auxotrophy of an MC4100 ppGpp0 but not an MG1655 ppGpp0 mutant.
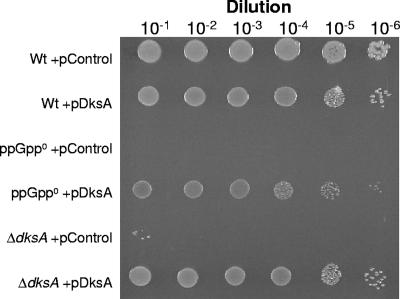
A ppGpp0 mutant is auxotrophic for many amino acids, and a ΔdksA mutant has multiple, but not identical, amino acid requirements (5). In view of the fact that DksA overproduction was, at least partially, able to suppress the attenuated expression of livJ (Fig. 1B) in the ppGpp0 mutant, we examined whether DksA overproduction could rescue ppGpp0 cells from amino acid auxotrophy. Indeed, it is possible to restore growth of the ppGpp0 mutant by overexpressing DksA in the MC4100 ppGpp0 background (Fig. 3) but not in the MG1655 background, in agreement with previously published results (5; also data not shown). The reason for the strain difference remains to be elucidated and was not linked to differential levels of RpoS in the two strain backgrounds since it was still possible to suppress amino acid auxotrophy in an MC4100 ppGpp0 rpoS mutant (data not shown). The amino acid auxotrophy of the ΔdksA strain could also be complemented by DksA overproduction (Fig. 3). Another case where overexpression of DksA cannot compensate for the loss of ppGpp even if the two molecules appear to have the same effect is in aminotriazole sensitivity of the ppGpp0 mutant in the MG1655 background (5).
FIG. 3.
DksA overproduction suppresses amino acid auxotrophy of an MC4100 ppGpp0 mutant. Suppression of the amino acid auxotrophy of the ppGpp0 and ΔdksA strains by overproduction of DksA. Ten microliters of serial dilutions of overnight cultures was spotted on minimal medium plates. Plates were incubated overnight at 37°C and for an additional 24 h at room temperature. The strains used are LM659, LM571, PJ13, PJ14, BG144, and PJ11. Relevant genotypes of the strains used are listed in Table 1.
DksA and ppGpp affect cell aggregation and adhesion.
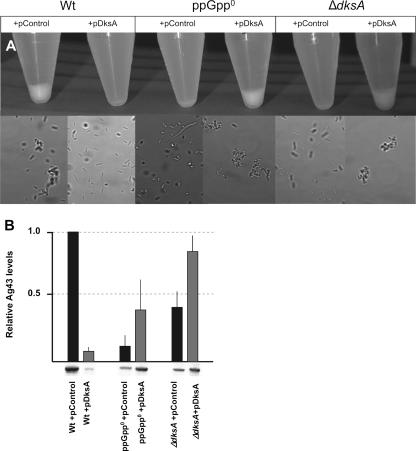
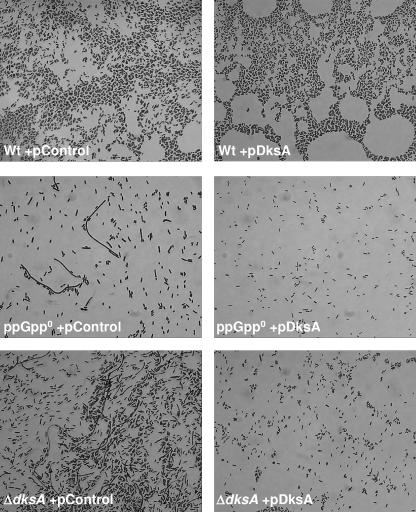
Upon entry into stationary phase, E. coli cells initiate a factor-dependent self-aggregation leading to sedimentation (20). During the course of working with the ppGpp0 mutant, we noticed that it failed to sediment in liquid culture even after hours of static incubation (Fig. 4A, upper panel). Microscopic observations of overnight cultures revealed that the ppGpp0 mutant did not form cell aggregates while the wild-type strain did (Fig. 4A, lower panel). We determined if the same was true for the ΔdksA mutant. Both the ppGpp0 strain and the ΔdksA strain are defective in the ability to sediment in liquid culture, and in agreement with the sedimentation phenotypes we could detect no aggregates in the two mutant strains (Fig. 4A, lower panel). Overproduction of DksA can complement the sedimentation defect of the ΔdksA strain as well as suppress the sedimentation defect of the ppGpp0 strain. Thus, as for gene expression, DksA appears to be able to work alone, in the total absence of ppGpp. Notably, overexpression of DksA in the wild-type context reduces the cell's ability to form aggregates. Sedimentation in liquid culture is partly associated with the expression of Ag43 (encoded by the flu gene) on the cell surface and has been shown to be counteracted by the presence of fimbriae (10). Therefore, we checked the levels of Ag43 in the different strains by Western blotting and detection using antibody raised against the α-domain of Ag43. The decreased autoaggregation in wild type upon DksA overexpression is correlated to lower levels of Ag43 (Fig. 4B). The ΔdksA strain expressed low levels of Ag43, and this is complemented by DksA overexpression (Fig. 4B). The ppGpp0 strain expressed very low levels of Ag43, and this is suppressed by overexpression of DksA (Fig. 4B).
FIG. 4.
ppGpp and DksA affect cellular autoaggregation. Overnight cultures were vigorously shaken and subsequently incubated statically at room temperature. (A) The top panel shows the cell settling state of each strain after 2 h. For the experiment shown in the lower panel, an aliquot of each culture was taken at the beginning of the assay for microscopic observation. Images were captured with a magnification of ×1,000, and representative data are shown. The strains used are PJ1, LM569, LM667, LM657, LM694, and LM624. Relevant genotypes of the strains used are listed in Table 1. (B) Ag43 production. For each strain, the equivalent of 0.2 OD600 unit of an overnight culture was analyzed by Western blotting and immunodetection using a polyclonal rabbit antiserum raised against the α-domain of Ag43. Quantification of the signal in the upper panel is normalized to the wild type, and the Western blot is shown below.
The Ag43-dependent aggregation of E. coli has been shown to be antagonized by the presence of type 1 fimbriae on the bacterial cell surface such that the presence of fimbriae abolishes auto aggregation regardless of the presence of Ag43 (10). Therefore, we also tested Fim-dependent adhesion of the ΔdksA and ppGpp0 mutants by scoring for their ability to agglutinate yeast cells, a fimbriae-dependent process (the FimH subunit binds to mannose residues on the yeast cell surface). To confirm that yeast agglutination was indeed due to the presence of bacterial fimbriae, we verified that the agglutination observed was abolished in the presence of d-mannose (data not shown). The ppGpp0 strain was found to be defective in yeast agglutination (Table 2), as has been shown previously (1). The ΔdksA strain, on the other hand, shows increased yeast agglutination compared to wild type (Table 2). This may explain why the ΔdksA strain failed to self aggregate in stationary phase despite expressing Ag43 (Fig. 4B), since elevated expression of fimbriae is known to physically antagonize Ag43-dependent aggregation (10). Overexpression of DksA decreased adhesion of both the wild-type and the ΔdksA strains, which is consistent with increased adhesion in the ΔdksA mutant. As expected, overproduction of DksA had no effect on adhesion in the ppGpp0 strain.
These results demonstrate that the two regulatory molecules have opposing roles in the cell, i.e., in adhesion (analyzed as yeast cell agglutination), where the loss of DksA increases while the loss of ppGpp abolishes adhesion (Table 2). ppGpp has previously been shown to affect adhesion by positively controlling the expression of the FimB recombinase (1). The ppGpp0 mutant exhibits lower expression of FimB and subsequently type 1 fimbriae and is therefore deficient in adhesion and biofilm formation (1). The increase in yeast cell agglutination of the DksA strain is most likely due to increased expression of type 1 fimbriae since it could be reversed by adding d-mannose (data not shown).
DksA overproduction rescues the motility defect of ppGpp0 cells.
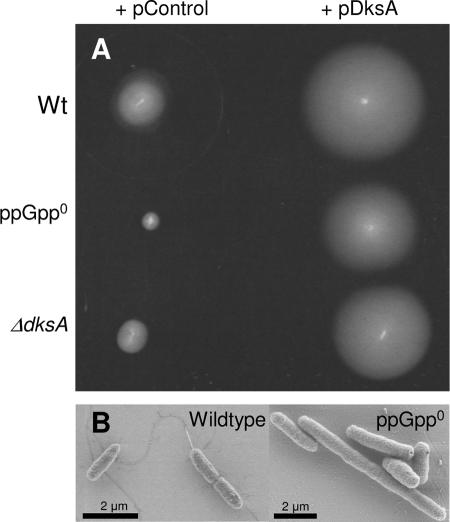
We also found that the ppGpp0 strain is defective in motility on 0.3% agar plates (Fig. 5A), and electron microscopy analysis shows that the ppGpp0 strain does not produce flagella (less than 1/10,000 cells have flagella) (Fig. 5B), which explains its failure to move. The ΔdksA strain is less motile than the wild type but more than the ppGpp0 strain (Fig. 5A). Overexpression of DksA complements the motility defect of the ΔdksA strain and suppresses the motility defect of the ppGpp0 strain (Fig. 5A). Overexpression of DksA also increases the motility of the wild-type strain (Fig. 5A).
FIG. 5.
DksA overproduction rescues the motility defect of ppGpp0 cells. Effect of dksA and relA spoT mutations as well as DksA overproduction on cell motility and flagella expression in wild type and ppGpp0. (A) Low-agar plates (0.3%) were inoculated with each strain as indicated. The halo of growing motile cells was observed after a 24-h incubation at room temperature. The strains used are PJ1, LM569, LM667, LM657, LM694, and LM624. (B) Detection of flagella by scanning electron microscopy. The relevant genotype of the observed strain is indicated in each panel; the strains used are MG1655 and KK371. Relevant genotypes of the strains used are listed in Table 1.
Reciprocal regulation of the two counterproductive activities, motility and adhesion (fimbriae), is logical, and direct cross talk between fimbrial gene products and bacterial motility genes has been shown (7). It is, however, argued that all cells within a population do not necessarily express the same surface-associated structures, and both structures have been observed on the bacterial surface at the same time (11). Both flagella and type 1 fimbriae have been shown to be important in biofilm formation, and within the population there is a dynamic interplay between adhesion and motility. Motility is controlled by the alternative sigma factor, σF (14), and it is possible that ppGpp can affect the competitiveness of this sigma factor, as it has been shown to do for other alternative sigma factors (12).
DksA overproduction can rescue filamentation and stationary phase morphology of the ppGpp0 mutant.
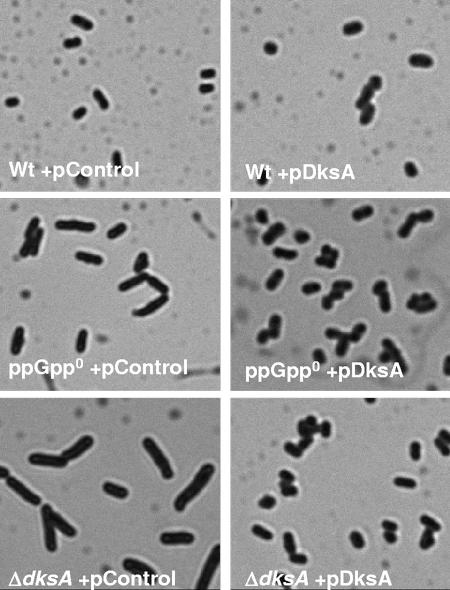
Cellular morphology was observed under the microscope after cells were stained with crystal violet. The ppGpp0 mutant forms filaments in log phase and fails to induce a spherical cell shape in stationary phase (6, 31) (Fig. 6 and 7). The ΔdksA strain also forms filaments, and the filamentation phenotype of the ΔdksA strain can be complemented by overproduction of DksA (Fig. 6). DksA has previously been shown to counteract the filamentation of a DnaK mutant (13). Overproduction of DksA also suppresses the filamentation of the ppGpp0 strain while overproduction of DksA in wild type has no obvious effect on cell morphology (Fig. 6). In addition, the cells that do not produce filaments in the heterogeneous population of the ppGpp0 culture also fail to become coccoid in stationary phase (Fig. 7), presumably due to problems in expressing BolA, a morphogene required for the cells to become coccoid (29); and therefore the cells look more or less like growing, rod-shaped cells. The same is true for the ΔdksA strain (Fig. 7). DksA overproduction complements the lack of DksA and suppresses the ppGpp0 strain with respect to stationary phase morphology (Fig. 7). Again, this suggests that DksA can act on its own without ppGpp also in respect to cellular morphology. The morphogene bolA is regulated by σs (16), and it is possible that the deficiency of the ppGpp0 strain and the ΔdksA strain in expressing RpoS can account for the failure in inducing sphere shape (5).
FIG. 6.
Filamentation of both ppGpp0 and ΔdksA mutants can be suppressed by overproduction of DksA. Cells were grown on LB plates overnight, resuspended in water, dried on glass slides, stained with crystal violet for 6 min, and destained with water. Photomicrography was performed with a Leica DMRXA microscope. Images were captured at a magnification of ×1,000. Representative panels are shown. In all the aliquots tested similar proportions of filaments were observed in the ppGpp0 and ΔdksA strains with control plasmids, while no filaments were observed in the corresponding strains overproducing DksA. The strains used are PJ1, LM569, LM667, LM657, LM694, and LM624. Relevant genotypes of the strains used are listed in Table 1.
FIG. 7.
Failure of both ppGpp0 and ΔdksA mutants to acquire the coccoid form in stationary phase can be suppressed by overproduction of DksA. Cells were grown 2 h into stationary phase in liquid LB medium, dried on glass slides, stained with crystal violet for 6 min, and destained with water. Photomicrography was performed with a Leica DMRXA microscope. Images were captured at a magnification of ×1,000, and representative panels are shown. Less pronounced morphological differences were observed also in overnight cultures (data not shown). The strains used are PJ1, LM569, LM667, LM657, LM694, and LM624. Relevant genotypes of the strains used are listed in Table 1.
Concluding remarks.
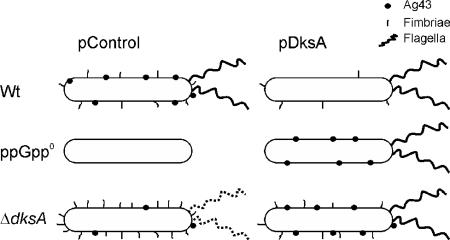
It has been suggested that ppGpp and DksA work together, and, indeed, it turns out that deleting dksA causes similar phenotypes as deleting relA and spoT. However, DksA and ppGpp seem to be able to function independently of one another, and DksA overproduction can compensate for the loss of ppGpp in many cases where the two molecules have the same effect. It therefore seems unlikely that the main role of DksA is to facilitate ppGpp binding to RNAP, as has been suggested (24). Notably, overexpression of DksA can suppress the amino acid auxotrophy of a ppGpp0 mutant. Even though deleting dksA can recreate parts of the relaxed phenotype at the same time as DksA overproduction can compensate for the loss of ppGpp, it turns out that the two mutants are different (Fig. 8), and the roles of ppGpp and DksA are not totally overlapping since the absence of ppGpp or DksA has opposite effects on adhesion. It has been shown that even though expression of rRNA is higher in the ΔdksA mutant, there is still a reduction in rRNA expression in stationary phase in the ΔdksA mutant of P. aeruginosa (25), indicating that ppGpp can also work independently of DksA; future experiments will include elucidating if ppGpp can, indeed, work in the absence of DksA in E. coli.
FIG. 8.
The surface features of wild-type, ppGpp0, and ΔdksA strains. A summary of the surface features (Ag43, fimbriae, and flagella) of the three strains and the effect of overproducing DksA are shown. Both the ppGpp0 and ΔdksA strains express lower levels of Ag43 than wild type and are nonmotile. The dotted flagella on the ΔdksA mutant indicate the failure to move on motility agar even though the presence or absence of flagella has not been confirmed. Overexpression of DksA can restore wild-type surface structures of the ΔdksA strain and also to some extent of the ppGpp0 strain except in fimbriation, where ppGpp and DksA have opposite effects.
It has been suggested that the role of ppGpp is to relocate cellular resources from growth-related activities to maintenance (21), and our results indicate that DksA also has its major function in this regulatory trade-off.
Acknowledgments
This work was funded by the Swedish Natural Science Research Council and an award from the Göran Gustafsson Foundation for Scientific Research in Molecular Biology.
We sincerely thank Ulf Nannmark for performing the electron microscopy, Mike Cashel for sharing strains and antibodies and discussing unpublished results, Peter Owen for sharing antibodies, and Rick Gourse and Kristian Kvint for sharing strains. Colleagues of the Nyström laboratory are gratefully acknowledged for discussion and suggestions on the manuscript.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Aberg, A., V. Shingler, and C. Balsalobre. 2006. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 60:1520-1533. [DOI] [PubMed] [Google Scholar]
- 2.Albertson, N. H., and T. Nystrom. 1994. Effects of starvation for exogenous carbon on functional mRNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol. Lett. 117:181-187. [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299-310. [DOI] [PubMed] [Google Scholar]
- 4.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 5.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC. [Google Scholar]
- 7.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godfrey, H. P., J. V. Bugrysheva, and F. C. Cabello. 2002. The role of the stringent response in the pathogenesis of bacterial infections. Trends Microbiol. 10:349-351. [DOI] [PubMed] [Google Scholar]
- 9.Gustavsson, N., A. Diez, and T. Nystrom. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107-117. [DOI] [PubMed] [Google Scholar]
- 10.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holden, N. J., and D. L. Gally. 2004. Switches, cross-talk and memory in Escherichia coli adherence. J. Med. Microbiol. 53:585-593. [DOI] [PubMed] [Google Scholar]
- 12.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang, P. J., and E. A. Craig. 1990. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 172:2055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kundu, T. K., S. Kusano, and A. Ishihama. 1997. Promoter selectivity of Escherichia coli RNA polymerase sigmaF holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 179:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvint, K., C. Hosbond, A. Farewell, O. Nybroe, and T. Nystrom. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35:435-443. [DOI] [PubMed] [Google Scholar]
- 16.Lange, R., and R. Hengge-Aronis. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 173:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236-242. [DOI] [PubMed] [Google Scholar]
- 18.Magnusson, L. U., T. Nystrom, and A. Farewell. 2003. Underproduction of sigma 70 mimics a stringent response. A proteome approach. J. Biol. Chem. 278:968-973. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Nachin, L., U. Nannmark, and T. Nystrom. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nystrom, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 22.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311-322. [DOI] [PubMed] [Google Scholar]
- 23.Paul, B. J., M. B. Berkmen, and R. L. Gourse. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 102:7823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perederina, A., V. Svetlov, M. N. Vassylyeva, T. H. Tahirov, S. Yokoyama, I. Artsimovitch, and D. G. Vassylyev. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118:297-309. [DOI] [PubMed] [Google Scholar]
- 25.Perron, K., R. Comte, and C. van Delden. 2005. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 56:1087-1102. [DOI] [PubMed] [Google Scholar]
- 26.Potrykus, K., D. Vinella, H. Murphy, A. Szalewska-Palasz, R. D'Ari, and M. Cashel. 2006. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 281:15238-15248. [DOI] [PubMed] [Google Scholar]
- 27.Rutherford, S. T., J. J. Lemke, C. E. Vrentas, T. Gaal, W. Ross, and R. L. Gourse. 2006. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. [DOI] [PMC free article] [PubMed]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Santos, J. M., M. Lobo, A. P. Matos, M. A. De Pedro, and C. M. Arraiano. 2002. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45:1729-1740. [DOI] [PubMed] [Google Scholar]
- 30.Wanner, B. L., R. Kodaira, and F. C. Neidhardt. 1977. Physiological regulation of a decontrolled lac operon. J. Bacteriol. 130:212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]