Abstract
The suborder Corynebacterianeae comprises bacteria like Mycobacterium tuberculosis and Corynebacterium glutamicum, and these bacteria contain in addition to the linear fatty acids, unique α-branched β-hydroxy fatty acids, called mycolic acids. Whereas acetyl-coenzyme A (CoA) carboxylase activity is required to provide malonyl-CoA for fatty acid synthesis, a new type of carboxylase is apparently additionally present in these bacteria. It activates the α-carbon of a linear fatty acid by carboxylation, thus enabling its decarboxylative condensation with a second fatty acid to afford mycolic acid synthesis. We now show that the acetyl-CoA carboxylase of C. glutamicum consists of the biotinylated α-subunit AccBC, the β-subunit AccD1, and the small peptide AccE of 8.9 kDa, forming an active complex of approximately 812,000 Da. The carboxylase involved in mycolic acid synthesis is made up of the two highly similar β-subunits AccD2 and AccD3 and of AccBC and AccE, the latter two identical to the subunits of the acetyl-CoA carboxylase complex. Since AccD2 and AccD3 orthologues are present in all Corynebacterianeae, these polypeptides are vital for mycolic acid synthesis forming the unique hydrophobic outer layer of these bacteria, and we speculate that the two β-subunits present serve to lend specificity to this unique large multienzyme complex.
The ubiquitous acetyl-coenzyme A (CoA) carboxylases are key enzymes of fatty acid synthesis (5). They carboxylate acetyl-CoA to provide malonyl-CoA required for the elongation of nascent fatty acid by a condensation reaction. Carboxylation proceeds in a multienzyme complex via a reaction sequence involving two steps and three enzyme domains. In the first step the ATP-dependent biotin carboxylase catalyzes the transfer of a carboxyl group to biotin, which is linked to the biotin carboxyl carrier protein. In the second step, catalyzed by carboxyltransferase (CT), the carboxyl group is transferred from carboxybiotin to acetyl-CoA to form malonyl-CoA. The enzyme domains may be fused, as is the case in mammals and most eukaryotes, to form one large polypeptide or separated, as is the case in Escherichia coli, with even the CT further spliced, thus resulting in four peptides to build the carboxylase complex. Although the structure of individual domains of acetyl-CoA carboxylase is known (2, 9, 34), the structure of the entire complex, which might have a size of approximately 800 kDa and is often labile (6, 16), is not. In Streptomyces and the related Corynebacterianeae like Mycobacterium tuberculosis and Corynebacterium glutamicum, biotin carboxylase and biotin carboxyl carrier protein form one polypeptide, termed the α-subunit, and the CT is represented by the separate β-subunit. In these bacteria paralogous α- and β-subunits are present also, with M. tuberculosis possessing as many as three α-subunits and six β-subunits (3). In addition, a small ɛ-subunit of 7.5 kDa, originally identified in Streptomyces (9), has been found to be associated with carboxylase proteins (25).
Interestingly, the Corynebacterianeae possess the linear fatty acids that require acetyl-CoA carboxylase activity for their synthesis, but in addition they have unique α-branched β-hydroxy fatty acids, called mycolic acids. These mycolic acids occur only in this group of bacteria, containing such important organisms like the pathogenic M. tuberculosis, Mycobacterium leprae, and Corynebacterium diphtheriae and also apathogenic bacteria like C. glutamicum used for industrial production of Na glutamate and other amino acids (11). The mycolic acids form a highly impermeable hydrophobic outer layer, providing the basis of the exceptional drug resistance of M. tuberculosis (24), and they may also influence amino acid excretion of C. glutamicum (10, 17). These mycolic acids originate from two linear fatty acids (28, 29), and their synthesis is still poorly understood. A systematic study on the paralogous β-subunits present in Corynebacterianeae revealed that three of the four β-subunits present in C. glutamicum are essential, and subunit AccD1 is a component of the acetyl-CoA carboxylase required for fatty acid synthesis, whereas AccD2 and AccD3 together are required for mycolic acid synthesis (15). This finding enabled the derivation of a new model for an acyl carboxylase reaction required to activate the α-carbon of a linear fatty acid by carboxylation. This activation enables the decarboxylative condensation with a second fatty acid and is mechanistically similar to carboxylation of the α-carbon of acetyl-CoA by the acetyl-CoA carboxylase during fatty acid synthesis. Carboxylation of a linear fatty acid was indeed demonstrated with extracts of Mycobacterium smegmatis, and this activity was absent in an AccD4 mutant (27). Furthermore, from M. smegmatis two different β-subunits, PccB and AccD5, together with an α-subunit, AccA1, were shown to coimmunoprecipitate (27). (The different annotations and names of the carboxylase constituents mentioned are given in Tables S2 and S3 in the supplemental material.) Moreover, an ɛ-peptide (AccE) was isolated recently from avidin affinity-enriched proteins from M. tuberculosis (25). However, there is considerable confusion about the native carboxylase complex involved in the synthesis of the unique branched mycolic acids, since for M. tuberculosis a complex consisting of AccD5, AccA, and AccE (ɛ-peptide) was shown to have activity without a second β-subunit present (14), and a hexameric crystal structure of AccD5 has been derived, suggesting that such a protein complex is involved in carboxylation of propionyl-CoA for the synthesis of multimethyl-branched fatty acids and not involved in mycolic acid synthesis (18, 21).
Thus, in part, detailed information—albeit often conflicting—on the carboxylase composition and function is available, mostly determined with isolated peptides. This prompted us to examine the composition and activity of acetyl-CoA carboxylase and the acyl carboxylase specific for mycolic acid synthesis in C. glutamicum, which is a model organism within the Corynebacterianeae and for which there is currently evolving a comprehensive picture on its cell wall synthesis (1, 15).
MATERIALS AND METHODS
Bacteria, plasmids, oligonucleotides, and cultivations.
All strains and plasmids used are given in Table 1, whereas oligonucleotides are given in Table S1 of the supplemental material. Copurifications were mostly done using C. glutamicum Δpyc, which is devoid of the nonessential pyruvate carboxylase representing, besides AccBC, the second biotinylated protein present in this bacterium (19). The minimal medium used for C. glutamicum was CGXII (12), which contained glucose as a carbon source, or Luria-Bertani (LB) broth (Difco). C. glutamicum was grown as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm with orbital shaking at a diameter of 12.5 cm. Growth was monitored by measuring the optical density at 600 nm (OD600). For gene expressions an overnight-grown preculture was used to inoculate 50 ml of LB medium in a 500-ml flask followed by overnight incubation. This was used to inoculate 5 liters of LB broth in a 7-liter Infors Labfors fermenter. Protein expression was induced at an OD of 1 by the addition of 100 μM isopropyl-β-d-thiogalactopyranoside. Cells were harvested at OD values of 3 to 4 by centrifugation and were frozen before use.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics or use | Reference or source |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Wild type | Culture collection |
| H6D1 strain | Chromosomal fusion of H6 codons at the N terminus of accD1 | This work |
| H6D2 strain | Chromosomal fusion of H6 codons at the N terminus of accD2 | This work |
| Δpyc strain | C. glutamicum deleted of biotinylated pyruvate carboxylase | 26 |
| paccBC p-H6D2-E strain | C. glutamicum with pJC1accBC and pEKEx3-H6D2-accE | This work |
| paccBC p-H6D3 strain | C. glutamicum with pJC1accBC and pEKEx3-H6D3 | This work |
| Δpyc paccBC p-H6D1 strain | C. glutamicum Δpyc with pJC1accBC and pEKEx3-H6D1 | This work |
| Δpyc paccBC p-H6D1-E strain | C. glutamicum Δpyc with pJC1accBC and pEKEx3-H6D1-accE | This work |
| Δpyc paccBC p-H6D2-E-D3 strain | C. glutamicum Δpyc with pJC1accBC and pEKEx3-H6D2-accE-D3 | This work |
| Plasmids | ||
| pJC1accBC | Overexpression of biotinylated α-subunit | This work |
| pEKEx3-H6D1 | Overexpression of H6-accD1 | This work |
| pEKEx3-H6D2-accE | Overexpression of H6-accD2, accE | This work |
| pEKEx3-H6D3 | Overexpression of H6-accD3 | This work |
| pEKEx3-H6D2-accE-D3 | Overexpression of H6-accD2, accE, and accD3 | This work |
| pEKEx3-H6D1-accE | Overexpression of H6-accD1 and accE | This work |
| pJC1 | Shuttle vector | 23 |
| pEKEx3 | Shuttle or expression vector | 13 |
| pK19mobsacB | Vector for integration or deletion | 31 |
| pK19mobsacB-H6accD1 | Allelic exchange of accD1 for H6-accD1 | This work |
| pK19mobsacB-H6accD2 | Allelic exchange of accD2 for H6-accD2 | This work |
Recombinant DNA work.
Standard protocols were applied for the generation of fragments via PCR, ligation, and restriction, with each plasmid finally verified by sequencing. The chromosomal mutations of C. glutamicum were made by introducing nonreplicative plasmids via electroporation. For in-frame deletions, the clones with integrated vector were subsequently selected for absence of vector due to the lethal sacB function of plasmid pK19mobsacB (31). Chromosomal deletions were verified by PCR analysis of the gene locus under consideration.
To enable the chromosomal fusion of accD1 with an N-terminal His tag, primer pairs accD1_H6_top_for and accD1_H6_top_rev were used as well as paccD1_H6_bot_for and accD1_H6_bot_rev with genomic DNA as a template. The purified amplificates served as templates for a second PCR with accD1_H6_top_for and accD1_H6_bot_rev as primers. The resulting fragment was ligated into SmaI-cleaved pK19mobsacB to give plasmid pK19mobsacB_H6accD1. Similarly, an accD2 fusion plasmid was made (see Table S1 in the supplemental material for primers). Plasmid pEKEx3-H6D1 was made by amplification of dtsR1 and its cloning in the BamHI/SmaI site of pEKEx3. To derive plasmid pEKEx3-H6D1-E, AccE was subcloned into pUC18, excised as an EcoRI fragment, and ligated with similarly cleaved pEKEx3-H6D1. The His-tagged AccD2 together with AccE was cloned as a BamHI/SmaI fragment into pEKEx3 to yield plasmid pEKEx3-H6D2-E. For construction of pEKEx3-H6D2-E-D3, accD3 was isolated as an EcoRI fragment from pUC18 and ligated into partially EcoRI-digested pEKEx3-H6D2-E. To derive pJC1-accBC a BamHI fragment containing accBC with its native promoter was cloned into pJC1. dtsR3 was amplified by use of primers CgdtsR3F52 and CgdtsR3R53, cloned into pQE30, excised as an AvaI fragment, and ligated with pEKEx3. All plasmids made were eventually verified by sequencing.
Protein isolation.
The C. glutamicum ATCC 13032 Δpyc paccBC p-H6D1-E strain (carrying plasmids pJC1accBC and pEKEx3-H6D1-accE) (Table 1) was grown on LB as described above but containing 25 mg/liter kanamycin and 150 mg/liter spectinomycin. Cells were suspended in wash buffer consisting of 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 10 mM imidazole, 2.5 mM MgCl2, and 0.5 mM dithiothreitol plus Complete protease inhibitor (Roche, Basel, Switzerland) and disrupted by sonication (Branson Sonifier S-250; 25% amplitude; 6 min, continuously). The cell extract was clarified by centrifugation at 20,000 × g for 30 min at 4°C, and the resulting supernatant was applied to a 1-ml Ni-nitrilotriacetic acid (NTA) column (QIAGEN, Hilden, Germany). The column was washed with the same buffer (wash buffer) but containing 20 mM imidazole and 5% glycerol, followed by elution of bound proteins with 1 ml of wash buffer containing 150 mM imidazole. Eluted protein was directly applied on a size exclusion column (HiLoad 16/60 Superdex G200; Pharmacia) equilibrated with 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5 mM MgCl2, 0.5 mM dithiothreitol, 5% glycerol, and Complete protease inhibitor. The size exclusion chromatography was performed at a flow rate of 1 ml/min at 4°C. The UV absorption of the column eluate was monitored at 260 nm, and 2.5-ml fractions were collected. A total of 100 μl of each fraction was analyzed by denaturating electrophoresis (NuPAGE 12% bis-Tris gel with morpholineethanesulfonic acid running buffer; Invitrogen).
Enzyme assays.
For acetyl-CoA carboxylase activity, two different assays were used. If not mentioned otherwise, the direct quantification of malonyl-CoA formation via high-performance liquid chromatography was performed as described previously (15). The quantification was by reversed-phase chromatography using a LiChrospher 1000 RP 18-EC-5μ column (125 by 4 mm; Merck) on an Agilent 1100 series high-performance liquid chromatography apparatus. Samples (12 μl) were automatically injected and separated by an increasing gradient consisting of 50 mM sodium phosphate buffer (pH 5.0) containing 2% acetonitrile, increasing up to 26% acetonitrile in the same buffer at a flow rate of 0.3 ml/min, and monitored at 254 nm. A coupled enzymatic assay was used to derive the enzyme kinetic data. In this assay the rate of ATP hydrolysis was measured by detecting the production of ADP using pyruvate kinase and lactate dehydrogenase and following the oxidation of NADH at 340 nm. Each reaction mixture contained 50 units of lactate dehydrogenase, 20 units of pyruvate kinase, 0.5 mM phosphoenolpyruvate, 0.2 mM NADH, 1 mM MgCl2, 65 mM potassium bicarbonate, and 60 mM Tris-HCl at pH 7.2. All assays were preincubated for 3 min at 30°C and initiated by the addition of substrate. The kinetic parameters Km and Vmax were determined by fitting the initial velocities versus the substrate concentration to the Michaelis-Menten equation using nonlinear regression analysis. All inhibitors were purchased from Sigma-Aldrich.
To assay acyl carboxylase activity, we adapted the method of Rainwater and Kolattukudy (30). Briefly, mixtures of purified enzymes were assessed for activity with various acyl substrates. Assay mixtures (100 μl) contained 100 mM potassium phosphate (pH 8.0), 2 mM ATP, 5 mM MgCl2, 2% Me2SO, 0.25 mM acyl substrate (acetyl-CoA, palmitoyl-CoA, or palmitic acid), 3 mg of bovine serum albumin, 50 mM NaH14CO3 (0.5 μCi; CFA421; Amersham Biosciences), and 30 μg of the enzyme. All of the components of the assay excluding the enzyme extract were premixed and then mixed with the enzyme extract to initiate the reaction, which was held at 30°C for 120 min. The reaction was quenched by the addition of 50 μl of concentrated HCl. This mixture was then held at 80°C under vacuum to remove nonfixed 14CO2. After evaporation to dryness, the residue was dissolved in 1 ml of water and 0.2 ml of butanol, and both extracts were added to 10 ml of Ecoscint A (National Diagnostics, Inc.) prior to liquid scintillation counting.
Protein identification by peptide mass fingerprinting.
For peptide mass fingerprinting, the protein spots of interest were excised from Coomassie blue-stained gels and subjected to in-gel digestion with trypsin. Peptides were extracted by sequential addition of 12 μl of water and 10 μl of 0.1% (vol/vol) trifluoroacetic acid in 30% (vol/vol) acetonitrile. The resulting peptide solution (0.5 μl) was mixed on a stainless steel sample plate with 0.5 μl of a saturated cyano-4-hydroxy-trans-cinnamic acid solution in 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. Samples were analyzed manually with an Applied Biosystems (Weiterstadt, Germany) Voyager STR matrix-assisted laser desorption ionization-time of flight mass spectrometer in the positive reflector mode at 20 kV and 63% grid voltage, and the delay time was set at 125 ns. External calibration was performed using calibration mixtures 1 and 2 of the Sequazyme peptide mass standard kit. Data analysis was performed using a Voyager Control Panel, version 5.0, and Voyager Data Explorer, version 3.5, software. The generated mass lists were used to search a local digestion product database of C. glutamicum proteins using ProteinProspector MS-Fit.
RESULTS
AccBC copurifies with AccD1.
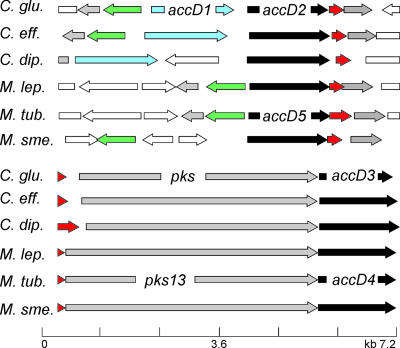
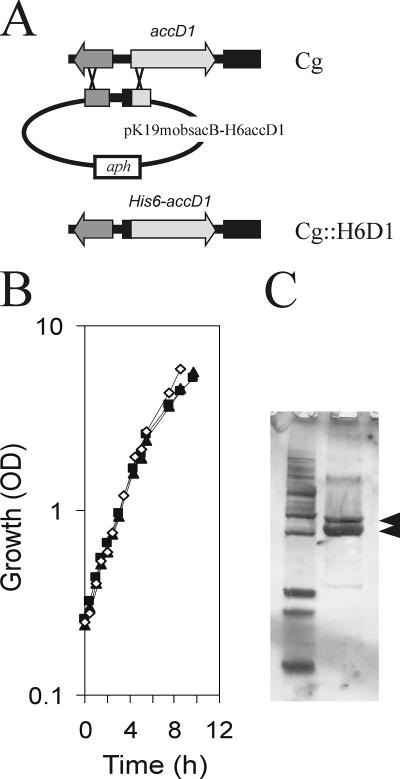
In the Corynebacterium species C. glutamicum, C. diphtheriae and Corynebacterium efficiens, the genes at the accD1 locus are syntenic, as is largely the case also with the Mycobacterium species (Fig. 1). We wanted to ensure that a His-tagged AccD1 does not disturb the function of the carboxylase, and we therefore studied growth of recombinant C. glutamicum with the wild-type allele replaced in the chromosome by six His codons attached to the 5′ end of accD1 (yielding H6-accD1) (Fig. 2A). As can be seen in Fig. 2B growth of this strain was indistinguishable from that of the wild type, with both strains exhibiting a growth rate of 0.42 ± 0.2 h−1; the same was true of a similar strain made with a His tag fused to accD2. An extract of the strain with the chromosomally encoded H6-AccD1 was prepared and subjected to Ni-NTA chromatography, which yielded two proteins with apparent molecular masses of 63 and 58 kDa (Fig. 1C). The lower band was identified by peptide mass fingerprinting as AccD1, and the upper copurified band was identified as AccBC, which represents the single acetyl-CoA carboxylase biotin-containing subunit (α-subunit) in the C. glutamicum genome (19, 20).
FIG. 1.
On top is shown the syntenic organization of accD1 as component of the acetyl-CoA carboxylase. Orthologous genes are colored similarly. Green is the biotin protein ligase birA; black is the paralogous accD2, which is named accD5 in M. tuberculosis (M. tub.); and red is the open reading frame encoding the ɛ-peptide. At the bottom is shown the syntenic organization of accD3, whose gene product together with accD2 of C. glutamicum is a component of the acyl carboxylase required for mycolic acid synthesis. The polyketide synthase, or condensase, ligating the two long-chain fatty acids to the mycolic acid backbone is pks in C. glutamicum (28) and pks13 in M. tuberculosis. C. glu, C. glutamicum; C. eff, C. efficiens; C. dip, C. diphtheriae; M. lep, M. leprae; M. tub, M. tuberculosis; M. sme, M. smegmatis.
FIG. 2.
Panel A shows the scheme to fuse in the chromosome of the wild-type of C. glutamicum (Cg) six His codons at the N-terminal end with accD1 to result in chromosomally encoded H6-AccD1 in the H6D1 strain. Panel B shows growth of the resulting strain on LB broth (⋄) compared to the wild type (▴). Also shown is growth of a similarly constructed strain (H6D2 strain) with six His codons fused in the chromosome at the 5′ end of accD2 (▴). Panel C shows the isolation of AccBC (upper arrow) together with H6-AccD1 via affinity chromatography (lower arrow).
AccBC was separately isolated from the C. glutamicum Δpyc strain where AccBC is the sole biotinylated protein in this strain by avidin chromatography. N-terminal microsequencing yielded the sequence SVETRKITKVLVAN that corresponds to Ser-2 to Asn-15 of the deduced polypeptide (Swiss-Prot accession no. Q79VI2_CORGL). This suggests removal of the first Met, as frequently observed in cases where this is followed by Ser (32). A peptide mass fingerprint analysis of tryptic digests of AccBC yielded a fragment of 3,256.5 Da. This corresponds to a peptide derived by cleavage after Lys-535 and Lys-563 but resistant to cleavage after the intermediate Lys-557 and additionally containing a fused biotin molecule. These data are consistent with attachment of biotin to the ɛ-amino group of Lys-557.
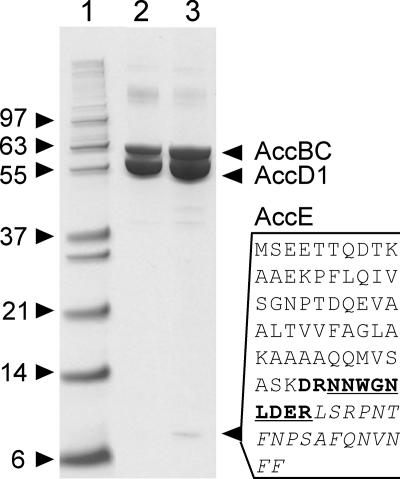
AccE copurifies with AccD1-AccBC.
The acetyl and propionyl-CoA carboxylases of Streptomyces coelicolor have recently been shown to contain a third small peptide, termed AccE or ɛ-peptide, which is different in the two carboxylases (9). A gene encoding a similar small peptide is present in C. glutamicum downstream of accD2 (Fig. 1). We overexpressed this gene together with accBC and H6-accD1 in C. glutamicum Δpyc and applied a cytoplasmic extract to Ni-NTA chromatography. Besides the expected AccBC and H6-AccD1, this yielded a small peptide of about 9 kDa (Fig. 3, lane 3). Albeit weaker, this band was also visible in extracts of an otherwise isogenic strain not overexpressing this gene in silver-stained gels (data not shown). The protein band was excised and subjected to digestion with trypsin, followed by peptide mass fingerprinting. The small peptide of 82 amino acids yields theoretically five fragments. Of these, fragments of 1,117.57 and 2,100.16 Da were identified, and by allowing a missed cleavage an additional one of 1,388.63 was found, thus identifying the copurified peptide as that encoded by NCgl0676.
FIG. 3.
Copurification of the ɛ-subunit AccE and α-subunit AccBC together with the His-tagged β-subunit AccD1 from C. glutamicum. Lane 3 shows the copurification from the C. glutamicum Δpyc paccBC p-H6D1-E strain. The arrowhead marks the small ɛ-peptide whose full sequence is given alongside. The peptides derived from the small ɛ-peptide and identified by mass fingerprinting are in bold, italics, and underlined. Lane 2 shows the copurification from an otherwise isogenic strain (the C. glutamicum Δpyc paccBC p-H6D1 strain) without accE overexpression. Silver-stained gels also confirmed in this strain AccE copurification (data not shown). Lane 1 shows standards with molecular masses in kDa.
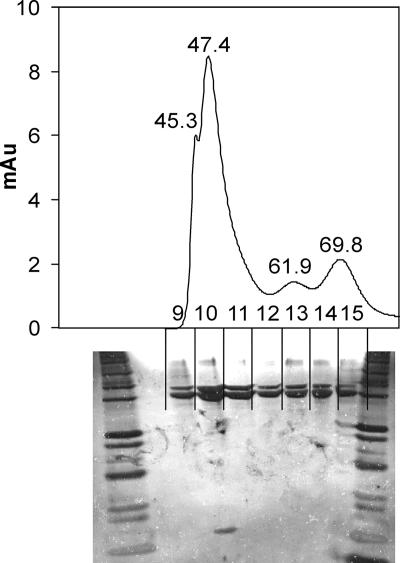
Molecular mass of acetyl-CoA carboxylase complex.
An extract of the C. glutamicum Δpyc paccBC p-H6D1 strain subjected to Ni-NTA and gel filtration chromatography yielded a major protein fraction at a retention time (RT) of 47.4 min, corresponding to a molecular mass of 812 ± 35 kDa. Additional fractions were obtained corresponding to masses of about 1,000 kDa (RT, 45.3), 225 ± 40 kDa (RT, 61.9 min), and 112 ± 40 kDa (RT, 69.8 min) (Fig. 4). The additional fractions preferentially contained AccD1, as evidenced from less sensitive Coomassie-stained gels (data not shown). This interesting observation shows that AccD1 with a molecular mass of 58.5 kDa tends to form multimers without AccBC present. However, additional isolations consistently showed the isolation of a protein complex corresponding to a mass of about 800 kDa. This particular fraction always contained the ɛ-subunit, as can be seen in Fig. 4, (whereas larger and smaller protein complexes were devoid of this subunit). Fractions without the ɛ-subunit were composed of AccD1 and AccBC in variable amounts and also prone to form large complexes upon dialysis (data not shown).
FIG. 4.
Gel filtration of acetyl-CoA carboxylase complex isolated from the C. glutamicum Δpyc paccBC p-H6D1-E strain. On top is shown the elution profile with retention times of peaks marked, as well as the individual fractions collected and marked 9 to 15. At the bottom the silver-stained gel is shown where the individual fractions were separated. The majority of the protein elutes with a retention time of 47.4 min, and only this fraction contains the ɛ-peptide. Au, arbitrary units.
Kinetic characterization of acetyl-CoA carboxylase.
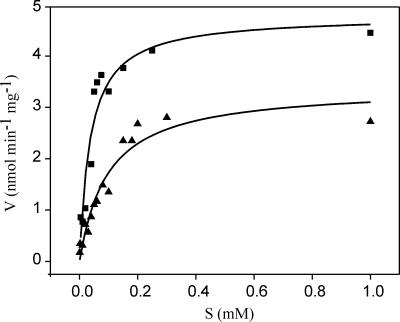
The acetyl-CoA carboxylase was kinetically characterized in terms of Km, Vmax, substrate specificity, and inhibition. The Km for acetyl-CoA was 37 ± 9 μM with a Vmax of 4.8 nmol min−1 mg−1 (Fig. 5), and the enzyme exhibited a weaker affinity for propionyl-CoA, with a Km of 97 ± 17 μm but a comparable Vmax of 3.4 nmol min−1 mg−1. With butyryl-CoA, malonyl-CoA, succinyl-CoA, or methylmalonyl-CoA, no significant activity could be detected (<0.05 mmol min−1 mg−1). A recently reconstituted carboxylase activity of M. tuberculosis yielded a comparable ratio for the affinities for acetyl and propionyl-CoA but with a remarkably lower affinity for the substrates, with a Km of 4,900 μM for acetyl-CoA (7). We also assayed for inhibition of the carboxylase. Medica 16 was an extremely efficient inhibitor since already at a concentration of 0.15 mM the activity was reduced to 16%, and at 0.5 mM no activity remained (Table 2). Haloxyfop also resulted in full inhibition, albeit at significantly higher concentrations, whereas with the structurally related aryloxyphenoxypropionates diclofop-methyl and clodinafop-propargyl, only partial inhibition could be obtained. Citrate and l-glutamate were also assayed for their influence on acetyl-CoA carboxylase activity, and stimulation was observed with l-glutamate, which could well be of physiological relevance considering that the cytosolic concentration in C. glutamicum is higher than 100 mM (22).
FIG. 5.
Substrate (S) saturation kinetics of acetyl-CoA carboxylase of C. glutamicum with either acetyl-CoA (▪) or propionyl-CoA (▴) as substrate.
TABLE 2.
Inhibition of acetyl-CoA carboxylase activity
| Inhibitor | Activity (%) at specified inhibitor concna
|
||
|---|---|---|---|
| 0.5 mM | 1 mM | 2 mM | |
| Haloxyfop | 79 | 62 | 0 |
| Dichlofop-methyl | 100 | 62 | 62 |
| Clodinafop-propargyl | 67 | 58 | ND |
| Alloxydim | 54 | 54 | ND |
| Medica 16 | 0 | 0 | ND |
| Glutamate | 110 | 110 | 140 |
| Citrate | 100 | 100 | 100 |
The total activity used in each assay (100%) was 3.9 μmol min−1 mg of protein−1.
AccD2, AccD3, AccBC, and AccE are constituents of the acyl carboxylase of mycolic acid synthesis.
As outlined in the introduction, there is considerable confusion about the native carboxylase complex involved in the synthesis of mycolic acids. In particular, a complex consisting of AccD5, AccA, and the ɛ-peptide is suggested to carboxylate propionyl-CoA preferentially for M. tuberculosis (14), although AccD2 of C. glutamicum, which is the orthologue of AccD5 of M. tuberculosis (Fig. 1), and AccD5 of M. smegmatis were both shown to be necessary for mycolic acid synthesis together with an additional second β-carboxylase subunit (15, 27).
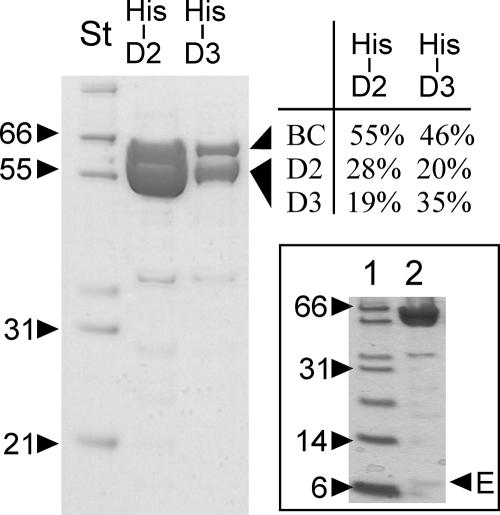
We constructed plasmids coding for N-terminal His-tagged AccD2 and His-tagged AccD3 of C. glutamicum. These were individually introduced into the C. glutamicum paccBC strain overexpressing the biotinylated α-subunit. Extracts were subjected to Ni-NTA chromatography and analyzed by polyacrylamide gel electrophoresis and peptide mass fingerprinting (Fig. 6). With H6-AccD2, peptides AccD3 and AccBC were isolated, and the three peptides were also isolated with H6-AccD3. AccD2 and AccD3 migrate together due to their close molecular masses of 57.9 kDa and 55.8 kDa, respectively, and also their sequences are very similar, exhibiting 48% identical amino acyl residues over their entire lengths (15, 20). Overexpression of different combinations of accBC, accD2, accD3, and accE, the latter encoded by NCgl0676 and shown here to be a constituent of the acetyl-CoA carboxylase (see above), revealed highest complex formation when all the genes were expressed together (data not shown), substantiating that these four components form a native acyl carboxylase complex. Extracts derived from the C. glutamicum Δpyc paccBC p-H6D2-E-D3 strain were also run on a separate gel, allowing for detection of small peptides in silver-stained gels (Fig. 6, inset). Under these conditions a peptide of about 9 kDa was detectable, whose analysis resulted in the identical three fragments as already derived for the coisolated AccE peptide in the acetyl-CoA carboxylase complex and given in Fig. 3 (data not shown).
FIG. 6.
Use of either H6-AccD2 or H6-AccD3 results in copurification of four peptides. In lane marked His-D2, peptides AccBC and AccD3 are copurified with AccD2 from an extract of the C. glutamicum paccBC p-H6D2 strain, and in the lane marked His-D3, AccBC and AccD2 are copurified with AccD3 from an extract of the C. glutamicum paccBC p-H6D3 strain. The copurification is evident from the peptide mass fingerprint analysis, with percentages given for the peptides recovered. The gel in the insert shows copurification of AccE with His-D3 (marked by E) from an extract of the C. glutamicum Δpyc paccBC p-D2-E-H6D3 strain. St, molecular weight standard.
Isolation of acyl carboxylase of mycolic acid synthesis.
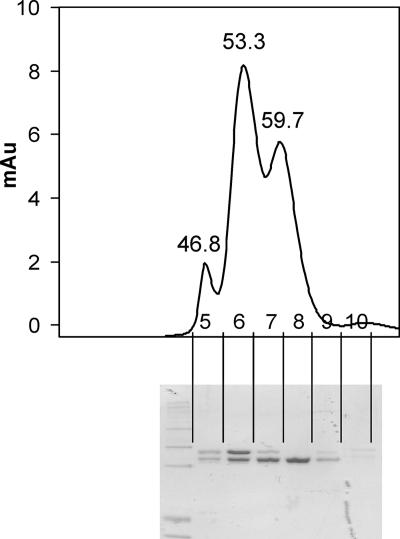
An extract of the C. glutamicum Δpyc paccBC p-H6D2-E-D3 strain was subjected to Ni-NTA and gel chromatography. This yielded three major fractions as given in Fig. 7. From three independent experiments the corresponding molecular masses assigned are 840 ± 18 kDa, 570 ± 54 kDa, and 370 ± 52 kDa. Peptide mass fingerprinting verified that the upper band visible in sodium dodecyl sulfate gels contained AccBC and that the lower band contained AccD2 together with AccD3. We assume that the native acyl carboxylase has a molecular mass of approximately 840 to 570 kDa and that it is also not possible to derive the numbers of the individual subunits present in the complex, which have molecular weights of 63.4 (AccBC), 57.99 (AccD2), 55.8 (accD3), and 8.9 (accE). Also, carboxylase isolated from either a strain with a chromosomally encoded His-fused AccD3 and no gene overexpressed or genes expressed in other combinations yielded no single peak in gel filtration experiments (data not shown), indicating that the acyl carboxylase complex is rather labile.
FIG. 7.
Isolation of acyl carboxylase of mycolic acid synthesis. Shown is the elution profile with retention times of peaks marked, as well as the individual fractions collected and marked 5 to 10. At the bottom the Coomassie-stained gel is shown where the individual fractions were separated. Au, arbitrary units.
Activity of acyl carboxylase of mycolic acid synthesis.
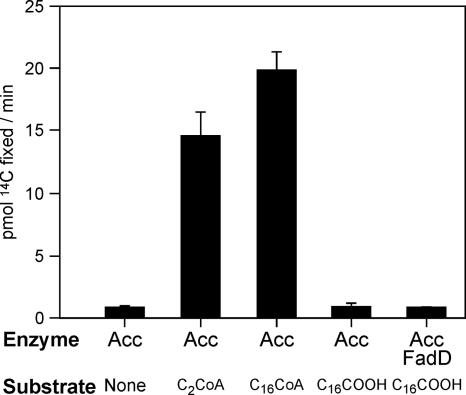
The isolated acyl carboxylase complex was assessed for activity using a fatty acyl substrate appropriate to the synthesis of mycolic acids; thus various C16 fatty acyl substrates were tested. As the precise mechanism for mycolic acid biosynthesis is not yet clear, palmitic acid and palmitoyl-CoA were used as substrates. Unlike M. tuberculosis, C. glutamicum does not possess a discrete acyl carrier protein, so acyl carrier protein thioesters could be disregarded. Recently, Trivedi et al. (33) demonstrated the generation of fatty acyl adenylates in mycobacteria and confirmed FadD32, encoded by the gene immediately upstream of pks13 in M. tuberculosis, as an adenylating enzyme involved in mycolic acid biosynthesis. Here, we also used the C. glutamicum orthologue FadD to generate palmitoyl adenylates. The activity of FadD was confirmed in vitro (data not shown). Portevin et al. (27) showed the accumulation of a tetradecylmalonic acid in a C. glutamicum fadD mutant, suggesting that acyl adenylation might occur after carboxylation.
The activity of the carboxylation complex was assessed by following the fixation of 14C from NaH14CO3. When no fatty acyl substrate was used in the assay, a low level of carbon fixation was observed, and this was presumably related to charging of the biotin moiety of the complex. Similar quantities of radiation were retained when palmitic acid was used as a substrate, regardless of whether a pretreatment with FadD had been incorporated into the assays (Fig. 8). These data suggest that neither the fatty acid nor its adenylates are efficiently activated in vitro. However, marked retention of 14C was observed when palmitoyl-CoA was used as acyl substrate. Interestingly, significant activity was also seen using acetyl-CoA, suggesting that this enzyme complex possesses a relaxed substrate specificity for acyl-CoAs but prefers the longer acyl chains appropriate for corynomycolic acid biosynthesis.
FIG. 8.
Substrate specificity of acyl carboxylase for mycolic acid synthesis. Carboxylation activity was followed by fixation of H14CO3− using various acyl substrates. Fixed radioactive carbon was measured by liquid scintillation counting after acidification of the reaction mixtures and evaporation to dryness.
DISCUSSION
C. glutamicum possesses genes for four paralogous β-subunits in its genome (15, 20), and the present study shows that the three essential β-subunits AccD1, AccD2, and AccD3 are components of two different multienzyme complexes which carboxylate straight chain ketoacyl derivatives. Consistent with experimental data on carboxyltransferase subunits in other Corynebacterianeae species (8, 15, 27), these are considered as the key enzymes to enable fatty acid and mycolic acid synthesis, the latter of which is required to build the protective hydrophobic outer lipid bilayer of this group of bacteria (24). The syntenic organization of the corresponding gene loci might be instructive to identify the relevant subunits of these complexes in other Corynebacterianeae, too, as we similarly used genome comparisons to identify further essential genes of arabinogalactan synthesis in Corynebacterianeae (1). In this respect, it is relevant that in the pathogenic M. leprae characterized by massive gene decay (4), only the two orthologues of corynebacterial accD2 and accD3 are fully retained (Fig. 1). This is in agreement with the view that they are required for mycolic acid synthesis since mycolic acids cannot be provided by the host. However, production of malonyl-CoA is likely to be necessary to extend host-derived acyl primers to meromycolic acids to afford the organisms mycolates (35). Interestingly, our analyses demonstrated that the acyl carboxylase possessed a significant carboxylation activity with acetyl-CoA that might allow some acetyl-CoA-dependent fatty acyl elongation reported in this organism (16).
The acetyl-CoA carboxylase of C. glutamicum is made up of AccD1, AccBC, and AccE, and the acyl-CoA carboxylase is made up of AccD2, AccD3, AccBC, and AccE. The biotinylated AccBC and the ɛ-subunit AccE are identical in both enzyme complexes. The fact that AccE is shared by the two different complexes is thus different from the acetyl-CoA carboxylase and propionyl-CoA carboxylase of S. coelicolor, where the ɛ-subunit has first been detected as an acetyl-CoA carboxylase component and where the enzymes have specific β- and ɛ-subunits and share only the biotinylated α-subunit (9). From M. tuberculosis several of the carboxylase peptides have been isolated, and reconstitution experiments with AccD5 of M. tuberculosis, which is the orthologue of corynebacterial AccD2 (Fig. 1), suggested that AccD5 is a component of a propionyl-CoA carboxylase (22, 25). However, biochemical data obtained with the orthologue of AccD5 of M. smegmatis clearly show that this forms a complex together with a second β-carboxylase subunit (27), and genetic data obtained with C. glutamicum also show that two carboxyltransferase β-subunits are involved in mycolic acid synthesis (15). The propionyl-CoA carboxylase activity of mycobacterial AccD5, in the presence of the β- and ɛ-subunits but without AccD4, may be a side activity in the artificial system used. Interestingly, the crystal structure of AccD5 was determined to form a homohexameric complex, where each monomer within the hexamer packs head to tail with a second copy to create a dimeric assembly, thus forming the carboxyltransferase active site (18, 21). Even more intriguing, the sequence identities of the different β-carboxylase paralogues are exceptional, with AccD4 and AccD5 of M. tuberculosis sharing as much as 47% identities and the least homologous pairs, which are AccD1 and AccD3, still sharing 22% identity (15). This enabled the superimposition of each of the six paralogues on the AccD5 crystal structure (18). Due to this high structural similarity together with the experimental evidence (15, 27), the two different β-carboxyltransferase domains, AccD4 and AccD5 in M. tuberculosis and AccD2 and AccD3 in C. glutamicum, could make up a heterohexameric complex, where two different monomers within the hexamer pack head to tail to create a dimeric assembly, thus forming a new carboxyltransferase active site. In this plausible scenario one of the carboxyltransferase domains would give specificity to the new complex. Our earlier work, in which the acyl carboxylation activities in extracts of C. glutamicum accD mutants and accD-overexpressing strains were characterized, and in vitro analyses of accD2 and accD3 overexpression strongly suggest that AccD3 might define the substrate specificity of such a complex. Furthermore, our sequence analysis of AccD paralogues supports this hypothesis, as AccD2 is more closely related to AccD1 than AccD3 (15). Specificity is a major issue for the carboxyltransferases in Corynebacterianeae, considering that M. tuberculosis has six β-carboxylase subunits (3).
Interestingly, the genes encoding AccD4 and AccD5 of M. tuberculosis are expressed at high levels during exponential growth, as is AccA3 (7), one of the three biotinylated α-subunits in M. tuberculosis, as well as AccD6, which is a third β-carboxyltransferase subunit not yet discussed. Since AccD6 has in a reconstituted system higher activity with acetyl-CoA than propionyl-CoA, this is likely to be a component of the acetyl-CoA carboxylase of M. tuberculosis. Thus, the acetyl-CoA carboxylases of M. tuberculosis and C. glutamicum, as well as the complex involved in mycolic acid synthesis, have in common that they share the biotinylated α-subunit. This is also the case for the ɛ-subunit that we coisolated for the C. glutamicum proteins and most probably also the case for the enzymes of M. tuberculosis (7, 25). A structure for fully assembled acetyl or propionyl-CoA carboxylase is not available yet, albeit this is known for individual domains of such enzymes (2, 8, 34). More than 20 years ago, Haase and coworkers suggested that a propionyl-CoA carboxylase of M. smegmatis consists of six biotin-containing α-subunits and six biotin-free β-subunits (16). Based on these data together with the crystallization of carboxyltransferase subunits as hexamers (8, 18, 21), it is reasonable to assume that acetyl-CoA carboxylase in C. glutamicum has an α6β6ɛ6 structure, which yields a theoretical mass of 784 kDa and agrees well with the experimentally determined value (Fig. 4). However, a similar consideration applied to the acyl carboxylase does not give a satisfying agreement with an α6β23β33ɛ6 or α4β22β22ɛ4 structure, for instance. This points to a high instability of the complex, accessible up to now only via several peptides (14, 18, 21, 25, 27). In summary, the present study shows that two structurally related carboxylase complexes reside in C. glutamicum. They share individual peptides, and both complexes carboxylate a β-carbon of an acyl derivative.
Supplementary Material
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Alderwick, L. J., E. Radmacher, M. Seidel, R. Gande, P. G. Hitchen, H. R. Morris, A. Dell, H. Sahm, L. Eggeling, and G. S. Besra. 2005. Deletion of Cg-emb in Corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280:32362-32371. [DOI] [PubMed] [Google Scholar]
- 2.Bilder, P., S. Lightle, G. Bainbridge, J. Ohren, B. Finzel, F. Sun, S. Holley, L. Al-Kassim, C. Spessard, M. Melnick, M. Newcomer, and G. L. Waldrop. 2006. The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme. Biochemistry 45:1712-1722. [DOI] [PubMed] [Google Scholar]
- 3.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. K. Badcoc, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 5.Cronan, J. E. 2003. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57:203-224. [DOI] [PubMed] [Google Scholar]
- 6.Cronan, J. E., and C. O. Rock. 1996. Biosynthesis of membrane lipids, p. 612-636. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 7.Daniel, J., T. J. Oh, C. M. Lee, and P. E. Kolattukudy. 2007. AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme A carboxylase in Mycobacterium tuberculosis. J. Bacteriol. 189:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diacovich, L., D. L. Mitchell, H. Pham, G. Gago, M. M. Melgar, C. Khosla, H. Gramajo, and S.-C. Tsai. 2004. Crystal structure of the beta-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry 43:14027-14036. [DOI] [PubMed] [Google Scholar]
- 9.Diacovich, L., S. Peiru, D. Kurth, E. Rodriguez, F. Podesta, C. Khosla, and H. Gramajo. 2002. Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem. 277:31228-31236. [DOI] [PubMed] [Google Scholar]
- 10.Eggeling, L., and H. Sahm. 2001. The cell wall barrier of Corynebacterium glutamicum and amino acid efflux. J. Biosci. Bioeng. 92:201-213. [Google Scholar]
- 11.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 12.Eggeling, L., and O. Reyes. 2005. Experiments, p. 535-565. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 13.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93-98. [DOI] [PubMed] [Google Scholar]
- 14.Gago, G., D. Kurth, L. Diacovich, S. C. Tsai, and H. Gramajo. 2006. Biochemical and structural characterization of an essential acyl coenzyme A carboxylase from Mycobacterium tuberculosis. J. Bacteriol. 188:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gande, R., K. J. Gibson, A. K. Brown, K. Krumbach, L. G. Dover, H. Sahm, S. Shioyama, T. Oikawa, G. S. Besra, and L. Eggeling. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279:44847-44857. [DOI] [PubMed] [Google Scholar]
- 16.Haase, F. C., K. P. Henrikson, D. H. Treble, and S. H. Allen. 1982. The subunit structure and function of the propionyl coenzyme A carboxylase of Mycobacterium smegmatis. J. Biol. Chem. 257:11994-11999. [PubMed] [Google Scholar]
- 17.Hashimoto, K., H. Kawasaki, K. Akazawa, J. Nakamura, Y. Asakura, T. Kudo, E. Sakuradani, S. Shimizu, and T. Nakamatsu. 2006. Changes in composition and content of mycolic acids in glutamate-overproducing Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 70:22-30. [DOI] [PubMed] [Google Scholar]
- 18.Holton, S. J., S. King-Scott, A. Nasser Eddine, S. H. Kaufmann, and M. Wilmanns. 2006. Structural diversity in the sixfold redundant set of acyl-CoA carboxyltransferases in Mycobacterium tuberculosis. FEBS Lett. 580:6898-6902. [DOI] [PubMed] [Google Scholar]
- 19.Jäger, W., P. G. Peters-Wendisch, J. Kalinowski, and A. Pühler. 1996. A Corynebacterium glutamicum gene encoding a two-domain protein similar to biotin carboxylases and biotin-carboxyl-carrier proteins. Arch. Microbiol. 166:76-82. [DOI] [PubMed] [Google Scholar]
- 20.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Pühler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 21.Lin, T. W., M. M. Melgar, D. Kurth, S. J. Swamidass, J. Purdon, T. Tseng, G. Gago, P. Baldi, H. Gramajo, and S. C. Tsai. 2006. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:3072-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marx, A., A. A. de Graaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotech. Bioeng. 49:121-129. [DOI] [PubMed] [Google Scholar]
- 23.Menkel, E., G. Thierbach, L. Eggeling, and H. Sahm. 1989. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ Microbiol. 55:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido, H., and P. J. Brennan. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 25.Oh, T. J., J. Daniel, H. J. Kim, T. D. Sirakova, and P. E. Kolattukudy. 2006. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J. Biol. Chem. 281:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters-Wendisch, P., G. B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Mockel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 27.Portevin, D., C. de Sousa-D'Auria, H. Montrozier, C. Houssin, A. Stella, M A. Laneelle, F. Bardou, C. Guilhot, and M. Daffe. 2005. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J. Biol. Chem. 280:8862-8874. [DOI] [PubMed] [Google Scholar]
- 28.Portevin, D., C. de Sousa-D'Auria, C. Houssin, C. Grimaldi, M. Chami, M. Daffe, and C. Guilhot. 2004. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. USA 101:314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radmacher, E., L. J. Alderwick, G. S. Besra, A. K. Brown, K. J. Gibson, H. Sahm, and L. Eggeling. 2005. Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum. Microbiology 151:2421-2427. [DOI] [PubMed] [Google Scholar]
- 30.Rainwater, D. L., and P. E. Kolattukudy. 1982. Isolation and characterization of acyl coenzyme A carboxylases from Mycobacterium tuberculosis and Mycobacterium bovis, which produce multiple methyl-branched mycocerosic acids. J. Bacteriol. 151:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 32.Schaffer, S., and A Burkovski. 2005. Proteomics, p. 99-120. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL.
- 33.Trivedi, O. A., P. Arora, V. Sridharan, R. Tickoo, D. Mohanty, and R. S Gokhale. 2004. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428:441-445. [DOI] [PubMed] [Google Scholar]
- 34.Waldrop, G. L., I. Rayment, and H. M. Holden. 1994. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry 33:10249-10256. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler, P. R., K. Bulmer, and C. Ratledge. 1990. Enzymes for biosynthesis de novo and elongation of fatty acids in mycobacteria grown in host cells: is Mycobacterium leprae competent in fatty acid biosynthesis? J. Gen. Microbiol. 136:211-217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.