Abstract
Three flagellar proteins, FliG, FliM, and FliN (FliGMN), are the components of the C ring of the flagellar motor. The genes encoding these proteins are multifunctional; they show three different phenotypes (Fla−, Mot−, and Che−), depending on the sites and types of mutations. Some of the Mot− mutants previously characterized are found to be motile. Reexamination of all Mot− mutants in fliGMN genes so far studied revealed that many of them are actually temperature sensitive (TS); that is, they are motile at 20°C but nonmotile at 37°C. There were two types of TS mutants: one caused a loss of function that was not reversed by a return to the permissive temperature (rigid TS), and the other caused a loss that was reversed (hyper-TS). The rigid TS mutants showed an all-or-none phenotype; that is, once a structure was formed, the structure and function were stable against temperature shifts. All of fliM and fliN and most of the fliG TS mutants belong to this group. On the other hand, the hyper-TS mutants (three of the fliG mutants) showed a temporal swimming/stop phenotype, responding to temporal temperature shifts when the structure was formed at a permissive temperature. Those hyper-TS mutation sites are localized in the C-terminal domain of the FliG molecules at sites that are different from the previously proposed functional sites. We discuss a role for this new region of FliG in the torque generation of the flagellar motor.
Many species of bacteria are propelled by flagella. The flagellar motor is a rotary motor powered by the proton gradient across the cell membrane (13). The mechanism of torque generation by the motor has not been determined. In general, torque in a rotary motor is generated between a stator and rotor. Mutations, even single amino acid substitutions, in any one of five flagellar proteins (MotA, MotB, FliG, FliM, and FliN) can give rise to a Mot− or paralyzed phenotype (22, 23). It is believed that a complex of MotA and MotB (MotAB complex) forms the stator, which is anchored to the rigid peptidoglycan layer. FliG, FliM, and FliN (FliGMN) form a cup-shaped structure called the C ring at the bottom of the flagellar basal body, which is thought to be the rotor (4, 6, 9, 10). Protons flow through a channel formed by the MotAB complex, which interacts with the rotor to generate torque.
Although all three components of the C ring (FliGMN) give rise to a Mot− phenotype, Lloyd et al. (11) previously argued that FliG but not FliM or FliN played a direct role in torque generation. The reason was their observation that some fliM and fliN mutants were nonmotile at normal expression levels but became motile when the mutant proteins were overexpressed, whereas fliG (Mot−) mutations abolish motility at all expression levels (17). Zhou and collaborators have extensively studied the interaction between the MotAB complex and FliG. Zhou et al. (26) found evidence that five conserved, charged residues in FliG were important for torque generation, and they are K90 and E98 in MotA and K264, R281, D288, D289, and R297. Those authors proposed a model of the torque generation mechanism based on the electrostatic interaction between charged residues in MotA and those in FliG. The model was supported by data reported previously by Lloyd et al. (12), who solved the structure of the C-terminal region of FliG and showed that the five conserved, charged residues formed a ridge on the surface of FliG's C-terminal domain; they proposed that the charge-bearing ridge faced a complementary line of charged residues in the MotA cytoplasmic domain. Brown and collaborators further analyzed interactions between FliG and FliM using the method of Trp replacement in FliG (3). A complete understanding of the motor's mechanism depends on having a detailed structure of the FliG-MotAB complex.
Using the Na+-driven motor of Vibrio alginolyticus, Yorimitsu et al. (24) tested the model described previously by Zhou et al., with different results. Surprisingly, even when any of the conserved charged residues in PomA, the ortholog of MotA, were replaced by neutral residues (either Ala or Asn), the mutant cells were still motile (24). They further constructed multiple substitutions of the five residues on the charge-bearing ridge of FliG with neutral ones and confirmed that cells with the mutant FliG were still functional (25). They suggested that the important charged residues might be located in different places from the charge-bearing ridge claimed in the previously reported model and that the electrostatic interactions detected in Escherichia coli therefore might not be a general feature of flagellar motors. Yakushi et al. (21) carried out a comparative study of chimeric motors in E. coli cells that were engineered to use V. alginolyticus stator components, rotor components, or both and concluded that the charged residues of MotA (PomA) and FliG were still important in both species but that the rotor-stator interface in V. alginolyticus was more robust and not disrupted by the mutations.
To try to resolve the discrepancies in the results from the two different groups, we reexamined the same mutants employed previously. Lloyd et al. (11) previously observed that populations of certain fliM (Mot−) mutants contain a small number of motile cells, and assuming that these mutations did not fully abolish torque generation, they thus concluded that FliM was not essential for torque generation. We observed the same phenomenon and found that not only fliM but also fliG and fliN (Mot−) mutants were actually temperature sensitive (TS). Temperature sensitivity of fliGMN (Mot−) mutants has been noticed previously (8) and partly analyzed (7, 16, 20) but not fully discussed. In this paper, we have carried out a thorough analysis of these TS mutants and proposed a new working unit for torque generation.
MATERIALS AND METHODS
Strains and growth conditions.
All strains used in this study were derived from Salmonella enterica serovar Typhimurium SJW1103. MY strains were kindly provided by May Macnab. Cells were cultivated in LB medium (1% peptone, 0.5% yeast extract, 1% NaCl [pH adjusted to 7]) at various temperatures by gentle shaking and harvested at around an optical density at 660 nm of 1.0. Incubation times were 15 h at 37°C and 35 h at 20°C. Reagents were of the first grade and purchased from Wako Pure Chemical Industries, Ltd. (Tokyo, Japan).
Dark-field microscopy and recording of images.
Cells were analyzed in a concaved well on a hole-slide glass (Toshinriko, Oosaka, Japan) observed with a phase-contrast microscope (Olympus CH-2) and recorded on a video tape recorder through a charge-coupled-device camera (Panasonic BL200) attached to the microscope. Swimming speeds were measured on a screen from frames at 1-s interval.
Analysis of rotating speeds of tethered cells.
Tethered cells were made by a conventional method using anti-flagellin antibody. Images of rotating cells were centered in the monitor screen and analyzed by software developed for this purpose (DAGA Graphics Co., Ltd., Utsunomiya, Japan). The basic procedure of measurements is to put the marker spot at a point where a rotating cell body passes by, measure the color density of the spot, take the difference of the densities from previous frames, find the highest peak, and count frames between two peaks for one rotation (this was done automatically, and data were stored in a comma separated values [CSV] file). To take the average time for one rotation, frames showing smooth rotation in one run (about 40 rotations for 5 s) were manually chosen.
Temperature shift experiments.
Cells grown at a given temperature were briefly incubated in a bath at the other temperature, and their motility was observed with the microscope, whose stage and glass slides were preheated at the desired temperature for observation. To prevent cell growth during the experiments, chloramphenicol was added into the medium to a final concentration of 25 μg/ml at 5 min before observation. During the experiments, for 1 h, the optical density of the culture did not increase, indicating that cell division was halted.
DNA sequencing of fliN mutants.
Mutation sites of some new strains of fliN mutants were analyzed by DNA sequencing. The genome of the fliN mutant was purified using a DNeasy tissue kit (QIAGEN). A PCR fragment including mutated fliN was amplified using the purified genome as a template, an ExTaq (TaKaRa) polymerase, and the following sets of primers: forward primer 5′-CATTATTGCTCATGTGGACGG-3′, whose sequence is located 129 bp upstream of the fliN start codon, and reverse primer 5′-ATCCCATACGCTTAATCACCC-3′, whose sequence is located 132 bp downstream of the fliN stop codon. The length of the PCR fragment was 675 bp. After ligation with the pGEM-T Easy vector (Promega), the plasmid was introduced into E. coli DH5α cells and selected according to instructions provided by the manufacturer. Objective plasmids were purified using a QIAprep Spin Miniprep kit (QIAGEN) and used to determine the sequence. To determine the sequence, a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) and an ABI PRISM 3100 genetic analyzer (Applied Biosystems) were used.
Building molecular models for FliG and FliN.
The data for coordinates of FliG, FliM, and FliN were obtained from the RCSB Protein Data Bank (PDB). Molecular models were created using the free software Chime from Elsevier MDL (http://www.mdl.com/index.jsp).
RESULTS
Some Mot− mutants that do not swarm on agar can swim in liquid.
We often found a small fraction of swimming cells in cultures of some Mot− mutants of fliG, fliM, and fliN. Since the fraction of motile cells increased when the culture was left overnight at room temperature, we suspected that these mutants might be TS. For example, the colonies from strain SJW1774 showed no swarms on agar at any temperature, as illustrated in Fig. 1, but when the cells were grown at 20°C, most cells were indeed swimming under the microscope (Tables 1 to 3). Thus, strain SJW1774 is actually a TS Mot− mutant and shows weak motility at the permissive temperature. Also, Mot− mutants SJW2827, SJW1770, SJW1771, and SJW1799 were originally selected on agar, since the cells produced no swarms at any temperature (Fig. 1). However, they turned out to be motile in liquid, and hence, their TS phenotypes were missed. Therefore, we reexamined all of the Mot− mutants in our stock in liquid media.
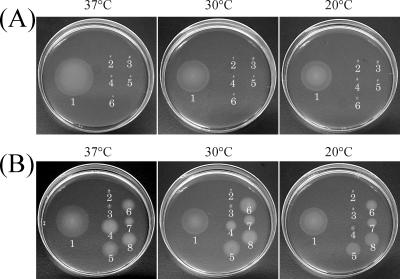
FIG. 1.
Swarm plates of TS switch (Mot−) mutants. Strains were inoculated on 0.3% agar plates and incubated at 37°C for 9 h, at 30°C for 15 h, and at 20°C for 20 h. (A) Swarm plates of TS switch (Mot−) mutants. 1, wild-type SJW1103; 2, SJW1774 [fliG(F236V)]; 3, SJW2827 [fliG(P127L)]; 4, SJW1770 [fliM(T147I)]; 5, SJW1771 [fliM(G132D)]; 6, SJW1799 [fliN(G103V)]. (B) Swarm plates of slow-swimming motAB mutants. 1, wild-type SJW1103; 2, MY6029 [motA(E33K)]; 3, MY6056 [motA(R90L)]; 4, MY6089 [motA(G176S)]; 5, MY6097 [motA(V207S)]; 6, MY6003 [motB(A37V)]; 7, MY6018 [motB(K28R)]; 8, MY6051 [motB(I29T)].
TABLE 1.
Data for fliG (Mot−) mutants
| Strain | Amino acid change | Motile fraction (%) at:
|
Groupa | |
|---|---|---|---|---|
| 20°C | 35°C | |||
| SJW1759 | Q128H | 70 | 0 | A |
| SJW1766 | F236S | 0 | 0 | C |
| SJW1768 | I308S | 10 | 0 | B |
| SJW1769 | Δ(294-297) | 0 | 0 | C |
| SJW1774 | F236V | 75 | 0 | A |
| SJW1777 | L239Q | 0 | 0 | C |
| SJW1778b | V254E | 39 | 0 | B |
| SJW1791 | Δ(077-126) | 0 | 0 | C |
| SJW1793c | E302D | 15 | 20 | D |
| SJW1798 | L250R | 0 | 0 | C |
| SJW2806 | R154P | 0 | 0 | C |
| SJW2810 | D244Y | 85 | 7 | A |
| SJW2814 | Δ(303-317) | 0 | 0 | C |
| SJW2816 | A217P | 0 | 0 | C |
| SJW2818 | Δ(099-146) | 0 | 0 | C |
| SJW2819 | T132P | 85 | 0 | A |
| SJW2820 | A200E | 0 | 0 | C |
| SJW2823 | A199E | 0 | 0 | C |
| SJW2826 | Δ(282-287) | 0 | 0 | C |
| SJW2827 | P127L | 93 | 15 | A |
| SJW2828 | L251P | 0 | 0 | C |
| SJW2829 | F236C | 0 | 2 | D |
| SJW2830 | K273E | 70 | 0 | A |
| SJW2831 | L259P | 0 | 0 | C |
| SJW2833 | L250H | 15 | 0 | B |
| SJW2834 | R160H | 7 | 0 | B |
Group A, motile fraction at 20°C was >60%; group B, motile fraction was between 10% and 60%; group C, nonmotile at all temperatures; group D, exceptions (see the text).
SJW1778 cells were most actively motile (95%) at 15°C.
SJW1793 cells were most actively motile (97%) at 30°C.
TABLE 3.
Data for fliN (Mot−) mutants
| Strain | Amino acid change | Motility fraction (%) at:
|
Groupa | |
|---|---|---|---|---|
| 20°C | 35°C | |||
| SJW1758 | Y104N | 2 | 0 | B |
| SJW1765 | L105Q | 3 | 7 | D |
| SJW1767 | L78Q | 0 | 0 | C |
| SJW1779 | L78P | 0 | 0 | C |
| SJW1799 | G103V | 25 | 0 | B |
| SJW1802 | V38A | 6 | 0 | B |
| SJW2406 | D98V | 12 | 0 | B |
Group B, motile fraction was between 10% and 60%; group C, nonmotile at all temperatures; group D, exceptions (see the text).
Identification of TS Mot− mutants.
There are 26 fliG (Mot−), 12 fliM (Mot−), and 7 fliN (Mot−) mutants in our stock (Tables 1 to 3), many of which were also used by other laboratories for functional assays (7, 11, 16, 19). Each of them was grown in LB medium either overnight at 35°C or for about 40 h at 20°C, and the motile fraction (percent) in a population was measured by microscopy. We found that many of the mutants were indeed TS: 12 strains of fliG mutants, 10 strains of fliM mutants, and 5 strains of fliN mutants. They were further divided into three groups (groups A, B, and C) depending upon the motile fraction at the permissive temperature (Tables 1 to 3). Group A cells were actively motile at 20°C, and the motile fraction was more than 60% of the population. This group contains six fliG mutants (SJW2827/P127L, SJW1759/Q128H, SJW2819/T132P, SJW1774/F236V, SJW2810/D244Y, and SJW2830/K273E) and six fliM mutants (SJW1762/F131C, SJW1764/F131L, SJW1771/G132D, SJW1783/G133D, SJW1770/T147I, and SJW1808/T147P) but no fliN mutants. Although most of the cells in group A were motile at 20°C and not motile at 35°C, small fractions from some strains (SJW2827/P127L, SJW2810/D244Y, SJW1783/G133D, SJW1770/T147I, and SJW1808/T147P) were still motile at 35°C.
Group B cells were not motile at 35°C and only poorly motile at 20°C. The percentage of motile cells was between 10% and 60%. Group B includes four fliG mutants (SJW2834/R160H, SJW2833/L250H, SJW1778/V254E, and SJW1768/I308S), four fliM mutants (SJW1805/F131S, SJW1760/T149K, SJW1796/L250R, and SJW1763/L272R), and four fliN mutants (SJW1802/V38A, SJW2406/D98V, SJW1799/G103V, and SJW1758/Y104N).
Group C contains the rest of the fliG mutants (14 strains), two fliM mutants, and two fliN mutants. Most of the mutant strains in this group were not motile at either temperature (Tables 1 to 3). Exceptions are SJW2829/F236C, where cells swam at 35°C but not at 20°C, and SJW1793/E302D and SJW1765/L105Q, where cells swam at both temperatures (group D) (Tables 1 to 3).
All TS mutants in groups A and B had point mutations and not deletions. It is interesting that some sites gave rise to two different properties depending on the substitutions: F236V of FliG gave rise to TS properties, but neither F236S nor F236C did. The same is true for L250H/L250R of FliG; F131C/F131L/F131S, T147I/T147P, and L250Q/L250R of FliM; and L78P/L78Q of FliN. Although there seems to be no rule in the pattern of amino acid changes (in terms of sizes and charges), there must be a particular combination of substituted amino acids and mutation sites (so-called hot spots) that show a TS phenotype. In the two instances (F131S/C and F236S/C), where a sharp reduction of motility has been observed, the neutral (aromatic) side chain is replaced by polar O-H or S-H groups capable of forming hydrogen bonds and thereby possibly resulting in an altered, nonfunctional conformation.
Mutation sites that give rise to the non-TS Mot− phenotype are not localized in one domain but are spread throughout the molecule. This suggests that the whole molecule is important for motor function. Perhaps a subtle conformational change almost anywhere can cause a distortion of the entire rotor (C ring) structure and cause a loss of function. The TS Mot− sites, in contrast, are localized. To understand these TS mutants, we turned our attention to the group A cells; group B cells, with their poorer motility, were left for future studies.
Reasons why some strains are motile but do not swarm.
Many of the group A strains that recovered motility in liquid at 20°C did not swarm even at the same temperature. Cells that swim smoothly (by counterclockwise [CCW] rotation of the motor) for too large a fraction of time or that swim tumbly (by clockwise [CW] rotation of the motor) for too large a fraction of time do not form swarm rings on agar. We therefore examined the switching probability of the flagellar motor rotation of the group A strains by using the conventional tethered-cell method.
(i) TS (Che−) mutants do not swarm.
As summarized in Table 4, many mutants of the group A strains were defective in their switching between CCW and CW rotations. Three strains of fliG mutants (P127L, Q128H, and T132P) and four strains of fliM mutants (F131L, F131C, G132D, and G133D) swam tumbly; that is, they rotated CW more often than CCW. An fliG(Q128H) mutant was exclusively CW biased. Two of fliM mutants (T147I and T147P) swam smoothly; that is, they rotated exclusively CCW. Therefore, these mutants cannot form swarm rings on agar due to defective switching. However, three strains of fliG mutants (F236V, D244Y, and K273E) swam like the wild type. What are the reasons for these TS mutants not to swarm?
TABLE 4.
Switching probability of group A TS mutants at 20°C
| Tethered cell type | Mutant(s) with swimming pattern of:
|
||
|---|---|---|---|
| Smooth | WT (shaky)a | Tumbly | |
| CCW biased | fliM(T147I, T147P) | ||
| WT | fliG(F236V, D244Y, K273E) | ||
| CW biased | fliG(P127L, Q128H, T132P) and fliM(F131L, F131C, G132D, G133D) | ||
These three mutant cells swim chemotactically but more shakily than the wild type (WT).
(ii) Slow-swimming cells do not swarm.
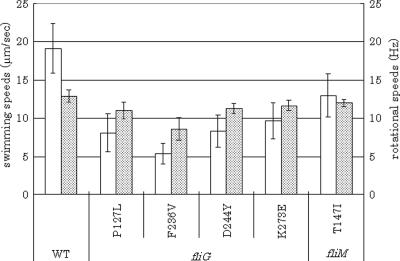
We determined swimming speeds and motor rotation rates in these three fliG strains. A fliG mutant (P127L) and a fliM mutant (T147I) were also examined for control and comparison. To make measurements easier, we eliminated switching by constructing cheY::Tn10 and fliGMN (Mot−) double mutants. In the video-recorded images, cells that were most actively swimming were chosen to measure the possible highest speeds. Swimming speeds of 10 to 20 active cells in each experiment were averaged. Tethered cells were prepared by the conventional method using anti-flagellin antibody. The rotational speeds of tethered cells were measured by the video image analyzer that we developed (see Materials and Methods). Both the swimming speed and the rotational speed of fliG(F236V) were the lowest among the mutants (Fig. 2). Thus, we conclude that torque produced by this mutant is too weak to make a swarm ring on agar. On the other hand, the other two fliG mutants (D244Y and K273E) showed swimming speeds that were a little weaker than that of the wild type but comparable to that of control strains, fliG(P127L) and fliM(G132D,T147I). Their rotational speeds were close within measurement errors to that of the wild type. Why were swarms not formed by these two fliG strains (D244Y and K273E) that can swim and produce torque at wild-type levels?
FIG. 2.
Swimming speeds and rotational speeds of group A TS mutants at 20°C. Empty columns show swimming speeds, and shaded columns show rotational speeds of wild-type SJW1103 (on the most left), fliG (Mot−) mutants (P127L, F236V, D244Y, and K273E), and a fliM (Mot−) mutant (T147I).
(iii) Short-hook mutants do not swarm.
When swimming cells of these two strains were observed by dark-field microscopy, the cells formed not tight but loose bundles of flagellar filaments. This loose bundle formation was previously observed in hook-shaped mutants (S.-I. Aizawa, unpublished observations). Many mutations in fliGMN produce short hooks (14). Thus, we isolated the hook-basal bodies from our two mutants and measured the hook length. The hook lengths of D244Y and K273E mutants were 47.1 ± 9.8 nm (n = 94) and 48.9 ± 8.4 nm (n = 121), respectively, which are shorter than that of wild type (55.0 ± 6.0 nm). We conclude that some cells had hooks that were too short to permit the proper bundling of flagellar filaments and thus failed to form swarm rings. Since some of the strains mentioned above also have short hooks (14), their inability to swarm might be caused by a combination of biased rotation and short hook length.
Temporal change of swimming speeds of TS mutants by temperature shifts.
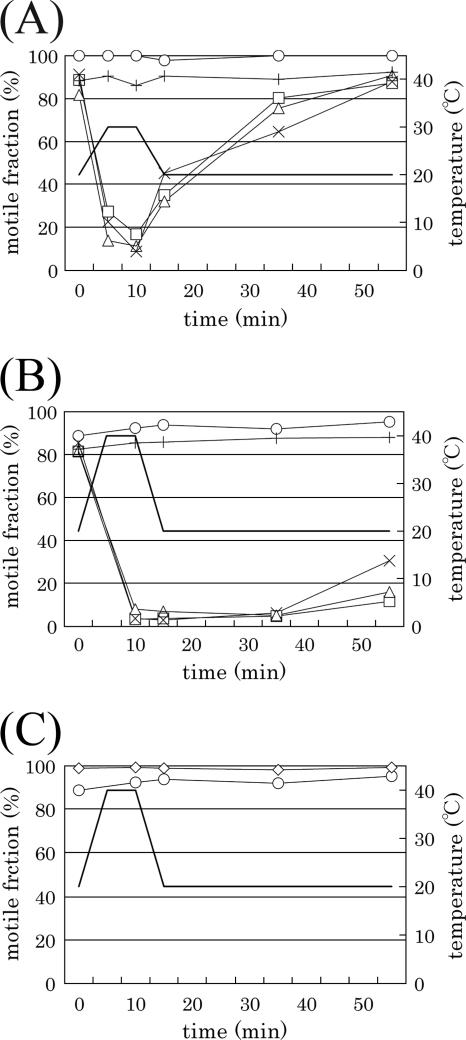
The TS mutants were tested for their response to temperature shifts. Three fliG mutants (F236V, D244Y, and K273E) showed a fast response to a temperature shift; when the temperature was shifted from 20°C to 30°C, the cells slowed down but quickly recovered full motility within 5 min after temperature was lowered back to 20°C (Fig. 3A). When the temperature was shifted from 20°C to 40°C, the swimming speeds of the cells slowed down within 5 min or stopped (Fig. 3B). With a shift to 40°C, the cells stopped and did not recover motility immediately after the temperature was lowered to 20°C. Only 10% of the population was motile after 20 min when returned to 20°C. Since the culture medium contains chloramphenicol, the recovery of motility is not because of the newly synthesized proteins. We term these strains temperature-hypersensitive (hyper-TS) strains because wild-type cells or even ordinary TS mutant cells that were grown at 20°C could swim normally up to 50°C. Moreover, neither the rest of fliG mutants (as represented by T132P) nor any of the fliM (as represented by T147P) or fliN (not shown) mutants changed their swimming behavior following temperature shifts (Fig. 3A, B, and C). Interestingly the mutation sites that give rise to the hypersensitive phenotype are localized to one surface region of FliG (see below and Discussion).
FIG. 3.
Swimming behaviors of TS mutants responding to temporal temperature shifts. Cells were grown at 20°C, shifted to the incubator at 30°C or 40°C for 10 min, and then returned to 20°C (thick line and measures on the right). Changes of motile fractions in group A TS mutants are shown. (A) Behavior of hyper-TS fliG (Mot−) mutants (F236V, D244Y, and K273E) caused by a shift from 20°C to 30°C shift; T132P represents ordinary TS fliG (Mot−) mutants. (B) Behavior of the same set of fliG (Mot−) mutants as described above (A) caused by a shift from 20°C to 40°C. (C) Behavior of ordinary TS fliM (Mot−) mutants caused by a shift from 20°C to 40°C. The motile fraction of each strain was measured as mentioned in the text. Signs in the graph are as follows: ○, wild type; +, fliG(T132P); □, fliG(F236V); ×, fliG(D244Y); ▵, fliG(K273E); ⋄, fliM(T147P).
Mot− mutants in motAB are not TS.
We found many TS Mot− mutants in fliGMN in this work. However, there are no Mot− mutants of either the motA or the motB gene in the literature, and we have failed to isolate TS mutants despite an exhaustive search. Perhaps MotAB may better tolerate temperature shifts. There is a report of Mot− TS mutants using a chimera of PomA and MotB (5). The explanation is not straightforward because of the complexity of the construct, and we regard this mutation as being an anomalous exception (see Discussion). There are slow-swimming mutants of motA and motB, which were isolated as repressors of an fliG (CW-biased Che−) mutant (19). Since the behavior of fliGMN TS mutants mentioned above resembles that of these mot mutants, we examined their temperature sensitivities (Fig. 1B). The swarm sizes of the slow-motile (motA or motB) mutants were larger than those of fliGMN TS mutants on soft agar plates, suggesting that the latter swim slower than the former. However, the swarm sizes of motAB slow-swimming mutants did not change at lower temperatures (Fig. 1B). Motile fractions of these mutants were also unchanged after a temperature shift (data not shown). Altogether, MotAB are rarely temperature sensitive and show functional stiffness, suggesting that the Mot complex may have a different role in torque generation (see Discussion).
Distribution of TS mutation sites on FliGMN.
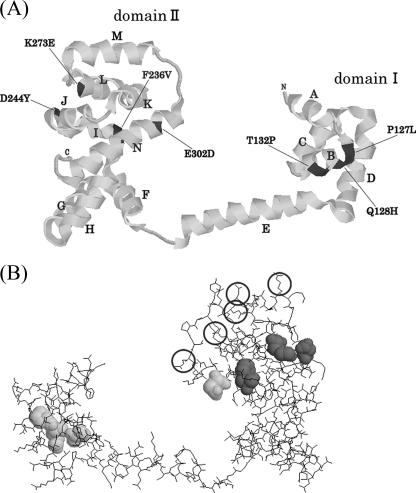
The atomic structures, albeit partial, of FliG, FliM, and FliN molecules have been known (1, 2, 3, 15). We have placed our TS mutational sites on the most recent model of FliGMN molecules proposed previously by Brown et al. (3) (Fig. 4).
FIG. 4.
Molecular models of FliG. (A) Ribbon diagram of FliG domain structures. Distinguishable strands are indicated by the strand names, and TS mutation sites are indicated by bars and the dark ribbon. (B) Back view of FliG (molecular wireframe model). Hyper-TS mutation sites are indicated by dark balls, ordinary TS sites are indicated by lighter balls, and the functional sites previously suggested are indicated by circles.
FliG.
The original Mot− mutation sites were scattered all over the FliG molecule, except at the N-terminal region in which neither Che− nor Mot− sites have been found. However, TS Mot− mutation sites were localized mainly in two domains, called domains I and II, as shown in Fig. 4A (1, 3). TS mutation sites in domain I containing the EHPQR motif have also been known to be responsible for the switching of the rotational direction (3).
TS sites in domain II locate on four helices, J, K, and L, that form the hydrophobic patch (3). The hyper-TS sites (F236V, D244Y, and K273E) that responded to temporal temperature shifts locate on the other side of the domain; residue F236V is hydrophobic and buried, and K273E has the aliphatic parts of its side chain partially buried. D244Y is hydrophilic and exposed on the surface (Fig. 4B, dark balls). Note that these sites are different from those on the charge-bearing ridge previously proposed for the functional region of torque generation (Fig. 4B, circles). Although the site that we identified is half buried in the current model, it is not clear what conformation the region would take when FliG binds FliM. We propose this region as a new candidate for the functional site (see Discussion). Interestingly, those residues (F236V, D244Y, and K273E) are not necessarily charged.
FliM.
The FliM middle domain has a pseudo-two-fold symmetric topology (15). TS sites of FliM were localized on a disordered loop between α3 and α1′, which connects the two pseudosymmetric halves of the molecule and probably interacts with FliG. The loop contains a highly conserved GGXG motif, and the first two G's were actually TS sites. The loop region was previously assigned to the CCW-biased region by Park et al. (15), which incidentally contradicts our data. Our TS CCW-biased mutants were localized at residue 147 on the α1′ helix, which is in their CCW-biased region.
FliN.
FliN forms a butterfly-shaped dimer (2). TS residues are localized in the central region. Since these residues sit near the hinge region of the dimer, a subtle conformational change of the region may cause a drastic change in the whole molecule. The role of this region in torque generation, however, awaits the uncovering of the whole motor structure.
In conclusion, point mutations in the fliGMN mutants gave rise to various phenotypes that affect motility: a defect in protein transport that results in short hooks, a defect in torque generation, and a defect in the switching of rotation. This versatility in function accompanies the fragility of the structure; many of the mutants are TS. Hyper-TS mutations are so subtle that they do not induce the conformational changes in the whole structure but only in a local area, which might be directly involved in torque generation.
DISCUSSION
We have reexamined the behavior of fliGMN (Mot−) mutants and found that many of them were TS mutants when tested for swimming in liquid but nonfunctional when tested for swarming on agar. The mutation sites were often separated on a protein [e.g., FliG(P127L) and FliG(F236V)]. Perhaps subtle conformational changes in a local area on FliG induce structural distortions in the torque-generating parts, resulting in a loss of function.
TS mutations are likely biphasic; the protein takes on one conformation at low temperatures and another at higher temperatures. Brown et al. (3) proposed a model for subunit organization in the switch complex, where two conformations between FliM and FliG are introduced. FliM can interact with two domains of FliG: one with the EHPQR motif (outboard location) and the other with the hydrophobic patch (inboard location). Incidentally, FliG(P127L) and FliG(Q128H) are on the EHPQR motif, and FliG(F236V) is in the vicinity of the hydrophobic patch. We suspect that those mutants that Brown et al. (3) previously analyzed might be also TS mutants, perhaps like the hyper-TS mutant that we report here. If it is the case, it will be interesting to see if they are hyper-TS or not.
Three residues on the FliG molecule (F236V, D244Y, and K273E) were hypersensitive to temperature shifts when cells were grown at the permissive temperature (20°C) and transferred to a higher temperature. When the temperature was raised to 30°C, cells lost the swimming speed but could recover full motility when the temperature was lowered again. However, when the temperature was raised to more than 35°C, the change was irreversible; cells could not recover motility. When cells were grown at a nonpermissive temperature (35°C) and transferred to lower temperatures, cells did not recover motility until 25 min (data not shown). In a narrow range of temperatures, the hyper-TS region affects the rotational speed.
Do we have evidence for direct interaction between two components?
Previous studies on the interaction between the stator and the rotor (for example, between MotA and FliG) were dependent mostly on genetic analyses of Mot− mutants or their suppressor mutants. One amino acid substitution in the interaction sites would naturally result in defective motility, which can be restored in some cases by a compensating change in charge on the other site. The genetic analysis was effective in identifying proteins directly involved in function. If the protein functions as a monomer or a homo-oligomer, genetic methods would still be useful to identify functional sites (such as hot spots) on the protein. However, if the components form a supramolecular structure like the C ring, it is not straightforward enough to identify the functional sites on each protein by the same genetic method. For example, one-amino-acid substitutions on one domain of a protein would induce conformational changes of the whole structure, consequently giving the same results as mutations on other domains. Here, we discuss how strong the evidence is for such interaction sites on a molecule determined by those analyses.
(i) Point mutations.
Mutations resulting in a Mot− phenotype are scattered in several separate domains of the FliG molecule (7, 11). There is no way to distinguish direct interactions from indirect ones from these mutants, as long as we judge the cause simply from the Mot− phenotype.
(ii) Suppressor mutations.
Revertants of a specific mutation in FliG gave rise to many suppressors in MotB (19). Since the suppressors spread in both the cytoplasmic and periplasmic domains of MotB, we see that both direct and indirect interactions are involved in the suppressor analysis. Even allele specificity does not necessarily guarantee the specific interaction between two proteins.
(iii) Dominance effects.
When wild-type and mutant proteins coexist, one of them may interact with some proteins more strongly than others. This simply indicates the differences of interaction strengths between wild-type and mutant proteins, because the mutation sites are not necessarily the interaction sites.
(iv) Dose effects.
The phenotype of a mutant may simply be a reflection of its poor binding to the rest of the structure. For example, a Mot− mutation in FliN can be rescued by overproduction of itself (11), indicating that the mutation site was not essential for torque generation but was essential for the construction of the FliN ring (2). However, FliN itself is somehow necessary for torque generation, because the motor does not function without the FliN ring structure.
Therefore, we conclude that there is no direct evidence of torque generation sites on FliGMN molecules deduced from genetic analyses. Any genetic or molecular biological manipulation of these proteins may affect the conformation of the whole supramolecular structure and consequently alter function. However, TS mutants, especially hyper-TS mutants, may indicate the interaction sites on the proteins more directly.
Meaning of temperature hypersensitivity.
There are at least two types of TS mutants: ordinary (or rigid) TS mutants and hyper-TS mutants. The ordinary TS mutants can form a correct conformation of the motor at permissive temperatures but form a wrong one at nonpermissive temperatures. Once the correct conformation is formed at permissive temperatures, the motor is functional even at nonpermissive temperatures. Therefore, this effect on torque generation is indirect.
On the other hand, the hyper-TS mutants are very sensitive to temperature shifts. The motor speed changes according to the temperature applied to the motor. This quick response to temperature indicates that the sites are not constrained in one conformation but retain freedom of movement. Remember that ordinary TS mutant proteins were constrained in one conformation and lost the freedom to move. Within a certain temperature range, the movement of the hyper-TS site is directly reflected by the torque. In either lower or higher temperature ranges, the motor speed slows down, indicating that a certain frequency of the thermal vibration of the site and the synchronized interaction of these sites might be important for optimal speed.
Model of torque generation.
None of the conventional motAB mutants has shown any temperature sensitivity. One exception is a TS mutant of pomA, a motA homolog of V. alginolyticus (5). One of those pomA mutants has multiple mutations (R88A, K89A, E962, E972, E992, L131F, and T132M). It is not straightforward to apply the results obtained from V. alginolyticus to E. coli, because there are several differences between the two flagellar systems: a sodium-driven versus a proton-driven motor (21). It is desirable to see if the same mutation in E. coli or Salmonella enterica serovar Typhimurium motA could result in a TS phenotype.
The current model claims that MotA faces FliG. Although this model is most plausible at the moment, it does not fully explain how the motor generates torque. Modification of the model will be helpful to solve the mystery of the motor. While FliG retains TS properties, MotA does not. Why do FliG and MotA not share the TS properties, if they face each other and work together to generate torque? Perhaps MotA does not face FliG and does not directly generate torque but simply acts as a proton conduit. This possibility does not disagree with previously obtained data. What, then, is the rotor, and what is the stator?
Consider the complex of FliGMN: all of the component proteins are involved in both torque generation and switching of rotational direction, which might be inseparable at the molecular level. Rotation might occur between the rings made by FliG and FliMN, as suggested previously by Thomas et al. (18). To test this hypothesis, we must observe the rotation of the C ring (a complex of FliM and FliN) and the M ring (a complex of FliF and FliG). The tertiary structures of parts of FliGMN have been solved. The quaternary structures of FliGMN, however, are surmised mostly from results of genetic analyses of many fliGMN mutants and their revertants. We stress again that the data from genetic analyses and molecular biological experiments do not necessarily indicate direct interactions between components of interest in a supramolecular structure. Data from our analysis of TS mutants are not easily interpreted by the current model of interactions between MotAB and FliG (12). We need a new model that satisfies all data.
TABLE 2.
Data for fliM (Mot−) mutants
| Strain | Amino acid change | Motile fraction (%) at:
|
Groupa | |
|---|---|---|---|---|
| 20°C | 35°C | |||
| SJW1760 | T149K | 20 | 0 | B |
| SJW1762 | F131C | 60 | 0 | A |
| SJW1763 | L272R | 13 | 0 | B |
| SJW1764 | F131L | 99 | 1 | A |
| SJW1770 | T147I | 98 | 20 | A |
| SJW1771 | G132D | 90 | 1 | A |
| SJW1783 | G133D | 70 | 10 | A |
| SJW1796 | L250R | 10 | 0 | B |
| SJW1804 | L250Q | 0 | 0 | C |
| SJW1805 | F131S | 15 | 0 | B |
| SJW1808 | T147P | 98 | 30 | A |
| SJW1813 | H106P | 0 | 0 | C |
Group A, motile fraction at 20°C was >60%; group B, motile fraction was between 10% and 60%; group C, nonmotile at all temperatures.
Acknowledgments
We thank N. Nishimichi, K. Miyamoto, M. Aosasa, H. Matsuda for DNA sequencing; May Macnab for MY strains; M. Kanna for critical reading of the manuscript; and David DeRosier for intensively revising the manuscript.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Brown, P. B., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, P. N., M. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, P. N., M. Terrazas, K. Paul, and D. F. Blair. 2007. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka, H., T. Yakushi, and M. Homma. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J. Bacteriol. 186:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garza, A. G., L. W. Harris-Haller, R. A. Stoebner, and M. D. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, C. J., R. M. Macnab, H. Okino, and S.-I. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 9.Katayama, E., T. Shiraishi, K. Oosawa, N. Baba, and S.-I. Aizawa. 1996. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J. Mol. Biol. 255:458-475. [DOI] [PubMed] [Google Scholar]
- 10.Khan, S., I. H. Khan, and T. S. Reese. 1991. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J. Bacteriol. 173:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472-475. [DOI] [PubMed] [Google Scholar]
- 13.Macnab, R. M. 1996. Flagella, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 14.Makishima, S., K. Komoriya, S. Yamaguchi, and S.-I. Aizawa. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291:2411-2413. [DOI] [PubMed] [Google Scholar]
- 15.Park, S. Y., B. Lowder, A. M. Bilwes, D. F. Blair, and B. R. Crane. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. USA 103:11886-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang, H., S. Billings, X. Wang, L. Sharp, and D. F. Blair. 1995. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J. Bacteriol. 177:3496-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Togashi, F., S. Yamaguchi, M. Kihara, S.-I. Aizawa, and R. M. Macnab. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yakushi, T., J.-H. Yang, H. Fukuoka, M. Homma, and D. Blair. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and N+-driven motors in Escherichia coli. J. Bacteriol. 188:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi, S., H. Fujita, A. Ishihara, S.-I. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yorimitsu, T., Y. Sowa, A. Ishijima, T. Yakushi, and M. Homma. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403-413. [DOI] [PubMed] [Google Scholar]
- 25.Yorimitsu, T., A. Mimaki, T. Yakushi, and M. Homma. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 334:567-583. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]