Abstract
Bacterial nonhomologous end joining (NHEJ) is a recently described DNA repair pathway best characterized in mycobacteria. Bacterial NHEJ proteins LigD and Ku have been analyzed biochemically, and their roles in linear plasmid repair in vivo have been verified genetically; yet the contributions of NHEJ to repair of chromosomal DNA damage are unknown. Here we use an extensive set of NHEJ- and homologous recombination (HR)-deficient Mycobacterium smegmatis strains to probe the importance of HR and NHEJ in repairing diverse types of chromosomal DNA damage. An M. smegmatis ΔrecA Δku double mutant has no apparent growth defect in vitro. Loss of the NHEJ components Ku and LigD had no effect on sensitivity to UV radiation, methyl methanesulfonate, or quinolone antibiotics. NHEJ deficiency had no effect on sensitivity to ionizing radiation in logarithmic- or early-stationary-phase cells but was required for ionizing radiation resistance in late stationary phase in 7H9 but not LB medium. In addition, NHEJ components were required for repair of I-SceI mediated chromosomal double-strand breaks (DSBs), and in the absence of HR, the NHEJ pathway rapidly mutates the chromosomal break site. The molecular outcomes of NHEJ-mediated chromosomal DSB repair involve predominantly single-nucleotide insertions at the break site, similar to previous findings using plasmid substrates. These findings demonstrate that prokaryotic NHEJ is specifically required for DSB repair in late stationary phase and can mediate mutagenic repair of homing endonuclease-generated chromosomal DSBs.
DNA repair pathways in all domains of life are intensely studied in light of their importance for organism viability, genome integrity, and oncogenesis. There is a growing recognition that DNA repair pathways are important in pathogenic bacteria to counteract DNA damage inflicted by mutagenic products of the host. In Mycobacterium tuberculosis, a widely successful human pathogen, recent evidence implicated nucleotide excision repair (NER) and an error prone UV-induced polymerase in the defense against host-derived DNA damage in the mouse (2, 8). Mycobacteria contain many of the canonical DNA repair pathways including NER, base excision repair, and homologous recombination (HR), but notably lack identifiable homologues of the mismatch repair machinery (21, 30).
Double strand breaks (DSBs) are a particularly lethal form of DNA damage produced experimentally by ionizing radiation (IR) and restriction endonucleases. In mammalian cells, yeast, and flies, DSBs can be repaired either by HR or nonhomologous end joining (NHEJ), the former being error free and the latter potentially error prone. The most intensely studied model of prokaryotic DNA repair is Escherichia coli, which repairs DSBs by RecA-dependent HR. Although E. coli lacks an NHEJ pathway, wider examination of sequenced bacterial genomes identified genes encoding putative NHEJ proteins in some bacteria, including mycobacteria and bacillus (10). This bioinformatic finding was followed by a biochemical demonstration that mycobacterial Ku binds DNA ends and recruits a polyfunctional DNA ligase/polymerase (LigD) to DNA ends in vitro (9, 39).
Definitive demonstration of a prokaryotic NHEJ pathway came from examination of mycobacterial null mutants in Ku and LigD, an ATP-dependent DNA ligase. Deletion of M. tuberculosis LigD reduced the efficiency of linear plasmid recircularization (14). Deletion of Mycobacterium smegmatis LigD or Ku also reduced plasmid recircularization and revealed that mycobacterial NHEJ is an error-prone pathway for the repair of plasmid substrates (13). This error-prone NHEJ pathway is genetically dependent on ku and ligD, and the Ku and LigD proteins interact physically (13), likely through the polymerase module of LigD (28). Replacement of wild-type LigD with a polymerase-defective mutant (D136A D138A) abolished nontemplated nucleotide additions, demonstrating that the polymerase activity of LigD is a direct catalyst of nucleotide addition during plasmid joining (40). The atomic structure of the polymerase domain revealed similarity to archaeal DNA primase (40). Recent evidence also demonstrated that LigD is required for genome circularization of mycobacteriophages (27).
Despite this extensive characterization of mycobacterial NHEJ using plasmid substrates, very little information is available about the role of mycobacterial NHEJ in repairing chromosomal DNA damage. In this study we constructed a comprehensive set of NHEJ-deficient and HR-deficient M. smegmatis strains and probed the contributions of these repair systems to a defense against chromosomal DNA damage of diverse types. We find that loss of NHEJ sensitizes mycobacteria to IR only in late stationary phase in minimal but not rich medium. In addition, NHEJ has an important role in the repair of site-specific chromosomal DSBs generated by the restriction endonuclease I-SceI.
MATERIALS AND METHODS
Media and growth conditions.
M. smegmatis strains were cultured at 37°C on Middlebrook 7H10 medium (Difco) containing 0.5% dextrose and 0.5% glycerol or on LB medium with 0.5% glycerol and 0.5% dextrose. Liquid medium was LB medium containing 0.5% glycerol and 0.05% Tween 80 or 7H9 medium with 0.5% dextrose, 0.5% glycerol, and 0.05% Tween 80. When appropriate, hygromycin B (Boehringer Mannheim) at 50 μg/ml or kanamycin (Sigma) at 20 μg/ml was used. A total of 50 ng/ml anhydrotetracycline (AHT; Sigma) was used to induce I-SceI expression.
Strain construction.
All strains used in this study are listed in Table 1. Gene names from the M. smegmatis genome sequence (available at www.tigr.org) are as follows: MSmeg_5580, ku; MSMEG_5570, ligD; MSMEG_2723, recA; MSMEG_1327, recB; MSMEG_1328, recC; and MSMEG_1325, recD. All null mutations were constructed using two-step sucrose counter-selection as previously described (13). Null mutations were introduced as truncations of the gene of interest while the reading frame of the native protein was maintained. All deletions were confirmed by Southern analysis of genomic DNA using flanking DNA as the probe. The M. smegmatis ku and ligD null mutants have been described previously (13).
TABLE 1.
Strains used in this study
| Strain name | Description | Genotypea | Source or reference |
|---|---|---|---|
| mc2155 | Wild type | ept1 | 35 |
| Mgm140 | ΔligD | mc2155 ligD::ligD(Δ202-2391) | 13 |
| Mgm153 | ΔligD Δku | mgm140 ku::ku(Δ90-947) | This study |
| Mgm154 | Δku | mc2155 ku::ku(Δ90-947) | 13 |
| Mgm177 | ΔrecBCD | mc2155 recBCD::recC(Δ76-3264) ΔrecB ΔrecD | This study |
| Mgm182 | Wild type; I-SceI | mc2155 attB::pMSG375 | This study |
| Mgm184 | ΔligD ΔrecBCD | Mgm140 recBCD::recC(Δ76-3264) ΔrecB ΔrecD | This study |
| Mgm199 | ΔrecA | mc2155 recA::recA(Δ49-1014) | This study |
| Mgm201 | ΔrecA Δku | Mgm154 recA::recA(Δ49-1014) | This study |
| Mgm213 | ΔligD I-SceI | Mgm140 attB::pMSG375 | This study |
| Mgm214 | ΔligD Δku I-SceI | Mgm153 attB::pMSG375 | This study |
| Mgm215 | Δku I-SceI | Mgm154 attB::pMSG375 | This study |
| Mgm222 | ΔrecA I-SceI | Mgm199 attB::pMSG375 | This study |
| Mgm223 | ΔrecA Δku I-SceI | Mgm201 attB::pMSG375 | This study |
Deleted residues are indicated in parentheses.
UV assays.
UV exposure was performed in a Stratalinker (Stratagene) fitted with 254-nm bulbs. Serial 10-fold dilutions of the strains to be tested in mid-logarithmic growth phase were spotted on LB plates and exposed to the UV doses indicated in Fig. 1. Immediately after exposure, the plates were wrapped in foil to prevent repair by photolyase. Surviving colonies were counted at 3 days, and survival ratios were calculated in comparison to the same dilutions of unexposed cells from the same culture.
FIG. 1.
UV resistance of NHEJ- and HR-deficient strains. Wild-type, NHEJ-deficient (Δku, ΔligD, or ΔligD ku), HR-deficient (ΔrecA or ΔrecBCD), or combined HR/NHEJ-deficient strains (ΔrecBCD ΔligD or ΔrecA Δku) were exposed to high-dose (A) and low-dose (B) UV irradiation, and survival was plotted on a log scale y axis as a percentage compared to unexposed cells of the same strain. Error bars ± standard errors of the means. WT, wild type.
Gamma irradiation killing assays.
IR killing experiments were performed using a 137Cs source that delivers a dose rate of 12 Gy per minute. Log-phase (A600 of 0.2 to 0.3), early-stationary-phase (A600 of 2 to 3), or late-stationary-phase (24 to 36 h after replication ceased) cultures were collected by centrifugation and resuspended in phosphate-buffered saline with 0.05% Tween 80. After 10 s of water bath sonication (Branson Ultrasonics) to disperse clumps, 200 μl of suspension (approximately 107 bacteria) was exposed on a rotating platform within the chamber of the cesium source to assure equal dosage between samples. After exposure, serial 10-fold dilutions were cultured on 7H9 plates, and survival was calculated compared to unexposed control cultures.
MMS.
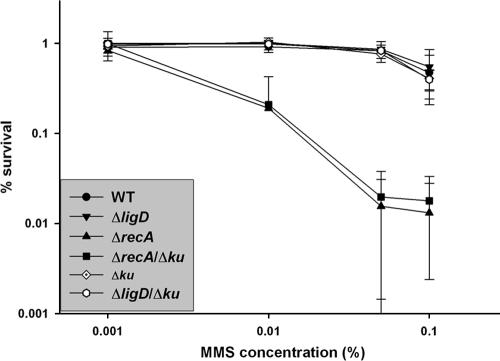
For methyl methanesulfonate (MMS) killing assays, fresh agar medium containing 0.001 to 0.1% MMS was prepared for each experiment due to rapid loss of MMS potency with storage. Serial 10-fold dilutions of each strain in log phase were cultured on plates containing no MMS or the concentration indicated in Fig. 2, and surviving colonies were counted after 3 days of incubation. Percent survival was calculated compared to unexposed cells.
FIG. 2.
NHEJ is required for resistance to IR in late stationary phase. Wild-type, NHEJ-deficient, HR-deficient, or HR/NHEJ-deficient strains were exposed to gamma irradiation from a 137Cs source, and survival was calculated by enumerating surviving colonies compared to a mock-exposed control sample of the same strain. Panel A depicts cells harvested from logarithmically growing cultures in LB medium. Panel B compares logarithmic- and early-stationary-phase cells of wild-type, ΔrecA, and ΔrecA Δku strains in LB medium. Panel C shows late-stationary-phase cells grown in LB medium. Panel D shows late-stationary-phase cells grown in 7H9 medium. For panels C and D, the symbols are identical to those in panel A. Error bars are ± standard errors of the means. WT, wild type.
I-SceI strain construction.
An I-SceI cDNA with an N-terminal hemagglutinin (HA) tag was cloned from plasmid pCBASceI (a kind gift of Maria Jasin and David Weinstock) as an SphI/MscI fragment into pTX-2M1X cut with SphI/PmeI to create pMSG371. Plasmid pTX-2M1X contains the tetracycline-inducible Pmyc1 TetO promoter and tetR. To create a ribosome binding site (RBS) in pMSG371, the oligonucleotides I-SceI RBS 1(5′-CAGAAAGGAGGCCATATGGG-3′) and I-SceI RBS 2 (5′-TCGTCCCATATGGCCTCCTTTCTGCATG-3′) were annealed and ligated into pMSG371 cut with SphI/BbsI to create pMSG373. To create a genomic I-SceI site, we annealed I-SceI 1 (5′-AGCTTAGGGATAACAGGGTAAT-3′) and I-SceI 2 (5′-AATTATTACCCTGTTATCCCTA-3′) (I-SceI sites in bold) and ligated this duplex into pMV306Kan cut with HindIII and EcoR1 to create pMSG372. pMV306kan integrates into the mycobacterial chromosome at the attB site, thereby incorporating all plasmid sequences into the chromosome. Finally, the tetracycline-inducible I-SceI cassette from pMSG373 as a NotI/SwaI fragment was moved into pMSG372 cut with NotI/EcoRV to make pMSG375. As a control, the NotI/SwaI fragment from pMSG373 was transferred into pMV306kan to create pMSG374, which integrates I-SceI into the chromosome without an I-SceI cutting site.
I-SceI expression in mycobacteria.
Expression of I-SceI endonuclease was induced by the addition of 50 ng/ml AHT. Samples were taken at different time points, followed by cell lysis and standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using mouse monoclonal anti-HA antibody (Covance). In parallel, samples from the same cultures were used to observe the genomic DSB at the I-SceI site. Genomic DNA was purified and digested with SmaI, followed by Southern blotting with a 32P-labeled DNA probe that spans the I-SceI site. I-SceI cleavage yielded an 890-bp band while SmaI digestion without I-SceI cleavage yielded a 4,501-bp band.
I-SceI killing assays and sequence analysis.
A sensitivity assay to I-SceI-induced chromosomal DSBs was performed as follows. Log-phase (A600 of 0.5) and stationary-phase (A600 of 2.0) cultures were collected by centrifugation and resuspended in phosphate-buffered saline with 0.05% Tween 80. Serial 10-fold dilutions of each strain were plated on LB plates supplemented with 20 μg/ml kanamycin and 50 ng/ml AHT. Colonies were counted after 3 days, and percent survival was calculated compared to cells grown in the absence of AHT. To determine the molecular outcome of the DSB repair event, we used PCR amplification directly from surviving colonies using the primers IS1a (5′-GTATGCCGCCATTATTACGAC-3′) and IS1b (5′-TCCCTGATCTGGCTACTTTC-3′), which anneal approximately 300 bp upstream and downstream of the I-SceI site in the chromosome. The 612-bp PCR product was gel purified and sequenced using the IS1a primer.
RESULTS
Mycobacterial RecA and NHEJ are dispensable for in vitro survival.
To survey the division of labor between HR and NHEJ systems in M. smegmatis for the repair of DNA damage, we attempted to delete canonical HR and NHEJ components, singly and in combination. The ΔligD and Δku null mutants were reported previously (13) and are deficient for NHEJ assayed by linear plasmid transformation. As is the case in most studied bacteria and as previously reported for M. smegmatis and Mycobacterium bovis BCG (12, 33), we found that recA is nonessential for survival of M. smegmatis in the absence of exogenous DNA damage. In addition, we found that a ΔrecA Δku double mutant is viable and displays no growth defects under laboratory conditions. We also deleted the HR component recBCD alone and in combination with core NHEJ genes.
UV sensitivity phenotypes.
We tested the relative contribution of HR and NHEJ to survival after UV irradiation. M. smegmatis cells deficient in NHEJ components either singly (Δku or ΔligD) or in combination (Δku ΔligD) and deficient in HR singly (ΔrecA or ΔrecBCD) or in combination (ΔrecA Δku or ΔrecBCD ΔligD) were exposed to escalating doses of UV irradiation, and survival was determined as a percentage of unexposed cells from the same culture. Wild-type cells exhibit dose-dependent killing after UV exposure at greater than 5 mJ/cm2 (Fig. 1A). At a dose of 30 mJ/cm2, wild-type survival is reduced by 10,000-fold compared to unexposed cells, similar to the previously reported sensitivity of this strain to UV (2). Deficiency of NHEJ components either singly or in combination (ΔligD Δku) does not affect survival after UV irradiation compared to wild-type organisms (Fig. 1A). M. smegmatis without recA is highly sensitive to killing by UV light. At a dose of 5 mJ/cm2, titers of ΔrecA cells were reduced 50-fold compared to wild-type cells, which were fully viable at this dose (Fig. 1B), consistent with prior reports in E. coli, Bacillus subtilis, and mycobacteria (33). ΔrecA Δku cells were no more sensitive to UV irradiation than ΔrecA cells (Fig. 1B). Surprisingly, and in sharp contrast to E. coli (5, 16), deletion of recBCD did not sensitize M. smegmatis to UV (Fig. 1A). In addition, deletion of ligD in a ΔrecBCD strain had no effect on UV sensitivity (Fig. 1A). We also tested cells grown to early stationary phase and also found no effect of NHEJ mutations on UV sensitivity (data not shown). Taken together, these results confirm a well-described role for recA in a defense against UV-induced DNA damage and indicate that NHEJ in mycobacteria is not required for defense against UV.
IR.
IR produces multiple types of DNA damage including base damage and strand breaks, the latter of which are repaired by either HR or NHEJ. We exposed log phase wild-type, NHEJ-deficient, HR-deficient, or HR- and NHEJ (HR/NHEJ)-deficient strains to escalating doses of IR from a 137Cs source and measured bacterial survival. Wild-type M. smegmatis suffers 10-fold killing at a dose of 732 Gy and 200-fold killing at 976 Gy (Fig. 2A and B). Deletion of ligD, ku, or both genes did not change IR sensitivity compared to wild-type cells (Fig. 2A). However, ΔrecA cells displayed ∼1,000-fold greater sensitivity than wild-type cells at a dose of 732 Gy (Fig. 2A). The ΔrecA Δku double mutant was no more sensitive to IR than the ΔrecA strain, indicating that NHEJ does not repair IR-induced DNA damage under the conditions tested, even when HR is disabled (Fig. 2A).
We considered the possibility that the lack of sensitization of the Δku ΔrecA strain could be due to active bacterial replication that provides a template for HR in the form of a replicated chromosome, as the experiments pictured in Fig. 2A used cells harvested from logarithmically growing cultures. In addition, we considered the possibility that nutritional availability in stationary phase might affect sensitivity to IR. To examine this possibility, we measured survival of wild-type and mutant cells to IR in early stationary phase (optical density at 600 nm of 3.0) and of cells in stationary phase for 24 to 36 h in both rich (LB) and minimal media (7H9). When grown in LB medium, log-phase wild-type and ΔrecA cells were more sensitive to IR than early-stationary-phase cells (Fig. 2B). This result could be consistent with a role for NHEJ in efficiently repairing DSBs in stationary phase cells. However, ΔrecA and ΔrecA Δku stationary phase cells had identical sensitivities, despite being more resistant than their respective log phase counterparts.
To test whether longer incubation in a nonreplicating state or nutrient availability would affect IR sensitivity of NHEJ mutants, we tested late-stationary-phase cultures (24 to 36 h without replication) in both LB and 7H9 media. Late-stationary-phase LB cultures of ku and ligD mutants were not sensitized to IR compared to wild-type cells (Fig. 2C). The ΔrecA Δku double mutant was somewhat more sensitive than the ΔrecA mutant, although only at the highest radiation dose (Fig. 2C). In contrast, late-stationary 7H9 cultures of Δku and ΔligD mutants were 8- and 20-fold more sensitive, respectively, than wild-type cells at 1,416 Gy and 2.8-fold more sensitive at 708 Gy (Fig. 2D). In comparison to the recA null strain, the ΔrecA Δku double mutant was 16-fold more sensitive at 708 Gy and 300-fold more sensitive at 1,416 Gy (Fig. 2D). These results demonstrate that NHEJ is required for defense against DNA damage caused by IR in late stationary phase in minimal but not rich medium.
DNA gyrase inhibitors.
DSBs are produced through poisoning of DNA gyrase by quinolone antimicrobial drugs, and prior studies have indicated that recombination-deficient E. coli strains are hypersensitive to quinolones (25). Similarly, NHEJ-deficient chicken cells are hypersensitive to the topoisomerase II poison VP-16 (1). We tested the sensitivity of HR- and NHEJ-deficient strains to killing by the gyrase poison ciprofloxacin both by determining the MIC using the broth dilution method and by using disk diffusion testing on solid medium. We found that ΔrecA strains were hypersensitive to ciprofloxacin, displaying 35-fold reduction in viable bacteria at a dose of 80 ng/ml on agar medium, a dose that does not affect wild-type M. smegmatis (data not shown). The ΔrecA Δku mutant phenocopied the quinolone sensitivity of the recA single mutant. Disk diffusion tests of growth inhibition also failed to demonstrate any effect of NHEJ mutations on quinolone susceptibility (data not shown).
MMS.
MMS is an alkylating agent that methylates nitrogen centers in DNA bases. Defense against MMS in E. coli and Saccharomyces cerevisiae is mediated predominantly by the NER pathway, but ku-deficient yeast and plants are sensitive to MMS-mediated killing (4, 20, 34). In the case of ku80 deficient yeast, MMS sensitivity was only observed in the absence of HR. We therefore tested the sensitivity of HR/NHEJ-deficient M. smegmatis to MMS. When an MMS dose ranging from 0.001% to 0.1% was used for testing, we found no effect of deleting NHEJ components on sensitivity to killing by alkylating agents. As was observed both for UV and IR, ΔrecA strains were highly sensitive to killing by MMS (Fig. 3).
FIG. 3.
MMS sensitivity of NHEJ- and HR-deficient strains. Serial dilutions of the indicated strains were cultured on agar medium containing an MMS dose ranging from 0.001 to 0.1%, and survival was calculated compared to unexposed control strains. Error bars are ± standard errors of the means. WT, wild type.
Conditional expression of I-SceI breaks the mycobacterial chromosome.
The various types of damage that are caused by IR and the different repair pathways involved in repairing these lesions, with a prominent role for base excision repair and NER, make it complicated to study repair of the few DSBs that appear. To minimize involvement of other repair pathways, we sought to create a controlled, single genomic DSB using the homing endonuclease I-SceI. I-SceI and HO endonuclease have been used extensively to study the molecular requirements and dynamics of chromosomal DSB in yeast (15) and mammalian cells (29, 31). As there is no I-SceI site in the M. smegmatis or M. tuberculosis genome, we attempted to adapt the I-SceI system for use in mycobacteria. We placed an I-SceI cDNA with an N-terminal HA epitope tag under the control of a tetracycline-inducible promoter (11). This I-SceI cassette was placed on a mycobacterial plasmid vector which integrates into the chromosome at the attB site through the action of the mycobacteriophage L5 integrase encoded on the plasmid (19). In addition, this vector (pMSG375) (Fig. 4A) has an I-SceI restriction site that provides a single site of chromosomal cleavage after plasmid integration. A control plasmid (pMSG374) encodes the I-SceI endonuclease under tetracycline control but lacks a chromosomal I-SceI site.
FIG. 4.
Conditionally expressed I-SceI efficiently cleaves the M. smegmatis chromosome at a supplied I-SceI site. (A) Plasmid map of pMSG375 which contains the I-SceI homing endonuclease with an N-terminal HA epitope tag under the control of Pmyc1 TetO, which confers tetracycline-inducible expression when tetR is coexpressed. The plasmid also contains a single I-SceI site and an integrase which catalyzes site-specific integration at the attB site in the mycobacterial chromosome. When integrated at attB, all plasmid sequences are contained on the chromosome, thereby creating a single unique I-SceI cleavage site. (B) AHT-dependent expression of HA-I-SceI. The indicated strains transformed with the tetracycline-inducible HA-I-SceI construct were cultured with or without 50 ng/ml AHT. Whole-cell extracts were analyzed by Western blotting with an anti-HA monoclonal antibody. The predicted size of the HA-I-SceI protein is 29.4 kDa. The anti-DlaT antibody serves as a loading control. (C) Map of the I-SceI expression cassette when integrated at the attB locus with probe location and predicted sizes of bands by Southern blotting. (D) I-SceI cleaves the mycobacterial chromosome. Southern analysis of genomic DNA prepared from M. smegmatis transformed with pMSG375 without (−) and with (+) AHT for 0.5, 1, and 2 h after AHT addition. The probe spans the I-SceI site at attB, yielding a band of 800 bp with I-SceI cleavage (Cut). wt, wild type.
We tested the utility of this system by monitoring the inducibility of I-SceI in wild-type M. smegmatis by immunoblotting using an antibody against HA. We observed HA-reactive protein of the predicted size for HA-I-SceI within 30 min after the addition of AHT. A faint band was visible without addition of the inducer in the recA mutant, suggesting slight basal expression of the endonuclease (Fig. 4B). Having confirmed AHT inducibility of I-SceI, we next asked whether the enzyme generates chromosomal breaks in mycobacterial cells. We analyzed chromosomal DNA by Southern blotting before and after AHT induction of I-SceI in strains with and without a chromosomal I-SceI site. The probe spans the I-SceI site and should anneal to two fragments if the I-SceI site is cut (Fig. 4C). AHT-dependent, site-specific chromosomal cleavage was visible by Southern blotting by 30 min after induction as detected by a 813-bp band (Fig. 4D). No chromosomal cleavage was detected in a strain lacking an I-SceI site (data not shown). These results indicate that I-SceI can create DSBs in the mycobacterial chromosome and that the basal level of I-SceI expression is sufficiently low to allow preservation of the I-SceI site in uninduced cells.
NHEJ components are required for resistance to I-SceI-mediated killing.
To test the role of NHEJ and HR in repair of an I-SceI-generated DSB, HR- and NHEJ-deficient strains were transformed with pMSG374 (I-SceI and no I-SceI site) or pMSG375 (I-SceI with an I-SceI site) and assayed for survival after I-SceI induction. Serial dilutions of each strain were plated on agar medium with or without AHT, and survival was calculated as the fraction of cells on AHT plates compared to no AHT. Expression of I-SceI in the absence of a chromosomal I-SceI site did not compromise viability in any strain background (data not shown). In wild-type M. smegmatis, constitutive I-SceI induction reduced viability by sixfold (Fig. 5A, WT) (average survival, 16.8%). Δku and ΔligD cells were additionally sensitized to I-SceI-mediated killing by 11.5- and 16.5-fold compared to uninduced cells (average Δku survival, 6%; average ΔligD survival, 8.7%) (Fig. 5A). These results indicate that constitutive induction of I-SceI leads to cell death in M. smegmatis and that deficiency of either Ku or LigD causes mild additional sensitivity. Surprisingly, the I-SceI sensitivity of the ΔligD Δku double mutant was similar to the wild-type strain. This surprising result is contrary to our expectation that the double mutant would phenocopy the ligD and ku single mutants but is consistent with observations of NHEJ mutants in other systems, discussed below. We next tested the M. smegmatis ΔrecA strain for sensitivity to I-SceI-mediated killing. In contrast to wild-type and NHEJ-defective strains, the ΔrecA strain was not substantially susceptible to I-SceI-mediated killing when plated on AHT. Specifically, the recA null strain displayed an average survival of 95% of its uninduced control. To control for possible premature mutation of the I-SceI site in the ΔrecA strain, we confirmed that the I-SceI site was intact in this strain after transformation with the I-SceI plasmid. Deletion of ku in the ΔrecA strain reduced the average survival by approximately 95-fold such that only 1% of ΔrecA Δku double mutant cells survived on AHT (Fig. 5A).
FIG. 5.
NHEJ is required for mutagenic repair and survival after I-SceI chromosomal DSBs. (A) The indicated wild-type or mutant M. smegmatis strains bearing pMSG375 were grown in the absence of tetracycline induction, and then serial dilutions were plated on 50 ng/ml AHT. Survival was calculated compared to control plates without AHT. Error bars are ± standard deviations. (B and C) Molecular outcomes of repair of I-SceI-induced chromosomal breaks in HR- and NHEJ-deficient strain backgrounds. (B) For each strain, the number of independent repair events analyzed (N) is noted in the first column. Numbers in each column are the number of wild-type or mutated sites, with percentages given in parentheses. For mutated sites, the number (percentages in parentheses) of deletions or additions is recorded, followed by the frequency of each addition event. The numbers (1 to 3) refer to the additions depicted in panel C. The top sequence in panel C is the wild-type I-SceI site. WT, wild type.
Molecular outcomes of I-SceI break repair in M. smegmatis.
To understand the mechanism of DSB repair at the I-SceI site, we analyzed the molecular outcomes of repair of I-SceI chromosomal breaks by determining the nucleotide sequence of the I-SceI site in surviving colonies with and without AHT exposure. These results are summarized in Fig. 5B. In all strains except for ΔrecA (see below), colonies from plates lacking tetracycline displayed intact I-SceI sites (data not shown). Of 41 repair events analyzed from wild-type cells that survived I-SceI induction, the majority (71%) had mutated I-SceI sites, with the predominant mechanism of mutation being deletions (88% deletions). A minority of repair events (12%) in wild-type cells were single-nucleotide additions to the I-SceI site. These single-nucleotide additions (pictured in Fig. 5C) most likely arose by nontemplated additions to the 3′ overhang of one of the I-SceI ends followed by ligation and gap filling.
In contrast, of 31 Δku survivors examined, all had wild-type I-SceI sites (Fig. 5B). To investigate whether these survivors were the result of efficient faithful repair or loss of I-SceI expression, we replated three of these Δku survivors on tetracycline and measured I-SceI-mediated killing. We found that these Δku survivors were not susceptible to I-SceI-mediated killing despite an intact I-SceI site. Western blotting confirmed loss of I-SceI enzyme expression (data not shown). These results demonstrate that the predominant mechanism of survival after constitutive I-SceI induction in the ku null strain is loss of I-SceI expression. In wild-type cells, site mutation is the common mechanism of survival.
I-SceI repair in the ΔrecA strain differed from wild-type and Δku cells in both timing and molecular outcome. In contrast to all other strains examined, the recA strain rapidly mutated the I-SceI site in the absence of tetracycline induction. Despite an intact I-SceI site at the beginning of growth, approximately eight generations of growth led to a 91% mutation rate of the I-SceI site (Fig. 5B). Fifty percent of these mutations were single-nucleotide insertions while 50% were deletions (Fig. 5B and C). Tetracycline induction led to 100% mutation of the site and strongly biased the repair events toward single-nucleotide insertions (78%; n = 32). This predominance of insertions contrasts with wild-type cells in which deletions are the predominant mechanism of repair. The most common repair event was a single A addition to the bottom strand (Fig. 5C, addition 1). Deletion of ku in the ΔrecA strain abolished mutation of the I-SceI site such that 100% (n = 24) of the surviving colonies had intact sites. These survivors had lost I-SceI expression (data not shown).
Analysis of deletion events at I-SceI breaks.
The data presented above indicate that mutation of the I-SceI site occurs only in NHEJ-proficient strains. In addition, data from the recA null strain indicate that, in the absence of HR, Ku-dependent NHEJ rapidly mutates the cut I-SceI site even without tetracycline induction. To better characterize this repair pathway, we analyzed the deletion events that occurred at repaired I-SceI sites. In both wild-type and ΔrecA strains, deletions universally used microhomology for repair (Fig. 6A, boxed nucleotides). We found no difference in the number of nucleotides used for microhomology between wild-type and ΔrecA strains (Fig. 6B). Deletions in wild-type cells tended to be longer than in ΔrecA cells (Fig. 6).
FIG. 6.
(A) Molecular characteristics of deletions in wild-type and recA-deficient strains. Nucleotides of the original I-SceI site are in blue, resected nucleotides are indicated by a line, and nucleotides of microhomology are boxed. To the right of the sequences is the number of events observed for each type of microhomology in wild-type and recA strains, with the number of deleted nucleotides in parentheses. (B) Statistical analysis of microhomology usage and deletion size in wild-type and recA-deficient strains. WT, wild type.
DISCUSSION
Bacterial NHEJ is a recently described DNA repair pathway, the characteristics and physiologic importance of which are only beginning to be understood. Prior characterization of mycobacterial NHEJ revealed a central role for M. tuberculosis and M. smegmatis Ku and LigD in plasmid recircularization (13, 14). Repair of linearized plasmids was mutagenic through the addition of templated and nontemplated nucleotides catalyzed by the polymerase module of LigD (40). However, the role of NHEJ in the defense against damage to the bacterial chromosome is not well characterized. Here, we report an analysis of DNA damage resistance in M. smegmatis using a comprehensive panel of HR- and NHEJ-deficient strains and a comprehensive panel of clastogens.
Clastogen resistance.
In eukarya, NHEJ-deficient cells are typically sensitive to IR, although the degree of this sensitization varies between organisms. In mammalian cells, NHEJ deficiency through loss of Ku80 confers mild radiosensitivity even when HR is active (7, 24). In budding yeast, ku or ligIV deficiency does not sensitize haploid cells to IR killing unless HR is disabled (3, 36, 37). Several recent papers have shown that ku-deficient Bacillus spores are sensitive to DNA damage including IR and dry heat (22, 38). Spores are metabolically inactive structures with a single chromosome and are therefore a logical physiologic state during which double-strand DNA break repair without a homologous template would be necessary. Mycobacteria are nonsporulating, but M. tuberculosis spends prolonged periods in the nonreplicating state of latency in the human host, during which DSB repair by NHEJ might be important. Our finding that NHEJ-deficient mycobacteria are sensitive to IR only in late stationary phase in minimal medium may indicate that NHEJ is important during nonreplicating states of starvation. Of critical importance in determining the role of NHEJ in repair is understanding how the chromosomal copy number of mycobacteria during different phases of the cell cycle affects the availability of homologous templates for homology-directed repair.
Our finding that the M. smegmatis recBCD null mutant is not sensitive to DNA damaging agents is surprising, given the central role of this enzyme complex in E. coli in the presynapsis stage of recombination (6). In E. coli, RecBCD generates a 3′ DNA end which is the substrate for RecA nucleofilament formation and strand invasion. The lack of phenotype of the M. smegmatis recBCD null mutant suggests that alternative pathways of presynapsis are active in M. smegmatis and can compensate for the loss of RecBCD activity. The prime candidate for this activity is the RecFOR system, which is present in mycobacteria but has not been examined genetically. In E. coli, RecFOR loads RecA onto gapped DNA (23) and is capable of fully compensating for recBCD when suppressor mutations at the sbc loci are present (18). However, in B. subtilis, which has an NHEJ system and AddAB in place of RecBCD, recF mutants are more sensitive to MMS killing than AddAB mutants (32), suggesting that the recFOR system has a more dominant role in presynapsis in this organism, at least for some forms of damage. In this context, the lack of obvious homologues of sbc genes in mycobacteria (30) provides a possible explanation for the lack of effect of the recBCD mutation on damage resistance. Further studies will be required to fully understand the relative contributions of recBCD and recFOR to recombination in mycobacteria and the relationship of these systems to NHEJ.
NHEJ in I-SceI resistance.
We have adapted the homing endonuclease I-SceI for use in mycobacteria as a chromosomal DSB-inducing agent. When I-SceI is expressed in M. smegmatis with a supplied I-SceI site, we observe efficient cutting of the mycobacterial chromosome. Wild-type M. smegmatis is susceptible to constitutive I-SceI chromosomal cleavage, displaying approximately 80% lethality. The predominant mode of survival for wild-type cells is through DNA deletion that encompasses the I-SceI site. As the I-SceI substrate used in this study does not contain a donor sequence for homologous recombination, it is likely that a DNA resection at the break site and recA nucleofilament formation cannot proceed to strand invasion if all chromosomal copies are cut, including those on sister chromosomes. Therefore, both the lethality and the surviving cells with deletions may be the result of an accumulation of toxic recombination intermediates, which in some cases can be rescued by Ku-dependent NHEJ. The results indicate that survival of mycobacterial cells following constitutive chromosomal cleavage requires mutation of the cut site, which is absolutely dependent on an intact NHEJ pathway. Surprisingly, Δku ΔligD cells were more resistant than either single mutant to I-SceI killing. The expectation was that the double mutant would phenocopy each single mutant due to loss of NHEJ activity. However, this finding is conceptually similar to the rescue of ligIV embryonic lethality by the addition of a ku86 mutation (17). As was proposed in mice, we hypothesize that the presence of either LigD or Ku without the other may inhibit the activity of another repair pathway whose activity can repair DSBs when both LigD and Ku are deleted. This effect could be due to blocking physical access to the DNA ends by LigD or Ku binding or to functional interactions between NHEJ components and alternative repair pathways.
The relative resistance of the recA null strain to I-SceI-mediated killing, although initially surprising, is the result of rapid inactivation of the I-SceI site through NHEJ-mediated modification. In contrast to all other strains examined, the recA null strain displayed rapid mutation of the I-SceI site in the absence of tetracycline. This may be due to low-level constitutive expression of I-SceI which, in wild-type cells, does not lead to mutagenesis as all I-SceI sites in wild-type cells are preserved before enzyme induction. However, this basal chromosomal cleavage rapidly leads to mutation when NHEJ is unopposed in the absence of HR, a phenotype that is suppressed by inactivation of ku. This dynamic interplay between HR and NHEJ is reminiscent of the more efficient homology-directed repair of I-SceI-induced DSBs in Ku70-deficient mammalian cells (26). More extensive analysis of I-SceI substrates with donor sequences for HR will be required to assess the relative contribution of HR and NHEJ in repair of a single transient DSB.
Analysis of repair events at the I-SceI site revealed that mutagenic repair of the site only occurs in strains in which NHEJ is active (wild type and ΔrecA). The molecular outcomes of I-SceI repair are dominated by single-nucleotide insertions when NHEJ is unopposed. These findings parallel our previously reported finding with plasmid substrates in which nucleotide insertion is a major mechanism of mutagenic NHEJ (13). Our prior work indicated that nucleotide insertion in plasmid NHEJ is directly catalyzed by the polymerase module of LigD (40), and we hypothesize that the NHEJ-dependent modifications at chromosomal I-SceI sites will be similarly catalyzed by the LigD polymerase domain.
In summary, we conclude that mycobacterial NHEJ is required for growth phase-specific defense against IR and plays an important role in the mutagenic repair of chromosomal double-strand DNA breaks induced by the homing endonuclease I-SceI.
Acknowledgments
This work was supported by NIH grant AI064693.
We thank Maria Jasin and David Weinstock for kindly sharing the I-SceI cDNA and Ruslana Bryk and Carl Nathan for sharing the anti DlaT antibody.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Adachi, N., H. Suzuki, S. Iiizumi, and H. Koyama. 2003. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 278:35897-35902. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 3.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bundock, P., H. van Attikum, and P. Hooykaas. 2002. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30:3395-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhury, A. M., and G. R. Smith. 1984. Escherichia coli recBC deletion mutants. J. Bacteriol. 160:788-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chedin, F., and S. C. Kowalczykowski. 2002. A novel family of regulated helicases/nucleases from gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43:823-834. [DOI] [PubMed] [Google Scholar]
- 7.Couedel, C., K. D. Mills, M. Barchi, L. Shen, A. Olshen, R. D. Johnson, A. Nussenzweig, J. Essers, R. Kanaar, G. C. Li, F. W. Alt, and M. Jasin. 2004. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 18:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darwin, K. H., and C. F. Nathan. 2005. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 73:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Della, M., P. L. Palmbos, H. M. Tseng, L. M. Tonkin, J. M. Daley, L. M. Topper, R. S. Pitcher, A. E. Tomkinson, T. E. Wilson, and A. J. Doherty. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306:683-685. [DOI] [PubMed] [Google Scholar]
- 10.Doherty, A. J., S. P. Jackson, and G. R. Weller. 2001. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 500:186-188. [DOI] [PubMed] [Google Scholar]
- 11.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frischkorn, K., P. Sander, M. Scholz, K. Teschner, T. Prammananan, and E. C. Bottger. 1998. Investigation of mycobacterial recA function: protein introns in the RecA of pathogenic mycobacteria do not affect competency for homologous recombination. Mol. Microbiol. 29:1203-1214. [DOI] [PubMed] [Google Scholar]
- 13.Gong, C., P. Bongiorno, A. Martins, N. C. Stephanou, H. Zhu, S. Shuman, and M. S. Glickman. 2005. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 12:304-312. [DOI] [PubMed] [Google Scholar]
- 14.Gong, C., A. Martins, P. Bongiorno, M. Glickman, and S. Shuman. 2004. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 279:20594-20606. [DOI] [PubMed] [Google Scholar]
- 15.Haber, J. E. 1995. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17:609-620. [DOI] [PubMed] [Google Scholar]
- 16.Hickson, I. D., and P. T. Emmerson. 1981. Identification of the Escherichia coli recB and recC gene products. Nature 294:578-580. [DOI] [PubMed] [Google Scholar]
- 17.Karanjawala, Z. E., N. Adachi, R. A. Irvine, E. K. Oh, D. Shibata, K. Schwarz, C. L. Hsieh, and M. R. Lieber. 2002. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst). 1:1017-1026. [DOI] [PubMed] [Google Scholar]
- 18.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milne, G. T., S. Jin, K. B. Shannon, and D. T. Weaver. 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizrahi, V., and S. J. Andersen. 1998. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 29:1331-1339. [DOI] [PubMed] [Google Scholar]
- 22.Moeller, R., E. Stackebrandt, G. Reitz, T. Berger, P. Rettberg, A. J. Doherty, G. Horneck, and W. L. Nicholson. 2007. Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 189:3306-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 24.Nussenzweig, A., K. Sokol, P. Burgman, L. Li, and G. C. Li. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. USA 94:13588-13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piddock, L. J., and R. N. Walters. 1992. Bactericidal activities of five quinolones for Escherichia coli strains with mutations in genes encoding the SOS response or cell division. Antimicrob. Agents Chemother. 36:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce, A. J., P. Hu, M. Han, N. Ellis, and M. Jasin. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitcher, R. S., L. M. Tonkin, J. M. Daley, P. L. Palmbos, A. J. Green, T. L. Velting, A. Brzostek, M. Korycka-Machala, S. Cresawn, J. Dziadek, G. F. Hatfull, T. E. Wilson, and A. J. Doherty. 2006. Mycobacteriophage exploit NHEJ to facilitate genome circularization. Mol. Cell 23:743-748. [DOI] [PubMed] [Google Scholar]
- 28.Pitcher, R. S., L. M. Tonkin, A. J. Green, and A. J. Doherty. 2005. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 351:531-544. [DOI] [PubMed] [Google Scholar]
- 29.Richardson, C., B. Elliott, and M. Jasin. 1999. Chromosomal double-strand breaks introduced in mammalian cells by expression of I-Sce I endonuclease. Methods Mol. Biol. 113:453-463. [DOI] [PubMed] [Google Scholar]
- 30.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLOS Genet. 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouet, P., F. Smih, and M. Jasin. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14:8096-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez, H., D. Kidane, P. Reed, F. A. Curtis, M. C. Cozar, P. L. Graumann, G. J. Sharples, and J. C. Alonso. 2005. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics 171:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sander, P., K. G. Papavinasasundaram, T. Dick, E. Stavropoulos, K. Ellrott, B. Springer, M. J. Colston, and E. C. Bottger. 2001. Mycobacterium bovis BCG recA deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. Infect. Immun. 69:3562-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siede, W., A. A. Friedl, I. Dianova, F. Eckardt-Schupp, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142:91-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 36.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo, S. H., and S. P. Jackson. 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 16:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 39.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. d'Adda di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 40.Zhu, H., J. Nandakumar, J. Aniukwu, L. K. Wang, M. S. Glickman, C. D. Lima, and S. Shuman. 2006. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl. Acad. Sci. USA 103:1711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]