Abstract
Histidine biosynthesis is one of the best studied metabolic pathways in bacteria. Although this pathway is thought to be highly conserved within and between bacterial species, a previous study identified a genetic region within the histidine operon (his) of nontypeable strains of Haemophilus influenzae (NTHI) that was more prevalent among otitis media strains than among throat commensal NTHI strains. In the present study, we further characterized this region and showed that genes in the complete his operon (hisG, -D, -C, -NB, -H, -A, -F, and -IE) are >99% conserved among four fully sequenced NTHI strains, are present in the same location in these four genomes, and are situated in the same gene order. Using PCR and dot blot hybridization, we determined that the his operon was significantly more prevalent in otitis media NTHI strains (106/121; 87.7%) than in throat strains (74/137; 54%) (prevalence ratio, 1.62; P < 0.0001), suggesting a possible role in middle ear survival and/or acute otitis media. NTHI strains lacking the his operon showed attenuated growth in histidine-restricted media, confirming them as his-negative auxotrophs. Our results suggest that the ability to make histidine is an important factor in bacterial growth and survival in the middle ear, where nutrients such as histidine may be found in limited amounts. Those isolates lacking the histidine pathway were still able to survive well in the throat, which suggests that histidine is readily available in the throat environment.
Haemophilus influenzae strains are gram-negative bacteria normally found in the pharynges of humans and are classified based on the presence and type of a polysaccharide capsule. Strains without a capsule are referred to as nontypeable, while those that possess a capsule are serotypeable with antisera raised against the six capsular types a to f. The most pathogenic capsule is serotype b, which facilitates invasive infections such as bacteremia, septic arthritis, cellulitis, and meningitis in nonimmune children. The incidence of disease caused by type b strains has been dramatically reduced in the United States since the introduction and wide spread use of the type b polysaccharide conjugate vaccine. While nontypeable H. influenzae (NTHI) colonizes the pharynges of many humans with no clinical symptoms, NTHI may cause significant infections, such as bronchitis, sinusitis, conjunctivitis, pneumonia, and acute otitis media, in infants, young children, immunocompromised individuals, and individuals with chronic pulmonary disorders.
NTHI is the bacterial agent isolated from 30 to 52% of acute otitis media episodes (19). Otitis media is one of the most common infectious diseases in infancy and childhood, as between 50 and 85% of children will have had at least one episode by the age of 3 years (11, 53). In addition, population-based studies in Finland and the United States suggest an increased incidence of otitis media within the past 10 to 20 years (33, 38). In addition to high morbidity, otitis media is a significant financial burden, as the annual cost for its prevention, diagnosis and treatment in the United States is approximately $3 billion to $5 billion (8, 27, 34).
By mechanisms that are unclear, NTHI strains that colonize the pharynx migrate through the Eustachian tubes into the middle ear, where they elicit an immune response leading to inflammation, effusion, and the disease acute otitis media. Several host and epidemiological factors play a role in otitis media pathogenesis, including genetic predisposition, preceding viral respiratory infections, attendance in day care centers, lack of breastfeeding, and young age (7, 12, 48, 55). Recent data suggest that bacterial virulence factors also play a role in otitis media (40), such as lic2B, which is involved in lipooligosaccharide biosynthesis, and the hmw genes, which encode high-molecular-weight adhesins (17, 46). These two genes were shown to be more prevalent in middle ear isolates than in throat strains from healthy children, suggesting their role in otitis media virulence.
In a previous study (61), using genomic subtraction and dot blot hybridization, we identified several other putative gene regions in NTHI that may contribute to otitis media pathogenesis, including several regions involved in nutrient transport and metabolism that are considered to promote housekeeping functions rather than virulence. One of these genetic regions (sJPX132) was located within hisD (encoding histidinol dehydrogenase), a gene associated with histidine biosynthesis. The sJPX132 genetic region was significantly more prevalent among a panel of NTHI strains isolated from the middle ears of children with otitis media (107/121; 88.4%) than among throat isolates from healthy children (76/137; 55.5%) (prevalence ratio, 1.59, P < 0.0001). As a follow-up to the previous study, we have, in the present study, further explored the genetic variation of the his operon in H. influenzae and the association of H. influenzae histidine biosynthesis with acute otitis media.
Histidine biosynthesis is one of the best-studied metabolic pathways in bacteria. The ubiquity of the his genes in many different bacterial species suggests that this pathway is highly conserved (20). In Escherichia coli, where it has been extensively studied, histidine biosynthesis results from an unbranched metabolic pathway composed of 10 steps involving eight enzymes. Besides the biosynthesis of histidine, an essential amino acid for bacterial growth and maintenance, part of this pathway is also involved in nitrogen metabolism and de novo synthesis of purine nucleotides (21) that can also contribute vital nutrient requirements for bacterial growth, such as thiamine (vitamin B1) and adenosine. The eight enzymes involved in histidine biosynthesis in E. coli are encoded by eight genes (hisG, hisD, hisC, hisNB, hisH, hisA, hisF, and hisIE) organized in a 7.4-kb operon, and these his genes appear to be highly conserved; the amino acid profiles of three completely sequenced genome strains of E. coli (K-12, CFT073, and O157H7) showed 97.3 to 100% identity (P. C. Juliao, unpublished). In addition, the operons mapped to the same position in the genome and contained the same gene order.
Although the histidine biosynthesis enzymes in H. influenzae have not been studied, orthologs for each gene have been found (24, 52). In the fully sequenced H. influenzae laboratory strain Rd KW20, the eight his genes are located in a 7.5-kb genomic region flanked by HI0467, which encodes a hypothetical protein, and tyrP, which encodes a tyrosine-specific transport protein. These orthologs have an amino acid percent identity ranging from 54.1% to 81.5% with their E. coli counterparts. In addition, HI1166, a homolog of hisH, has been found elsewhere in the Rd genome (21, 24), but it has not been found to participate in any other pathway besides histidine biosynthesis (21).
In this study, we identified and characterized the extent of genetic diversity within the his operon in NTHI isolates. Our preliminary analysis revealed two distinct his operon profiles, i.e., presence of the complete his operon or absence of the complete his operon. To explore the role of the his genes in pathogenesis, we compared their prevalences among NTHI isolates from the middle ears of children with otitis media with those among isolates from the throats of healthy children. Finally, we examined NTHI growth characteristics in the presence and absence of exogenous histidine.
MATERIALS AND METHODS
Sample collection.
The initial panel of NTHI strains used for this study consisted of 129 isolates, 81 obtained from throat swabs of healthy children and 48 obtained from the middle ears of children with acute otitis media. These isolates were collected between 1980 and 2000 from the following sites: Ann Arbor, MI (22, 51), Battle Creek, MI (51), Minnesota (29), St. Louis, MO (37), Pittsburgh, PA (E. Wald and A. Hoberman, unpublished), and Bardstown, KY (S. Block, unpublished). Only 5 of the 137 throat isolates and none of the 122 middle ear strains were obtained from the same child. The isolates were identified as H. influenzae by colonial morphology during growth on chocolate agar with bacitracin, X and V factor dependence, porphyrin negativity, and failure to hemolyze horse red blood cells (10, 22, 35). These isolates were further screened to identify putative variant strains based on unique iga and P6 profiles (43, 47). Fourteen (10.9%) of the isolates did not possess iga by DNA probe analysis and/or did not react with monoclonal antibody 7F3, which recognizes the common epitope of P6. These isolates were classified as nonhemolytic Haemophilus haemolyticus and were excluded from the study in order to minimize possible selection bias. All isolates were previously genotyped by pulsed-field gel electrophoresis and are significantly different by at least seven band differences (22, 51).
To increase the power of the analysis, we expanded the initial set of isolates by adding 69 throat isolates from healthy children and 74 middle ear isolates that were collected from the sites previous mentioned as well as from Finland (36) and Israel (15, 39) and were shown not to be nonhemolytic H. haemolyticus strains. Additional strains used in this study included H. influenzae laboratory strain Rd KW20, obtained from the American Type Culture Collection (ATCC) (Manassas, VA).
Isolation of genomic DNA.
Isolates were subcultured on chocolate agar plates (BD Diagnostics, Sparks, MD) and incubated overnight in a 5% CO2 incubator at 37°C. The subcultured bacterial cells were removed from the plate, suspended in 1 ml phosphate-buffered saline, and centrifuged at 14,000 rpm for 3 min to pellet the cells. The DNA of each isolate was extracted using the Wizard genomic extraction kit (Promega Corp, Madison, WI). Agarose gel electrophoresis was performed on each genomic DNA sample to assess the presence and integrity of the DNA.
Characterization of the his operon.
Two strains, previously identified as possessing the hisD genetic region (middle ear strain G622) or lacking the region (throat strain 23221) (61), were used for characterizing the his operon deletion and its location. Internal oligonucleotide primers were designed for each of the eight his genes, as well as the two flanking genes of the his operon, based on data from the sequenced strain Rd (Table 1) (Fig. 1a) (24). PCR-based genomic walking was used to define the complete his regions of both strains, with strain Rd as a positive control, as follows. Using DNA from each strain as template, a 25-μl PCR mixture was prepared for each primer pair using 0.2 mM deoxynucleoside triphosphates, 3 mM MgCl2, 1 μM forward primer, 1 μM reverse primer, 2.5 μl 10× buffer, 1U of Taq DNA polymerase, and 1 μl of genomic DNA. PCR reagents were purchased from Invitrogen Corp (Carlsbad, CA). The PCR mixtures were first incubated at 95°C for 2 min in a Peltier Thermal Cycler-100 (Bio-Rad, Hercules, CA) to completely denature the DNA. Next, 30 cycles of PCR amplification were performed with the following conditions: 95°C for 30 seconds, 58°C for 20 seconds, and 72°C for 3 min. With each PCR, a negative water control was used to identify possible PCR contamination. Separately, we also PCR amplified pepN (a gene ubiquitous in H. influenzae) to serve as a DNA template positive control. PCR products were then separated by agarose gel electrophoresis, gel purified using the QIAquick gel extraction kit (QIAGEN Inc, Valencia, CA), and sequenced at the University of Michigan DNA Sequencing Core (Ann Arbor, MI) using an Applied Biosystems model 3730 DNA sequencer to confirm gene specificity. Sequence results were analyzed using Megalign (Lasergene version 6; DNASTAR, Madison, WI).
TABLE 1.
Oligonucleotide primer sequences for PCR and probe preparations
| Region amplified | Primera | Size (bp) | Nucleotide sequence in NTHI strain Rd |
|---|---|---|---|
| HI0467 | HI0467-F | 755 | 5′-TGACTGCCTTTGCACGCCTTG-3′ |
| HI0467-R | 5′-GAATCGGGAATCCAATACCCTTG-3′ | ||
| hisG | hisG-F | 856 | 5′-CAATGCAACCAAACCGCCTTCG-3′ |
| hisG-R | 5′-AAAATTGAACTAGCTCCTGCCTC-3′ | ||
| hisD | hisD-F | 1,226 | 5′-GCAAAGTCATTATGCGTCCAGTCC-3′ |
| hisD-R | 5′-TCGCCAATCTTACACTTACTGCC-3′ | ||
| hisC | hisC-F | 1,037 | 5′-GACAATCACAACTTTATCCCGAC-3′ |
| hisC-R | 5′-GCTTCAACAACTTTCTCACACTC-3′ | ||
| hisB | hisB-F | 1,029 | 5′-GACGGCACATTAATTGATGAACC-3′ |
| hisB-R | 5′-TTCCTTCAATCCGAATCGCCTG-3′ | ||
| hisH | hisH-F | 572 | 5′-GATTATAGACACAGGTTGTGCC-3′ |
| hisH-R | 5′-CGTTCAGGATGAAATTGTACACC-3′ | ||
| hisA | hisA-F | 656 | 5′-CTTATCAATGGTCAAGTTGTGCG-3′ |
| hisA-R | 5′-TCGACCTACAATTACACCTGAC-3′ | ||
| hisF | hisF-F | 749 | 5′-TTGTTTGGATGTACGTGATGGAC-3′ |
| hisF-R | 5′-TCACTCCGAATCTCGATGGCTG-3′ | ||
| hisIE | hisIE-F | 580 | 5′-AAGTACTCGTGAGGTACTAATGC-3′ |
| hisIE-R | 5′-GATGTAATCCAATCCCTTGATGG-3′ | ||
| tyrP | tyrP-F | 1,150 | 5′-CACTTCTTGTTGCTGGTACGATG-3′ |
| tyrP-R | 5′-ACGAATCGCAAATGGAATGGATG-3′ | ||
| pepN | pepN-F | 1,037 | 5′-GATTACAAACAACCAGATTTTA-3′ |
| pepN-R1 | 5′-GACATTGGGCTTGCATCTTCG-3′ |
F, forward primer; R, reverse primer.
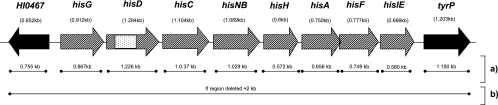
FIG. 1.
Genomic map of the his operon in strain Rd. □, location of sJPX132;▒, genes deleted in 23221 (throat strain) but present in G622 (middle ear strain) and Rd; ▪, genes present in 23221 (throat strain), G622 (middle ear strain), and Rd. (a) PCR protocol for each individual his gene and the flanking genes to determine size of insertion/deletion; (b) PCR protocol spanning the his region, used to verify size and location of the his indel. If an isolate is missing the complete his operon, a 2-kb product is amplified. If an isolate has the complete his operon, then a negative PCR will result, because the genomic region is too large to amplify (10.2 kb) using traditional PCR techniques.
To confirm the size and location of the his deletion previously observed in strain 23221, isolated from the throat of a healthy child (61), a PCR amplification spanning the complete his region was performed using the primers from the two opposite flanking genes of the his operon (primers HI0467R and tyrPR) (Fig. 1b). DNA sequencing was performed on the PCR product to verify the location of the deletion.
Sequenced genome comparison.
The genomic locus associated with the his operon was explored in four sequenced strains of H. influenzae and compared to those of throat strain 23221 and middle ear strain G622, where applicable. Rd KW20, the first sequenced free-living organism, is a nonpathogenic, no-longer-encapsulated laboratory strain passaged several times from a type d isolate (24). Strain 86-028NP is an NTHI isolate obtained from the nasopharynx of a child with otitis media (31). R2846 (also known as strain 12) is an NTHI isolate obtained from the middle ear of a child with acute otitis media (3). R2866 is an NTHI isolate obtained from the blood of a child with meningitis (44, 60). DNA comparison and percent identities were obtained using Megalign, SeqMan, and EditSeq (Lasergene v6; DNASTAR). The complete genome sequences of Rd (24) and 86-028NP (31) were obtained from the National Center for Biotechnology Information (NCBI) website. The genome sequences of R2846 and R2866 were obtained from Arnold Smith and Alice Erwin at the Biomedical Research Institute, Seattle, WA.
Screen of H. influenzae isolates for presence of his genes.
The presence or absence of each of the eight his genes within H. influenzae isolates was determined by dot blot hybridization as previously described (46, 49, 61, 62).
Design and preparation of oligonucleotides probes for use in dot blot assay.
Using middle ear strain G622 as a DNA template, PCR amplification, gel purification, and DNA sequencing, as detailed above, were performed on each of the eight his genes. Each purified PCR product was labeled with alkaline phosphatase using the AlkPhos direct labeling and detection system (GE Healthcare Life Sciences, Piscataway, NJ) and immediately used as a probe for DNA hybridization.
Preparation of cell lysates.
NTHI isolates were subcultured on chocolate agar plates and incubated overnight in 5% CO2 at 37°C. The subcultured cells were removed from the plate and transferred to 800 μl of Levinthal liquid medium containing sterile brain heart infusion (BD Diagnostics, Sparks, MD), supplemented with Levinthal base and NAD (Sigma-Aldrich Co., St. Louis, MO), and incubated overnight at 37°C (41). The suspensions were then centrifuged for 20 min at 3,000 rpm to pellet the cells, and the supernatant was removed. Lysates were made by resuspending the cells in 800 μl of lysis solution (0.4 M NaOH, 10 mM EDTA) and incubating at 70°C for 30 min.
Membrane preparation.
Forty microliters of each lysate was blotted onto a Hybond N+ positively charged nylon membrane (GE Healthcare Life Sciences, Piscataway, NJ) using a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories, Hercules, CA) to assure no cross contamination between dots. In addition, lysates from middle ear strain G622 (his positive control) and throat strain 23221 (his negative control), as well as water, served as controls. Membranes were then washed with 0.4 M NaOH, soaked in 2× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min, and then dried overnight. When dried, membranes were exposed to UV light (wavelength, 10 to 400 nm) for 3 min to cross-link the DNA with the membrane.
Hybridization and detection of his probes.
Duplicate blots were hybridized with each of the probes. The hybridization signals were detected using a chemifluorescence-based ECF detection system (GE Healthcare Life Sciences, Piscataway, NJ), measured using the STORM 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA), and analyzed with ImageQuant version 5.0 (Molecular Dynamics, Sunnyvale, CA). The signal intensity for each dot (i.e., each strain) was analyzed by correcting for the background and then expressed as the normalized signal intensity compared to that of the positive control strain (middle ear strain G622) (62). Discrepancies in strains for specific his probes were resolved with PCR amplification of the specific his gene, using the primers detailed in Table 1.
Statistical analysis.
All statistical analyses were performed using SAS (version 9.1) (SAS, Cary, NC). Prevalence ratios were calculated as the ratio of the proportion of middle ear isolates possessing the complete his operon to the proportion in the referent group, i.e., isolates from throats of healthy children. A χ2 analysis was used to determine the level of significance for all prevalence ratios; a P value of ≤0.05 was considered significant.
A pairwise analysis comparing the concordance between dot blot and PCR procedures in detecting the presence of the complete his operon was performed using the simple kappa statistic (κ). The statistic can be used to measure the level of agreement between the two methods and its level of significance (59). The κ coefficient range is from −1 (completely disagree) to 1 (completely agree).
Histidine-dependent growth of H. influenzae isolates.
Three different liquid media were prepared for the growth experiment. Levinthal liquid medium (41) was used for the initial seeding of bacteria and served, as well, as a control medium during the growth experiment. Two separate H. influenzae minimal media were prepared for the growth experiment (32): medium 1 (His positive) used the standard Herriot medium, which contained l-histidine, while medium 2 (His negative) lacked the addition of l-histidine.
Six NTHI isolates were selected for the growth experiment; three possessed the complete his operon, while three lacked the entire his operon. The six isolates were subcultured from frozen stocks to chocolate agar plates and incubated overnight in a 5% CO2 incubator at 37°C. The subcultures were each transferred into 1 ml of Levinthal liquid medium and incubated overnight on a shaker at 37°C. A uniform NTHI seeding density for the growth experiment was ensured by measuring the optical density (OD) at a wavelength of 600 nm in a Turner SP-830 spectrophotometer (Barnstead/Thermolyne, Dubuque, IA). The measured OD after overnight growth in Levinthal broth ranged from 0.6 to 0.8 for different isolates in the growth experiment. Ten microliters of overnight Levinthal liquid culture from each isolate was added to 1 ml of His-positive medium, 1 ml of His-negative medium, and 1 ml of Levinthal control medium and incubated on a shaker at 37°C.
To measure the growth of the bacteria, the OD was measured for each tube every 30 min for 12 h starting from initial seeding. For each isolate, a growth curve was constructed, plotting the incubation time points on the x axis and the OD reading on the y axis for each of the three media. The growth experiment for all six isolates was repeated three separate times to ensure reproducibility of the results.
RESULTS
Characterization of the his operon.
To characterize the his operon and its location, each gene in the his operon as well as the flanking genes (HI0467 and tyrP) from middle ear strain G622 (described in preliminary experiments as possessing the hisD genetic region) and throat strain 23221 (lacking the hisD region) were PCR amplified, with strain Rd as a positive control (Fig. 1a). In the PCR, both middle ear strain G622 and strain Rd demonstrated bands of the predicted sizes for each of the eight genes in the his operon. Throat strain 23221 was PCR negative for each of the eight genes in the his operon. All three isolates were PCR positive for both genes flanking the his operon, with products of the correct sizes, suggesting that the deletion in these strains may only span the his operon. All PCR products were confirmed by DNA sequence analysis. To confirm the location of the his deletion in the genome of throat strain 23221, PCR amplification from the beginning of gene HI0467 to the end of tyrP, thus spanning the his operon, was performed (Fig. 1b). A 2-kb PCR product, the expected band size if the operon was deleted, was found in throat strain 23221, and the sequence corresponded to the Rd sequence from nucleotide position 489497 (beginning of gene HI0467) to 499496 (end of tyrP) but lacked the his operon, thus validating the 7.5-kb deletion and its location.
Comparison of the his operons in four sequenced H. influenzae strains.
In silico analysis was also performed for the hisD genetic region in the four sequenced strains of H. influenzae (Rd KW20, R2846, R2866, and 86-028NP). The results showed that the four strains had each of the eight genes (hisG, -D, -C, -NB, -H, -A, -F, and -IE) in the same order and in the same genome location. The DNA and amino acid percent identities of the eight genes ranged from 99.2% to 100% between these strains.
Presence of each of the eight genes in the his operon among NTHI isolates.
To examine the content of the his operons among NTHI strains, we screened each isolate in our panels of ear and throat strains for the presence of the eight his genes by using dot blot hybridization. Our initial dot blot analysis, which excluded the 14 Haemophilus haemolyticus strains, probed 115 NTHI isolates (47 isolated from middle ears of children with otitis media and 68 isolated from throats of healthy children) for the presence of each of the eight his genes (Table 2). Eighty-one (70.4%) of the isolates had all his genes present, 30 isolates (26.1%) had complete deletion of all the his genes, and 4 of the 115 isolates (3.5%) were initially weakly positive by dot blot hybridization. PCR amplification of each of the individual genes in the four indeterminate isolates demonstrated the absence of all his genes in each isolate. Furthermore, the PCR results were confirmed negative by Southern blot analysis (data not shown). In summary, in this preliminary analysis, 34/115 (29.6%) of NTHI isolates lacked the complete his operon.
TABLE 2.
Distribution and prevalence of the complete his operon in NTHI isolates
| Site of collection | Initial set
|
Expanded set
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | % Positive (n)a | Prevalence ratio | P valuec | n | % Positive (n) | Prevalence ratio | P value | |
| Middle ear | 47 | 85.1 (40) | 1.41 | 0.0042 | 121 | 87.7 (106) | 1.62 | <0.0001 |
| Throatb | 68 | 60.3 (41) | 137 | 54.0 (74) | ||||
| Total isolates | 115 | 258 | ||||||
Percentage of isolates that possessed the complete his operon.
Referent group for prevalence ratio.
P value based on χ2 test.
Confirmation of his operon size and location in NTHI isolates.
To verify the location of the his deletion within the genomes of isolates identified as his negative by dot blot analysis (n = 30), we performed PCR spanning the his operon region using the two flanking genes (HI0467 and tyrP) as the primer sites (Fig. 1b). All 30 isolates tested amplified a PCR product of 2 kb, which is the predicted PCR band size if the his operon is deleted from the region.
Association between the his operon and otitis media.
To determine the association of his genes with otitis media, we determined the prevalence of the complete his operon among NTHI isolates from the middle ears of children with otitis media and from the throats of healthy children as determined from the dot blot results of our initial set of NTHI isolates (n = 115) (Table 2). χ2 analysis revealed a higher prevalence of the his operon in middle ear isolates (85.1%) than in throat isolates (60.3%) at a statistically significant level (prevalence ratio, 1.41; P = 0.0042). Specifically, middle ear isolates were 1.4 times more likely to possess the his operon than throat isolates from healthy children, suggesting that the presence of the his operon may contribute to survival in the middle ear leading to otitis media.
To strengthen the power of our analysis, we increased the sample size of our collection by adding 143 NTHI isolates (74 from the middle ears of children with otitis media and 69 from the throats of healthy children). To identify the his operon profile in this second set, the PCR amplification procedure spanning the his operon region (primers HI0467R and tyrPR) (Fig. 1b) was chosen. To ensure that the PCR procedure would give results concordant with those of the dot blot technique used to test the initial panel of strains, we compared results of both methods in our initial isolate set (n = 113) by using a kappa test (κ). Results showed a high percent agreement between the two methods at a statistically significant level (κ, 0.916; P < 0.0001), suggesting that the PCR method is sufficient to screen for the presence of the his operon.
Association between the his operon and otitis media: expanded set.
To improve the power of our analyses to detect an association between the presence of the complete his operon and otitis media, data from both the initial and second strain sets were merged, giving us a final expanded set of 258 NTHI isolates (137 isolates from the throats of healthy children and 121 isolates from the middle ears of children with otitis media). Again, the his operon was significantly more prevalent among middle ear strains (87.7%) than among throat strains (54.0%), with a prevalence ratio of 1.62 favoring the middle ear strains (P < 0.0001) (Table 2). In other words, middle ear isolates are 1.6 times more likely to possess the complete his operon than throat isolates from healthy children at a statistically significant level, demonstrating a more robust association than that for the initial set with a smaller sample size.
Histidine-dependent growth experiment with H. influenzae isolates.
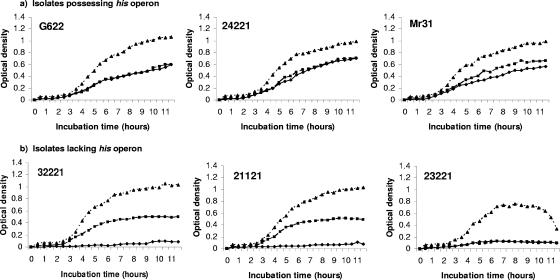
To determine if the deletion of the his operon interferes with histidine biosynthesis or if these isolates possess an alternate mechanism for synthesizing histidine, bacterial growth experiments were conducted using histidine-restricted medium (Fig. 2). Since histidine is critical to bacterial growth, a failure of NTHI isolates to grow well in histidine-restricted medium reflects their inability to synthesize histidine. Three NTHI isolates possessing the complete his operon and three lacking the entire his operon were tested. These isolates were also grown in Levinthal liquid medium (which possesses histidine and supports the growth of all known H. influenzae strains) to serve as a control for proper seeding. A growth curve over 12 h was plotted for each of the six isolates in each of the three media. As expected, the his-positive isolates grew well in both the histidine-depleted and histidine-rich minimal media, as the presence of the his operon allows them to synthesize the histidine needed for growth. Two of the three isolates lacking the his operon grew well in histidine-rich minimal medium but grew poorly in histidine-depleted minimal medium, confirming that these strains are his negative auxotrophs. The third isolate lacking the his operon grew poorly in both histidine-rich and histidine-deplete minimal media as well as in the Levinthal control medium, suggesting that this isolate may be missing other vital nutrient and metabolic pathways needed for efficient growth. This isolate has also shown unusual phenotypic properties not characteristic of H. influenzae, such as slow growth and clumping, which may explain these results. Indeed, subsequent testing of this isolate identified it as a nonhemolytic H. haemolyticus strain, as described by Xie et al. (61) and Murphy et al. (42). Figure 2 shows that all strains grew better in Levinthal (control, enriched) agar than in either the histidine-supplemented or histidine-restricted minimal medium and that the his-negative isolates achieved stationary growth earlier during incubation than his-positive isolates, which most likely reflects limited histidine availability in the supplemented media compared to Levinthal medium.
FIG. 2.
Growth curves of selected NTHI isolates grown in specific media for 12 h. ⧫, growth in histidine-restricted medium; ▪, growth in histidine-supplemented medium, ▴, growth in Levinthal control medium. (a) Growth curves of isolates possessing the complete his operon; (b) growth curves of isolates missing the his operon. Isolate 23221 (throat isolate) gave inconsistent results in the histidine medium as well as the Levinthal control medium.
DISCUSSION
H. influenzae strains are obligate human specific bacteria. They naturally reside in the human throat which is a highly competitive and rapidly changing environment. To guarantee survival, many bacteria, such as E. coli, use two-component regulatory mechanisms to monitor environmental changes and then adapt accordingly (5). H. influenzae, however, lacks many of these two-component systems (24, 56). Instead, it relies on inter- and intrastrain diversity that leads to an assortment/combination of traits advantageous in certain niches (5). It is this diversity that may drive the virulence characteristics of H. influenzae.
Many NTHI genes exhibit inter- and intrastrain diversity (30, 31); these include, among others, hifA and hifE (13, 28), hmw (16-18), hia (18, 50), and lipooligosaccharide-modifying genes (46, 57, 58). Interestingly, our previous study (61) identified an assortment of conserved genes associated with energy metabolism that displayed interstrain genetic diversity, including hisD, a gene involved in the histidine biosynthesis pathway. In the present study we further explored the interstrain diversity of the histidine biosynthetic pathway and identified a 7.5-kb deletion associated with the complete histidine biosynthesis operon, which may be important in NTHI survival and virulence in the middle ear.
The ubiquity of the histidine genes in many bacterial species suggests a strict requirement of this pathway for survival (21). Within H. influenzae, this conservation appears to be maintained, since all eight genes were present in the four sequenced strains (Rd, 86-028NP, R2846, and R2866), with a high percent identity at the DNA and amino acid levels. Few bacterial species have been reported to exhibit partial or full deletion of the histidine biosynthetic genes. Fani et al. reported that 47 out of 54 proteobacterial genomes examined contained the complete set of histidine biosynthetic genes, while the remainder had partial or complete gene deletions (20). Bacteria lacking the his genes, however, are still able to maintain persistence in the host. For example, Mycobacterium genitalium G-37 (25) and Helicobacter pylori strains 26695 and J99 (2, 54) lack complete histidine biosynthetic genes but are still able to colonize and persist in their specific host niche. In addition, many studies that have explored the presence of the his genes are limited to only fully annotated sequenced strains of bacteria that underrepresent the true diversity within the bacterial species under study. These observations suggest that the true importance and essential nature of the histidine biosynthetic genes to bacterial growth and survival may not be as conserved as previously thought and may be defined by their environment.
Since histidine is an essential nutrient in bacterial growth and survival, how are his-negative bacteria able to survive and persist? One possible explanation is that these bacteria may possess an alternative pathway for synthesizing histidine. Our preliminary results, however, show that NTHI isolates lacking the his operon do not grow well in histidine-depleted media and presumably fail to synthesize histidine necessary for growth, confirming that they are his-negative auxotrophs. Therefore, we suggest that these auxotrophs must obtain nutrients from the surrounding environment. Since the majority of the his-negative auxotrophic strains identified in our collection are commensals isolated from throat specimens rather than disease isolates from middle ear specimens, we suggest that the throat environment may be nutritionally rich in amino acids such as histidine, which the auxotrophic bacteria can then take up and use for nourishment. Such nutrients, however, may be found in limited amounts in the middle ear, which is considered to be a normally sterile environment in the absence of infection. Thus, the ability to synthesize histidine may play an important role in survival in the middle ear.
Several studies have explored the nutrient environment in certain niches, such as the blood (6, 26), human sweat and skin (14, 23), and sputa from cystic fibrosis patients (4, 45), and suggest varying degrees of nutrient availability in different niches. In particular, niches where auxotrophs appear to thrive, such as the sputa of patients with cystic fibrosis, have a relatively high concentration of amino acids and small peptides compared to more sterile regions, such as the blood.
Natural auxotrophs, although niche limited, may have a tremendous advantage for bacterial persistence through energy conservation. Histidine biosynthesis, for example, is one of the most energy-depleting metabolic pathways in bacteria, with the consumption of 41 molecules of ATP per histidine molecule synthesized (9). In addition, several complex regulatory pathways can contribute to energy consumption associated with histidine synthesis (1). This enormous metabolic cost might explain why bacteria are able to accommodate loss of this essential amino acid synthesis pathway, even though it would then restrict them to histidine-rich environments. Since the throat environment is a highly competitive niche in which an assortment of bacteria compete for survival, it may be that an increase in bioenergy capacity may give auxotrophs an adaptive advantage that allows for longer persistence.
While this study showed that NTHI strains from the middle ears of children with acute otitis media were significantly more likely to possess the complete his operon than those from the pharynges of healthy children, the role of the his genes in otitis media virulence remains unclear. The differential his prototrophy of organisms from these two sites may merely reflect nutrient availability, or the presence of the his operon may be a marker for an unrelated and unidentified virulence factor.
In summary, interstrain diversity, rather than strict essentiality, involving histidine biosynthesis may result in phenotypic characteristics that offer H. influenzae survival advantages in certain environmental niches. Since most middle ear isolates in our collection possessed the histidine biosynthetic pathway, we suggest that the ability to make histidine may be important in survival in the middle ear. This pathway, previously thought to be essential, appears to be absent in about half of commensal throat strains. We suggest that H. influenzae has evolved a way to bypass energy-exhaustive pathways such as histidine biosynthesis by utilizing already-synthesized histidine from its surrounding environment. The cost of this strategy for the bacteria is that auxotrophic organisms may be limited to very specific niches of high nutrient availability. If such an event, however, results in bacterial persistence in the host, it may be a sacrifice worth making. Future studies will explore other natural auxotrophs of NTHI to better understand the relationship of natural auxotrophy with persistence and virulence.
Acknowledgments
We thank Timothy Murphy, Buffalo, NY, for supplying us with P6 monoclonal antibody 7F3 and for his helpful discussions about Haemophilus influenzae variant isolates. In addition, we thank Ellen Wald and Alejandro Hoberman, University of Pittsburgh, and Stan Block, Bardstown, KY, for otitis media strains. Kirk McCrea provided valuable insight into these findings.
This study was funded by R01 award DC05840 from the National Institute on Deafness and Other Communication Disorders awarded to J.R.G.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Alifano, P., R. Fani, P. Lio, A. Lazcano, M. Bazzicalupo, M. S. Carlomagno, and C. B. Bruni. 1996. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 60:44-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, A. L., and T. L. Pitt. 1995. Auxotrophy of Burkholderia (Pseudomonas) cepacia from cystic fibrosis patients. J. Clin. Microbiol. 33:2192-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayliss, C. D., D. Field, X. de Bolle, and E. R. Moxon. 2000. The generation of diversity by Haemophilus influenzae: response. Trends Microbiol. 8:435-436. [DOI] [PubMed] [Google Scholar]
- 6.Benton, B. M., J. P. Zhang, S. Bond, C. Pope, T. Christian, L. Lee, K. M. Winterberg, M. B. Schmid, and J. M. Buysse. 2004. Large-scale identification of genes required for full virulence of Staphylococcus aureus. J. Bacteriol. 186:8478-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman, S. 1995. Otitis media in children. N. Engl. J. Med. 332:1560-1565. [DOI] [PubMed] [Google Scholar]
- 8.Bondy, J., S. Berman, J. Glazner, and D. Lezotte. 2000. Direct expenditures related to otitis media diagnoses: extrapolations from a pediatric medicaid cohort. Pediatrics 105:E72. [DOI] [PubMed] [Google Scholar]
- 9.Brenner, M., and B. N. Ames. 1971. The histidine operon and its regulation, vol. 5. Metabolic regulation. Academic Press Inc, New York, NY.
- 10.Campos, J. M. 1999. Haemophilus, p. 604-613. In P. R. Murray, E. Baron, M. A. Fallen, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 11.Casselbrant, M. L., and E. M. Mandel. 1999. Epidemiology, p. xi. In R. M. Rosenfeld and C. D. Bluestone (ed.), Evidence-based otitis media. Decker, Saint Louis, MO.
- 12.Casselbrant, M. L., E. M. Mandel, P. A. Fall, H. E. Rockette, M. Kurs-Lasky, C. D. Bluestone, and R. E. Ferrell. 1999. The heritability of otitis media: a twin and triplet study. JAMA 282:2125-2130. [DOI] [PubMed] [Google Scholar]
- 13.Clemans, D. L., C. F. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coltman, C. A., Jr., N. J. Rowe, and R. J. Atwell. 1966. The amino acid content of sweat in normal adults. Am. J. Clin. Nutr. 18:373-378. [DOI] [PubMed] [Google Scholar]
- 15.Dagan, R., A. Hoberman, C. Johnson, E. L. Leibovitz, A. Arguedas, F. V. Rose, B. R. Wynne, and M. R. Jacobs. 2001. Bacteriologic and clinical efficacy of high dose amoxicillin/clavulanate in children with acute otitis media. Pediatr. Infect. Dis. J. 20:829-837. [DOI] [PubMed] [Google Scholar]
- 16.Dawid, S., S. J. Barenkamp, and J. W. St. Geme III. 1999. Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA 96:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ecevit, I. Z., K. W. McCrea, C. F. Marrs, and J. R. Gilsdorf. 2005. Identification of new hmwA alleles from nontypeable Haemophilus influenzae. Infect. Immun. 73:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecevit, I. Z., K. W. McCrea, M. M. Pettigrew, A. Sen, C. F. Marrs, and J. R. Gilsdorf. 2004. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 42:3065-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eskola, J., and T. Kilpi. 2000. Potential of bacterial vaccines in the prevention of acute otitis media. Pediatr. Infect. Dis. J. 19:S72-78. [DOI] [PubMed] [Google Scholar]
- 20.Fani, R., M. Brilli, and P. Lio. 2005. The origin and evolution of operons: the piecewise building of the proteobacterial histidine operon. J. Mol. Evol. 60:378-390. [DOI] [PubMed] [Google Scholar]
- 21.Fani, R., E. Mori, E. Tamburini, and A. Lazcano. 1998. Evolution of the structure and chromosomal distribution of histidine biosynthetic genes. Orig. Life Evol. Biosph. 28:555-570. [DOI] [PubMed] [Google Scholar]
- 22.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 23.Farrior, J. W., and W. E. Kloos. 1976. Sulfur amino acid auxotrophy in Micrococcus species isolated from human skin. Can. J. Microbiol. 22:1680-1690. [DOI] [PubMed] [Google Scholar]
- 24.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenny, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Philips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 25.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 26.Garg, M. B., J. C. Sevester, J. A. Sakoff, and S. P. Ackland. 2002. Simple liquid chromatographic method for the determination of uracil and dihydrouracil plasma levels: a potential pretreatment predictor of 5-fluorouracil toxicity. J. Chromatogr. B 774:223-230. [DOI] [PubMed] [Google Scholar]
- 27.Gates, G. A. 1996. Cost-effectiveness considerations in otitis media treatment. Otolaryngol. Head Neck Surg. 114:525-530. [DOI] [PubMed] [Google Scholar]
- 28.Geluk, F., P. P. Eijk, S. M. van Ham, H. M. Jansen, and L. van Alphen. 1998. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect. Immun. 66:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilsdorf, J. R., H. Y. Chang, K. W. McCrea, L. J. Forney, and C. F. Marrs. 1992. Comparison of hemagglutinating pili of type b and non-typeable Haemophilus influenzae. J. Infect. Dis. 165(Suppl. 1):S105-S106. [DOI] [PubMed] [Google Scholar]
- 30.Gilsdorf, J. R. 1998. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect. Immun. 66:5053-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol. 187:4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herriott, R. M., E. Y. Meyer, M. Vogt, and M. Modan. 1970. Defined medium for growth of Haemophilus influenzae. J. Bacteriol. 101:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joki-Erkkila, V. P., P. Laippala, and J. Pukander. 1998. Increase in paediatric acute otitis media diagnosed by primary care in two Finnish municipalities-1994-5 versus 1978-9. Epidemiol. Infect. 121:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan, B., T. L. Wandstrat, and J. R. Cunningham. 1997. Overall cost in the treatment of otitis media. Pediatr. Infect. Dis. J. 16:S9-S11. [DOI] [PubMed] [Google Scholar]
- 35.Kilian, M. 2005. Genus III. Haemophilus Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 561AL. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 36.Kilpi, T., E. Herva, T. Kaijalainen, R. Syrjanen, and A. K. Takala. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654-662. [DOI] [PubMed] [Google Scholar]
- 37.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanphear, B. P., R. S. Byrd, P. Auinger, and C. B. Hall. 1997. Increasing prevalence of recurrent otitis media among children in the United States. Pediatrics 99:E1. [DOI] [PubMed] [Google Scholar]
- 39.Leibovitz, E., L. Piglansky, S. Raiz, D. Greenberg, K. A. Hamed, J. M. Ledeine, J. Press, A. Leiberman, R. M. Echols, P. F. Pierce, M. R. Jacobs, and R. Dagan. 2003. Bacteriologic and clinical efficacy of oral gatifloxacin for the treatment of recurrent/nonresponsive acute otitis media: an open label, noncomparative, double tympanocentesis study. Pediatr. Infect. Dis. J. 22:943-949. [DOI] [PubMed] [Google Scholar]
- 40.Melhus, A., A. Hermansson, A. Forsgren, and K. Prellner. 1998. Intra- and interstrain differences of virulence among non-typeable Haemophilus influenzae strains. APMIS 106:858-868. [PubMed] [Google Scholar]
- 41.Michaels, R. H., F. E. Stonebraker, and J. B. Robbins. 1975. Use of antiserum agar for detection of Haemophilus influenzae type b in the pharynx. Pediatr. Res. 9:513-516. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 43.Murphy, T. F., C. Kirkham, and D. J. Sikkema. 1992. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect. Immun. 60:2016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nizet, V., K. F. Colina, J. R. Almquist, C. E. Rubens, and A. L. Smith. 1996. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173:180-186. [DOI] [PubMed] [Google Scholar]
- 45.Ohman, D. E., and A. M. Chakrabarty. 1982. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect. Immun. 37:662-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 70:3551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rovers, M. M., A. G. Schilder, G. A. Zielhuis, and R. M. Rosenfeld. 2004. Otitis media. Lancet. 363:465-473. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.St. Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatusov, R. L., A. R. Mushegian, P. Bork, N. P. Brown, W. S. Hayes, M. Borodovsky, K. E. Rudd, and E. V. Koonin. 1996. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol. 6:279-291. [DOI] [PubMed] [Google Scholar]
- 53.Teele, D. W., J. O. Klein, and B. Rosner. 1989. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J. Infect. Dis. 160:83-94. [DOI] [PubMed] [Google Scholar]
- 54.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 55.Uhari, M., K. Mantysaari, and M. Niemela. 1996. A meta-analytic review of the risk factors for acute otitis media. Clin. Infect. Dis. 22:1079-1083. [DOI] [PubMed] [Google Scholar]
- 56.Weiser, J. N. 2000. The generation of diversity by Haemophilus influenzae. Trends Microbiol. 8:433-435. [DOI] [PubMed] [Google Scholar]
- 57.Weiser, J. N., D. J. Maskell, P. D. Butler, A. A. Lindberg, and E. R. Moxon. 1990. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J. Bacteriol. 172:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, S. A., and N. R. van den Broek. 2004. Methods for assessing reliability and validity for a measurement tool: a case study and critique using the WHO haemoglobin colour scale. Stat. Med. 23:1603-1619. [DOI] [PubMed] [Google Scholar]
- 60.Williams, B. J., G. Morlin, N. Valentine, and A. L. Smith. 2001. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie, J., P. Juliao, J. R. Gilsdorf, D. Ghosh, M. Patel, and C. F. Marrs. 2006. Identification of 10 new genetic regions found more frequently in strains of nontypeable H. influenzae that cause otitis media. J. Clin. Microbiol. 44:4316-4325, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44:225-233. [DOI] [PubMed] [Google Scholar]