Abstract
During maturation, heterocysts form an envelope layer of polysaccharide, called heterocyst envelope polysaccharide (HEP), whose synthesis depends on a cluster of genes, the HEP island, and on an additional, distant gene, hepB, or a gene immediately downstream from hepB. We show that HEP formation depends upon the predicted glycosyl transferase genes all4160 at a third locus and alr3699, which is adjacent to hepB and is cotranscribed with it. Mutations in the histidine kinase genes hepN and hepK appear to silence the promoter of hepB and incompletely down-regulate all4160.
The filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 produces terminally differentiated cells, called heterocysts, when it is deprived of fixed nitrogen. These specialized cells are internally microoxic as a result of (i) cessation of oxygen production; (ii) intensification of respiration; and (iii) deposition of a thick envelope, comprising an outer polysaccharide layer and an inner glycolipid layer, that impedes the entry of oxygen (35). The glycolipid layer is the principal barrier to the entry of oxygen, whereas the polysaccharide layer is evidently required for the integrity of the glycolipid layer. The homogeneous layer of heterocyst envelope polysaccharide (HEP) has been chemically analyzed in two other Anabaena sp. strains and in a Cylindrospermum sp. strain. In those strains, HEP is composed of oligosaccharide repeating units whose mannose-(glucose)3 backbone is decorated with side chains that are different in the different strains. In the Anabaena sp. strains, a highly similar polysaccharide was found in the envelopes of akinetes, a kind of spore. It was crudely estimated that, exclusive of regulatory genes, on the order of 20 genes are required for synthesis of the concatenated subunits (33).
Through DNA microarray analysis and near-saturation transposon mutagenesis, numerous Anabaena sp. strain PCC 7120 open reading frames in the HEP island (genes alr2825 through alr2841) of the chromosome were found to be required specifically for formation of the envelope polysaccharide layer (7, 17). Mutants with mutations in the presumptively glycosyl transferase-encoding genes hepB (alr3698) (23, 36), alr3699, and all4160 (this study) that are distant from the HEP island in the genome also lack the polysaccharide layer. Regulation of HEP deposition depends upon signal transduction systems that comprise, at least, the histidine kinase HepN (11, 26), the response regulator HenR (11), the serine/threonine kinase HepS (11), and the interacting two-component regulatory elements HepK and DevRA (39, 40). In addition, proteins Abp2 and Abp3 bind to the DNA sequence upstream of hepC, and inactivation of abp2 and abp3 greatly reduces the expression of hepA and hepC (20). abp2 and abp3 mutants, however, lack the glycolipid layer of the heterocyst envelope rather than the polysaccharide layer.
In the 1,076 sites of insertion mapped in a transposon mutagenesis project screening for Fox− mutants (i.e., mutants unable to fix dinitrogen in the presence of oxygen) (9, 10, 11, 17), two insertions of transposon Tn5-1063 were found in hepB, four insertions were found in the downstream neighboring gene alr3699, and nine insertions were found in all4160. Previously, we showed that there was up-regulation of hepB in heterocysts using gfp (encoding green fluorescent protein [GFP]) as a transcriptional reporter gene (32). By screening random promoter:gfp fusions for fusions that are regulated during heterocyst differentiation, we also found that the promoter of all4160, like that of the HEP island gene hepA (34), is up-regulated specifically in proheterocysts and heterocysts. In this paper, we show that all4160 and alr3699 are required for synthesis of the HEP layer and that whereas hepB and alr3699, like genes of the HEP island (17, 22, 26, 40), are strongly down-regulated in hepK and hepN mutants, all4160 is only modestly down-regulated by mutations in hepK and hepN.
Methods.
Anabaena sp. strain PCC 7120 and its derivatives (Table 1) were grown in BG-11 medium or in AA/8 plus nitrate as previously described (16, 26). All mutants, complemented mutants, and strains containing pHB912-derived plasmids were grown in the presence of appropriate antibiotics. To induce heterocyst formation, rapidly growing cultures were washed twice with nitrogen-free medium, resuspended, and incubated on a shaker in the presence of light (∼30 microeinsteins m−2 s−1). Molecular cloning was performed by standard protocols. Plasmids (Table 1) were introduced into Anabaena strains by conjugation (8). Bright-field microscopy, fluorescence microscopy, and Alcian blue staining for observation of the envelope polysaccharide layer were performed as described previously (14, 17, 32). Images showing the expression of a gene in different strains were taken in parallel during one session of microscopic observations. To minimize GFP bleaching, an image viewed with a microscope equipped with a JVC3 charge-coupled device color video camera was fixed immediately, assisted by the software 10moons-2000/PRO (version 4.5). Multiple alignments of DNA sequences were performed using CLUSTAL X (30) and the SEAVIEW alignment editor (13). Aligned sequences were analyzed using the neighbor-joining method (28) assisted by MEGA 3.1 (21). The sequence divergence between species pairs was calculated using the p-distance model. Branch support was assessed using 1,000 bootstrap replicates. Four glycosyl transferases, ExoY of Sinorhizobium meliloti (25), Atu3560 of Agrobacterium tumefaciens strain C58 (GenBank accession no. NP 534056), and WcaJ and WcaL of Escherichia coli (29), were included in the phylogenetic analysis of the predicted glycosyl transferases in cyanobacteria.
TABLE 1.
Cyanobacterial strains, plasmids, and primers used
| Strain, plasmid, or primer | Derivation and/or relevant characteristicsa | Reference(s) or source |
|---|---|---|
| Anabaena sp. strains | ||
| 1017 | Emr, alr0117 (hepN)::Tn5-1087b | 26 |
| ABP2 | Nmr | 20 |
| DRHB2429 | Emr, all4160::C.CE2 | This study |
| FQ689 | Bmr Nmr Smr, alr3699::Tn5-1063 at chromosomal bp 4467495 | This study |
| FQ749 | Bmr Nmr Smr, alr3699::Tn5-1063 at chromosomal bp 4467724 | This study |
| FQ792 | Bmr Nmr Smr, all4160::Tn5-1063 at chromosomal bp 5007183 | This study |
| FQ873 | Bmr Nmr Smr, all4160::Tn5-1063 at chromosomal bp 5006680 | This study |
| FQ1351 | Bmr Nmr Smr, alr3699::Tn5-1063 at chromosomal bp 4468098 | This study |
| FQ1421 | Bmr Nmr Smr, all4160::Tn5-1063 at chromosomal bp 5007278 | This study |
| PCC 7120 | Wild type | R. Haselkorn, University of Chicago |
| Y7 | Bmr Nmr Smr, hepK::Tn5-1065 | 9 |
| Plasmids | ||
| anc0304 | Cmr Emr, sequencing bridging clone, bp 5002439 through bp 5019236 (for comparison, all4160 extends from bp 5007439 to bp 5006384) | 19 |
| anc2015 | Cmr Emr, sequencing bridging clone, bp 4462687 through bp 4477773 (for comparison, alr3699 extends from bp 4467059 to bp 4468207) | 19 |
| anp01253 | Apr, sequencing clone, bp 4998732 through bp 5007158 | 19 |
| anp02990 | Apr, sequencing clone, bp 4466571 through bp 4474909 | 19 |
| pHB912 | Kmr Smr Spr, GFP-reporting vector | 32 |
| pHB1726 | Kmr Smr Spr, pHB912 derivative bearing chromosomal bp 5009091 to bp 5007155b | This study |
| pHB2416 | Apr, 2.4-kb fragment containing all4160, amplified with PCR using primers all4160-1 and all4160-2, cloned into pMD18-T | This study |
| pHB2427 | Apr Cmr Emr; cassette C.CE2 (Cmr Emr) excised with BamHI from plasmid pRL598 was blunted with T4 DNA polymerase and ligated to pHB2416 that had been linearized at the ClaI site within all4160 and blunted | 3; this study |
| pHB2429 | PstI-SmaI fragment, from pHB2427, that bears all4160::C.CE2 was blunted with T4 DNA polymerase and ligated with XbaI-cut, blunted pRL277 | 2; this study |
| pMD18-T | Apr, cloning vector | http://www.takara.com.cn/bioxiew/bioview02_14.pdf |
| pRL2881b | Kmr, 2,866-bp HinP1I fragment of anp02990 ligated to AccI-cut pK18 | 27; this study |
| pRL2890 | Smr Spr, insert of pRL2881b, containing alr3700 and alr3701 as its only intact open reading frames, excised with BamHI and PstI, ligated between the unique BamHI and NsiI sites of pRL2831a | 17; this study |
| pRL3169a | Kmr, 1,267-bp, alr3699-bearing fragment of RsaI- plus HaeIII-cut anp02990 ligated to SmaI-cut pK18 | 27; this study |
| pRL3176 | Cmr Emr, 1,338-bp fragment of anp01253, excised with BsrGI plus DraI and containing all4159 as its only intact open reading frame, ligated between the BsiWI and StuI sites of the polylinker of pRL2833a | 10; this study |
| pRL3177 | Cmr Emr, insert of pRL3169a, bearing alr3699 as its only intact open reading frame, excised with SacI plus SphI and inserted between the SacI and SphI sites of pRL2833b | 17; this study |
| pRL3182 | Smr Spr, 1,375-bp fragment, excised from anc0304 with Asp718I and XbaI and bearing all4160 as its only intact open reading frame, inserted between the BsiWI site and the Dam-insensitive XbaI site of pRL2831a | 17; this study |
| Primers | ||
| 7120pool-1 | 5′-AGCGTGCATAATAAGCCCTACA-3′ | |
| 7120pool-2 | 5′-CGTCTTGTAGTTCCCGTCATC-3′ | |
| all4160-1 | 5′-CGATGAGAGTGGTTAGAGGGA-3′ | |
| all4160-2 | 5′-CAGCTTGG AGCATTTCCATCGT-3′ | |
| hepB-1 | 5′-CATACAGTACTAGGAGATAAGAGTGGA-3′ | |
| hepB-2 | 5′-GTCGGCTGATAGAACAAACCACA-3′ | |
| hepB-3 | 5′-ACTGCTGGCGAGCTTGTTG-3′ | |
| hepB-4 | 5′-TGTCGCCAGTATGCTGTGAC-3′ | |
| alr3699-1 | 5′-CTGGTTTCCCACTCTGGTCT-3′ | |
| alr3699-2 | 5′-CTGTTTGACTGGCTTGGGA-3′ | |
| IDT141 | 5′-ACTAGCCAACCCACAGAGGTAA-3′ | |
| IDT142 | 5′-CATCAAGCTCCAGTTTTTCTGA-3′ | |
| IDT641 | 5′-TTGAAGGTTTTGGGTTAGCAAT-3′ | |
| IDT642 | 5′-GGAGTCTGTCAGGAAGGTTGTC-3′ |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance; Nmr, neomycin resistance; Bmr, bleomycin resistance.
See Fig. 1.
Identification of DNA fragments that can up-regulate expression of GFP in heterocysts of Anabaena sp. strain PCC 7120.
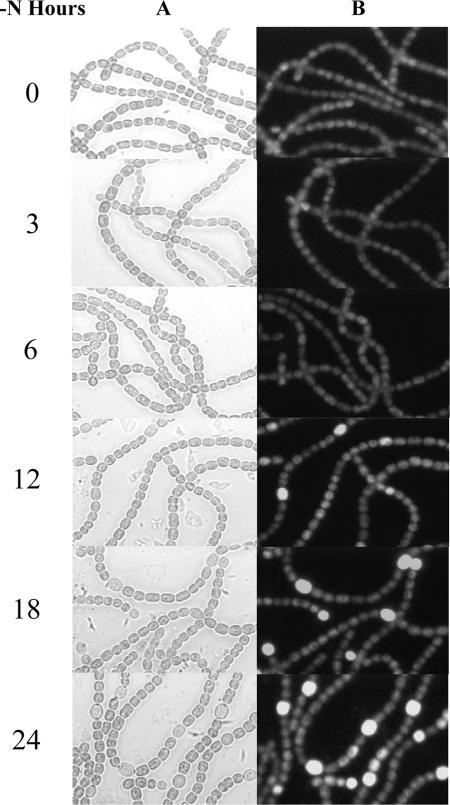
To identify genes up-regulated in developing heterocysts, as visualized at the level of single cells, we constructed an E. coli library of Anabaena sp. strain PCC 7120 genomic DNA in plasmid pHB912 (Table 1). About 80% of the 50,000 colonies of the library carried, at the BglII site of the vector, ca. 2- to 9-kb fragments derived by partial digestion of genomic DNA with Sau3AI. The library was introduced into Anabaena sp. strain PCC 7120 by conjugation. Twenty thousand resulting exconjugants were grown in petri dishes on agar with spectinomycin, heterocyst differentiation was induced in medium lacking fixed nitrogen, and the exconjugants were examined individually by fluorescence microscopy. Exconjugants in which GFP was observed to be up-regulated in proheterocysts or heterocysts were grown and treated as follows. Plasmids were extracted from the exconjugants and reintroduced into Anabaena sp. strain PCC 7120, and the resulting derivatives were examined. The cloned DNA fragments were identified by sequencing their ends with primers 7120pool-1 and 7120pool-2 (Table 1) and localizing the ends in the genome using the Kazusa database (www.kazusa.or.jp/cyano/cyano.html). Forty-nine DNA fragments clearly up-regulated gfp and 39 fragments clearly down-regulated gfp in developing heterocysts. We report here on a plasmid, pHB1726, whose chromosomal insert overlaps the 5′ end of all4160 and up-regulates gfp in developing heterocysts (Fig. 1 and 2). By following the expression of GFP, we found, consistent with the results of Ehira and Ohmori (6), that up-regulation of all4160 began between 6 and 12 h after nitrogen stepdown (Fig. 2). Because all4160 and the hepB gene cluster had both been shown by use of microarrays to be up-regulated during heterocyst development, were shown by use of gfp to be up-regulated in developing heterocysts, and were identified in a screen for Fox− mutants, we analyzed the physiological function and regulation of these genes.
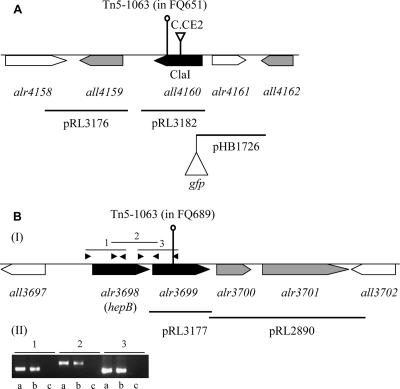
FIG. 1.
Two genomic regions involved in the formation of the HEP layer. (A) Map of the all4160 region, showing the locations of Tn5-1063 and C.CE2, complementing DNA fragments, and the cloned upstream region that is present in pHB1726. (B) Locations of Tn5-1063 and complementing DNA fragments in the hepB-alr3699 region (I) and cotranscription of hepB and alr3699 as visualized with RT-PCR (II). The arrowheads in panel B-I indicate primers for PCR whose products are shown in panel B-II. The combinations of primers used are as follows: lanes 1, hepB-1/hepB-3; lanes 2, hepB-2/alr3699-1; and lanes 3, hepB-4/alr3699-2. Lanes a, b, and c show electrophoretograms of products of PCR obtained using genomic DNA, cDNA, and RNA as the templates, respectively.
FIG. 2.
Time course of the up-regulation of all4160 in developing heterocysts after nitrogen stepdown, as reported by the GFP-based fluorescence of Anabaena sp. strain PCC 7120 bearing plasmid pHB1726. (A) Bright-field images; (B) images of fluorescence of GFP.
all4160 and alr3699 are predicted to encode glycosyl transferases.
Amino acids 163 to 340 of the 351-amino-acid protein All4160 show substantial similarity (58% positive; E value, 5 × 10−33) to amino acids of WcaJ, a putative glycosyl transferase. WcaJ may transfer glucose-1-phosphate to a lipid carrier, probably undecaprenylpyrophosphate, during biosynthesis of the colanic acid exopolysaccharide of E. coli (29). All4160 is most closely related to homologues in other heterocyst-forming species; is more distantly related to a homologue in a non-heterocyst-forming, filamentous, nitrogen-fixing species, Trichodesmium erythraeum; and is related even more distantly to other predicted glycosyl transferases (e.g., All2285) of Anabaena sp. strain PCC 7120 and to glycosyl transferases from unicellular cyanobacteria (see Fig. S1 in the supplemental material). A 64-amino-acid sequence (amino acids 42 to 105) close to the N terminus of All4160 shows similarity to SpoIIAA (62% positive; E value, 10−6), the antagonist of an anti-sigma factor, SpoIIAB, in Bacillus subtilis (1). The presence of an N-terminal, anti-sigma factor antagonist-like region in All4160 suggests that in addition to its glycosyl transferase activity, All4160 may act as a regulatory factor. As predicted with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html), All4160 is probably a transmembrane protein. This is consistent with its function as a glycosyl transferase and does not necessarily contradict the prediction that it may be a regulatory factor (38).
All4160, HepB (Alr3698), and Alr3699 are predicted to be glycosyl transferases. All4160 and All3699 are required for formation of the envelope polysaccharide layer (see below), and HepB may be required for formation of this layer. HepB and Alr3699 are more closely related to each other than either one is to All4160 (see Fig. S1 in the supplemental material). HepB shows some similarity to WcaL (45% of 247 amino acids positive in a total of 389 amino acids; E value, 2 × 10−9), another glycosyl transferase that is active in the biosynthesis of colanic acid (29). Proteins similar to Alr3699 are found in heterocyst-forming species and in a non-heterocyst-forming species, Lyngbya sp. strain PCC 8106 (see Fig. S1 in the supplemental material). After the suggested WcaJ-catalyzed sugar transfer to the lipid carrier, WcaL and other enzymes may catalyze the sequential transfer of additional sugars (29). HepB, Alr3699, and WcaL are members of glycosyl transferase family 4 (5), while Alr4160 and WcaJ are not included in the currently posted 89 families of glycosyl transferases (http://www.cazy.org/fam/acc_GT.html).
Effects of hepK and hepN on the up-regulation of all4160, alr3698 (hepB), and alr3699 during heterocyst differentiation.
In the microarray analysis of Ehira and Ohmori (6), alr3699 was clearly shown to be up-regulated upon N stepdown, and a reevaluation of the data of these workers (37) showed strong statistical support also for up-regulation of hepB (alr3698) but not for up-regulation of alr3700. Our previous analyses (22, 26, 40) showed that with few exceptions, HEP island genes depend very significantly on both hepK and hepN for up-regulation during heterocyst differentiation, and the same was observed for hepB at 14 h after nitrogen stepdown. Our report on the effect of regulatory genes on the expression of open reading frames of Anabaena sp. strain PCC 7120 (22) inadvertently omitted alr3699; data for hepK and hepN mutants showed reduced expression of alr3698, alr3699, and alr3700 with high statistical support. We have identified no stem-loop structure that might terminate transcription in the 62-bp sequence between hepB and alr3699.
We tested whether hepB and alr3699 are cotranscribed in filaments after nitrogen stepdown using reverse transcription-PCR (RT-PCR) (Fig. 1B), as reported previously (10). RNA was extracted from wild-type Anabaena sp. strain PCC 7120 after 24 h of N deprivation and freed of DNA until no PCR products could be detected with the RNA sample. We then used primers that started upstream from the coding region of hepB and extended to near the middle of alr3699, with the second RT-PCR overlapping the first and third RT-PCRs (Fig. 1B-I). The PCR products obtained with cDNA as templates matched the products predicted for, and obtained with, DNA as templates, whereas the use of RNA as templates resulted in no products (Fig. 1B-II). Our results support the idea that in response to nitrogen deprivation, the hepB and alr3699 genes are not just both up-regulated with a dependence on hepK and hepN but are jointly transcribed.
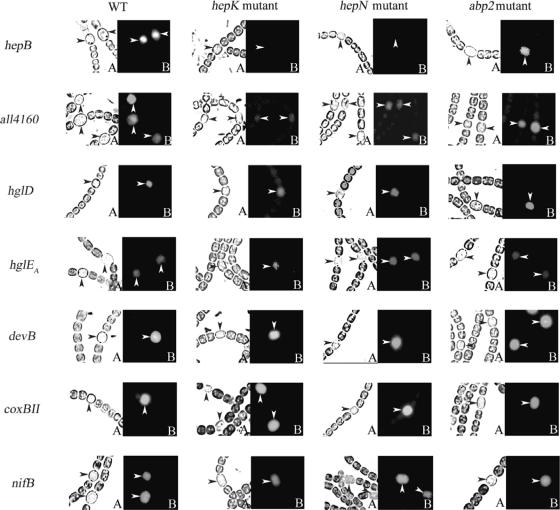
To examine at a single-cell level whether the transcription of all4160 and of the hepB-alr3699 cluster is affected by hepK and hepN, we introduced pHB912-based plasmids bearing presumptive promoter regions of all4160 and hepB (32) into strains Y7 (hepK::Tn5-1065) (9) and hepN::Tn5-1087b (26). As controls to compare the specificity of the regulation of all4160 and hepB with the regulation of other genes, we also introduced fusions of gfp, as a reporter, to the presumptive promoter regions of hglD, hglEA, devB, coxBII, and nifB (32) into the same hepK and hepN strains. hglD and hglEA are required for synthesis of heterocyst envelope glycolipids (10), devB is thought to be involved in transport of those lipids (12), coxBII encodes a component of a cytochrome oxidase that helps to maintain microoxic conditions in heterocysts (18, 31), and nifB encodes a protein on which a precursor of the FeMo-co prosthetic group of dinitrogenase is assembled (15). For further comparison, the same plasmids were introduced also into an abp2 mutant that forms the polysaccharide layer, but not the glycolipid layer, of the heterocyst envelope (20).
As shown in Fig. 3 and as previously reported (32), hepB, hglD, hglEA, devB, coxBII, and nifB were up-regulated in heterocysts of wild-type Anabaena sp. strain PCC 7120. Consistent with the results of microarray analyses, hepB was shown here to be up-regulated in heterocysts of wild-type Anabaena sp. strain PCC 7120 but not in heterocysts of hepK and hepN mutants (Fig. 3). However, we observed no significant decrease in expression of hglD, hglEA, devB, coxBII, or nifB in hepK and hepN mutants, which in general is consistent with the results of microarray analyses (22). Although the expression of all4160 was more reduced in the hepK mutant than in the hepN mutant, in neither of these mutants was all4160 expression reduced nearly as greatly as the expression of HEP island genes was (Fig. 3). In contrast, both all4160 and hepB were expressed similarly in the wild type and in the abp2 mutant; despite the lack of a glycolipid layer in the abp2 mutant (Fig. 3), so were hglD, hglEA, and devB.
FIG. 3.
Expression of fusions of gfp to presumptive promoter regions of all4160 and of other genes in Anabaena sp. strain PCC 7120 and its Y7 (hepK::Tn5-1065), hepN::Tn5-1087b, and abp2 derivatives at 24 h after nitrogen stepdown. The arrowheads indicate heterocysts. (A) Bright-field images; (B) images of fluorescence of GFP.
all4160 and alr3699 are involved in the formation of the HEP layer in Anabaena.
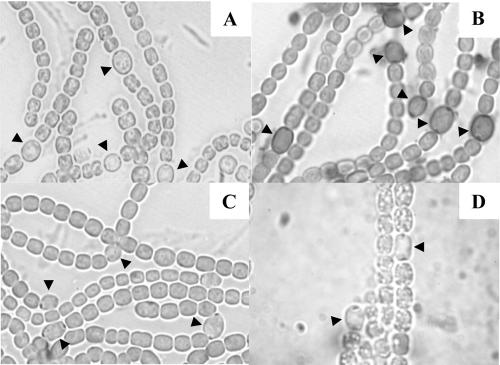
Transposon mutants and targeted insertional all4160 or alr3699 mutants were defective in the formation of the HEP layer. Plasmid pHB2429 (Table 1) was used to inactivate all4160, as described previously (4), by insertion of cassette C.CE2 into the ClaI site of this gene through homologous double recombination (Fig. 1). Complete segregation of the resulting mutant, DRHB2429, was confirmed by PCR with primers all4160-1 and all4160-2 (Table 1; data not shown). Similarly, complete segregation of the transposition-generated all4160 mutants FQ873 and FQ1421 was confirmed by PCR with primers IDT141 and IDT142, and complete segregation of the transposition-generated alr3699 mutant FQ689 was confirmed by PCR with primers IDT641 and IDT642 (Table 1; data not shown). Mutants DRHB2429 and FQ689 lacked the polysaccharide layer of the heterocyst envelope, as determined by staining with Alcian blue (Fig. 4), and were unable to grow aerobically in the absence of fixed nitrogen.
FIG. 4.
Micrographs of unstained Anabaena sp. strain PCC 7120 (A) and, after attempted staining with Alcian blue, of the wild type (B), mutant all4160::C.CE2 (C), and alr3699 mutant FQ689 (D). The arrowheads indicate heterocysts.
Because the independent transposon mutants and the targeted insertion mutants had the same phenotype, the lack of a HEP layer in the all4160 and alr3699 mutants was very unlikely to be attributable to the presence of additional mutations elsewhere in the genome. If all4160 and all4159 are cotranscribed, a mutation in all4160 might exert a polar effect on all4159. However, because these genes are separated by 674 bp of noncoding sequence, it is very improbable that they are cotranscribed. Moreover, the genes 3′ from alr3699 presumptively encode a defective PstI methylase and endonuclease (24), for which no role in synthesis of HEP is evident. Nonetheless, we sought to test experimentally whether all4160 and alr3699 play essential roles in heterocyst development or whether mutation of these genes has only a polar effect.
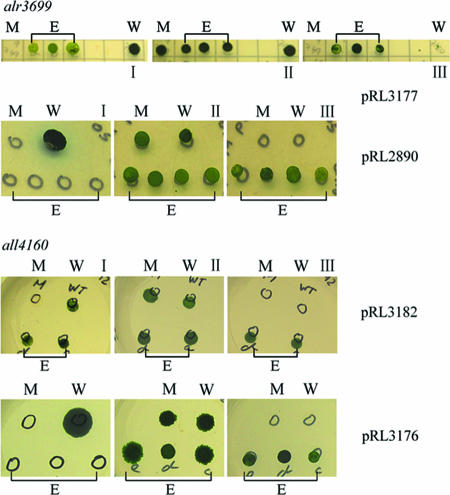
all4160 mutants FQ873 and FQ1421 were first successfully complemented with BAC clone anc0304, and alr3699 mutants FQ689 and FQ1351 were first complemented with BAC clone anc2015 (data not shown). In addition, complementation of all4160 mutant FQ792 was sought with plasmids pRL3182 and pRL3176, whose only intact Anabaena sp. strain PCC 7120 genes are all4160 and all4159, respectively, and complementation of alr3699 mutant FQ749 was sought with plasmids pRL3177 and pRL2890, which contain al3699 and both alr3700 and alr3701, respectively, as their only intact Anabaena sp. strain PCC 7120 genes (Fig. 1 and Table 1). The cloned fragments were all positioned to have their genes driven in the sense orientation by the glnA promoter in their vectors. pRL3182 complemented the mutation in FQ792, but pRL3176 did not, and pRL3177 complemented the mutation in FQ749, but pRL2890 did not (Fig. 5). These results support the idea that all4160 and alr3699 are required for complementation of their respective mutations, i.e., that the phenotypes of mutations in these genes are not due only to polar effects on their downstream genes.
FIG. 5.
Complementation of alr3699 mutant FQ749 and all4160 mutant FQ792. E, plasmid-bearing exconjugants; M, mutant; W, wild-type Anabaena sp. strain PCC 7120. Petri dishes contained agar-solidified medium AA (I), AAN (II), and AAN plus neomycin (to which the transposon in the mutant confers resistance) and either erythromycin (pRL3176, pRL3177) or spectinomycin (pRL2890, pRL3182), to which the potentially complementing plasmid confers resistance (III). Petri dishes were photographed at the end of the following periods of incubation: 4 weeks (pRL3177 series), 6 weeks (pRL2890 series), 2 weeks (pRL3182 series), and 2 months (pRL3176 series). All strains spotted grow in the presence of nitrate and the absence of antibiotics, only the exconjugant strains grow in the presence of neomycin plus a second antibiotic, and only the complemented mutant strains and wild-type Anabaena sp. strain PCC 7120 grow in the absence of fixed nitrogen. There was complementation of the mutants (i.e., growth of exconjugants in the absence of fixed nitrogen) by clones bearing a wild-type copy of the transposon-mutated gene (pRL3177, pRL3182), and there was no complementation by clones bearing a copy of the downstream gene(s) whose transcription may be affected as a polar effect of the transposon insertion. Especially when there is no spot of growth, the circles on some petri dishes show the approximate positions of spots.
This study, together with previous reports (23, 32, 36), shows that at least three predicted glycosyl transferase genes—all4160, hepB, and alr3699—located outside of the HEP island are up-regulated in developing heterocysts and that at least all4160 and alr3699 are required for the formation of HEP. The up-regulation of HEP island genes and of the hepB-alr3699 cluster appears to be highly dependent on hepK and hepN during heterocyst differentiation, and the up-regulation of all4160 is only partially dependent on these genes. Our unpublished results also indicate that other predicted glycosyl transferase genes (for example, all0919) are located outside of the HEP island and are up-regulated in developing heterocysts but are not essential for diazotrophic growth under aerobic conditions.
Supplementary Material
Acknowledgments
Work in the Xu laboratory was supported by the National Natural Science Foundation of China under grants 30170009 and 30570022. Work in the Wolk laboratory was supported by U.S. National Science Foundation grant MCB-0090232 and U.S. Department of Energy grant DE-FG02-91ER20021.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arabolaza, A. L., A. Nakamura, M. E. Pedrido, L. Martelotto, L. Orsaria, and R. R. Grau. 2003. Characterization of a novel inhibitory feedback of the anti-anti-sigma SpoIIAA on Spo0A activation during development in Bacillus subtilis. Mol. Microbiol. 47:1251-1263. [DOI] [PubMed] [Google Scholar]
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. (Printer's correction, 10:1153, 1994.) [DOI] [PubMed] [Google Scholar]
- 3.Black, T. A., and C. P. Wolk. 1994. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176:2282-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 6.Ehira, S., and M. Ohmori. 2006. NrrA, a novel nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 7.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 8.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, A., T. Black, Y. Cai, J. M. Panoff, D. N. Tiwari, and C. P. Wolk. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J. Bacteriol. 174:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, Q., G. Huang, S. Lechno-Yossef, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58:227-243. [DOI] [PubMed] [Google Scholar]
- 11.Fan, Q., S. Lechno-Yossef, S. Ehira, T. Kaneko, M. Ohmori, N. Sato, S. Tabata, and C. P. Wolk. 2006. Signal transduction genes required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6688-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193-1202. [DOI] [PubMed] [Google Scholar]
- 13.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 14.Hebbar, P. B., and S. E. Curtis. 2000. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J. Bacteriol. 182:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez, J. A., R. Y. Igarashi, B. Soboh, L. Curatti, D. R. Dean, P. W. Ludden, and L. M. Rubio. 2007. NifX and NifEN exchange NifB cofactor and the VK-cluster, a newly isolated intermediate of the iron-molybdenum cofactor biosynthetic pathway. Mol. Microbiol. 63:177-192. [DOI] [PubMed] [Google Scholar]
- 16.Hu, N. T., T. Thiel, T. H. Giddings, and C. P. Wolk. 1981. New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology 114:236-246. [DOI] [PubMed] [Google Scholar]
- 17.Huang, G., Q. Fan, S. Lechno-Yossef, E. Wojciuch, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, K. M., and R. Haselkorn. 2002. Newly identified cytochrome c oxidase operon in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 specifically induced in heterocysts. J. Bacteriol. 184:2491-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 20.Koksharova, O. A., and C. P. Wolk. 2002. Novel DNA-binding proteins in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 184:3931-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lechno-Yossef, S., Q. Fan, S. Ehira, N. Sato, and C. P. Wolk. 2006. Mutations in four regulatory genes have interrelated effects on heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:7387-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldener, I., S. Hannus, and M. Kammerer. 2003. Description of five mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst differentiation and identification of the transposon-tagged genes. FEMS Microbiol. Lett. 224:205-213. [DOI] [PubMed] [Google Scholar]
- 24.Matveyev, A. V., K. T. Young, A. Meng, and J. Elhai. 2001. DNA methyltransferases of the cyanobacterium Anabaena PCC 7120. Nucleic Acids Res. 29:1491-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller, P., M. Keller, W. M. Weng, J. Quandt, W. Arnold, and A. Puhler. 1993. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol. Plant-Microbe Interact. 6:55-65. [DOI] [PubMed] [Google Scholar]
- 26.Ning, D., and X. Xu. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447-453. [DOI] [PubMed] [Google Scholar]
- 27.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 47:1239-1249. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., and X. Xu. 2005. Regulation by hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, DC.
- 34.Wolk, C. P., J. Elhai, T. Kuritz, and D. Holland. 1993. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol. Microbiol. 7:441-445. [DOI] [PubMed] [Google Scholar]
- 35.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Wolk, C. P., and K. Zarka. 1998. Genetic dissection of heterocyst differentiation, p. 191-196. In G. Subramanian, B. D. Kaushik, and G. S. Venkataraman (ed.), Cyanobacterial biotechnology. Proceedings of the International Symposium, Sept. 18-21, 1996. Oxford and IBH Publishing Co., New Delhi, India.
- 37.Xu, X., J. Elhai, and C. P. Wolk. 2007. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 387-426. In T. Herrero and E. Flores (ed.), Cyanobacteria: molecular biology, genomics and evolution. Horizon Scientific Press, Norwich, United Kingdom.
- 38.Zhou, R., and L. Kroos. 2004. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 101:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, R., and C. P. Wolk. 2003. A two-component system mediates developmental regulation of biosynthesis of a heterocyst polysaccharide. J. Biol. Chem. 278:19939-19946. [DOI] [PubMed] [Google Scholar]
- 40.Zhu, J., R. Kong, and C. P. Wolk. 1998. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J. Bacteriol. 180:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.