Abstract
Penicillin-binding protein 2 (PBP 2) has long been known to be essential for rod-shaped morphology in gram-negative bacteria, including Escherichia coli and Pseudomonas aeruginosa. In the course of earlier studies with P. aeruginosa PBP 2, we observed that E. coli was sensitive to the overexpression of its gene, pbpA. In this study, we examined E. coli overproducing both P. aeruginosa and E. coli PBP 2. Growth of cells entered a stationary phase soon after induction of gene expression, and cells began to lyse upon prolonged incubation. Concomitant with the growth retardation, cells were observed to have changed morphologically from typical rods into enlarged spheres. Inactive derivatives of the PBP 2s were engineered, involving site-specific replacement of their catalytic Ser residues with Ala in their transpeptidase module. Overproduction of these inactive PBPs resulted in identical effects. Likewise, overproduction of PBP 2 derivatives possessing only their N-terminal non-penicillin-binding module (i.e., lacking their C-terminal transpeptidase module) produced similar effects. However, E. coli overproducing engineered derivatives of PBP 2 lacking their noncleavable, N-terminal signal sequence and membrane anchor were found to grow and divide at the same rate as control cells. The morphological effects and lysis were also eliminated entirely when overproduction of PBP 2 and variants was conducted with E. coli MHD79, a strain lacking six lytic transglycosylases. A possible interaction between the N-terminal domain of PBP 2 and lytic transglycosylases in vivo through the formation of multienzyme complexes is discussed.
Penicillin-binding proteins (PBPs) are a class of enzymes that catalyze the final stages of peptidoglycan biosynthesis within the periplasm and are essential for the growth and division of bacterial cells (recently reviewed in references 3, 26, 34, and 47). All bacteria so far characterized produce a number of PBPs for peptidoglycan metabolism (i.e., synthesis and turnover). Owing to the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) technique that was developed for their discovery and characterization (36), they are broadly classified according to their molecular size as either high-molecular-weight (HMW) or low-molecular-weight (LMW) PBPs (12). Thus, Escherichia coli produces 13 known PBPs: five HMW (PBPs 1a, 1b, 1c, 2, and 3) and eight LMW PBPs (PBPs 4, 5, 6, 7, and 8, DacD, AmpC, and AmpH). Less is known about Pseudomonas aeruginosa, but different studies have accounted for the production of eight PBPs by this opportunistic pathogen. Based on sequence alignment similarities, these are proposed to be homologs of E. coli PBPs 1a, 1b, 2, 3, 4, 5, and 7 (25, 32, 35).
PBPs have been demonstrated to associate with the cytoplasmic membrane as either integral or peripheral membrane proteins, and as their name implies, all possess a penicillin-binding (PB) module which extends into the periplasm (see Fig. 1). The PB module functions to catalyze either the transpeptidation (with HMW PBPs) or hydrolysis (with LMW PBPs) of the stem peptides associated with the muramoyl residues of the newly incorporated peptidoglycan subunits. The HMW PBPs also possess an N-terminal, non-penicillin-binding (n-PB) module which serves to anchor the proteins to the cytoplasmic membrane via a noncleavable signal-sequence peptide. This n-PB module of the class A HMW PBPs 1a, 1b, and 1c also catalyzes the transglycosylation of bactoprenol-linked peptidoglycan precursors into the growing sacculus, thus making them both multimodular and bifunctional. The n-PB module of the class B HMW PBPs 2 and 3 does not catalyze transglycosylase activity, but it has been demonstrated to play a critical role in directing the proper folding of the C-terminal PB domain, at least for E. coli PBP 3 (11, 31). Although not well established, it is believed that the n-PB module of E. coli PBP 3 is also involved in protein-protein interactions (27, 33, 45). Nothing was known previously about the n-PB module of PBP 2.
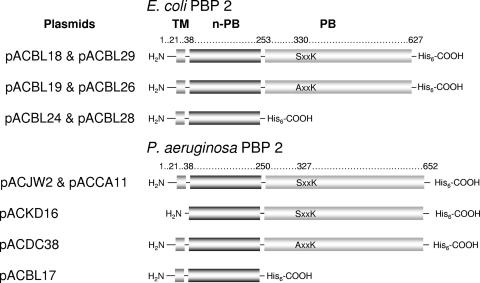
FIG. 1.
Modular structures of E. coli and P. aeruginosa PBP 2 constructs. Each is composed of an N-terminal transmembrane anchor (TM), followed by an n-PB module and then a C-terminal PB module. The position of the consensus SxxK sequence, corresponding to the catalytic Ser of the PB module, is identified. The names of plasmids encoding the wild-type and mutant derivatives of the PBPs are listed at left.
Inhibition (reviewed in reference 30) and in vitro deletion studies targeting the gene encoding PBP 2 in E. coli (29, 39, 40) and P. aeruginosa (23) have shown that this PBP functions in the elongation and maintenance of the rod shape of these bacteria. For this to occur, however, new sites within the cylindrical regions of the sacculus are required for integration of new peptidoglycan precursors. These sites are thought to be provided by the action of lytic transglycosylases, endogenous bacterial enzymes that cleave peptidoglycan during the process of biosynthesis and turnover (18). To prevent their uncontrolled activity resulting in autolysis, these lytic enzymes have been proposed to form complexes in vivo with PBPs (17). In vitro experiments involving affinity chromatography have demonstrated an association between E. coli PBP 2 and both membrane-bound lytic transglycosylase A (MltA) and soluble lytic transglyosylase 70 (Slt70) (42, 43). Our preliminary examination of the associations of PBPs with lytic transglycosylases in P. aeruginosa have indicated that its PBP 2 forms a complex with the lytic transglycosylase SltB1 (22).
In the course of our studies with P. aeruginosa PBP 2, we observed that E. coli was sensitive to the overexpression of its gene, pbpA (23). It has also been reported elsewhere that insertion of the E. coli pbpA gene in high-copy umber plasmids resulted in deleterious effects on bacterial growth and that the plasmids could not be stably maintained (7, 37). However, no details of these effects were reported in the latter studies, and so this prompted us to investigate further. Herein we describe the effect of overproduction of both E. coli and P. aeruginosa PBP 2 on the growth and morphology of E. coli.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
All plasmids and bacterial strains used in this study are listed in Table 1. Cultures of E. coli were routinely grown in Luria-Bertani (LB) broth (1% tryptone [Difco, Detroit, MI], 0.5% yeast extract [Difco], 0.5% NaCl) at 37°C with agitation. For overexpression of cloned genes, cultures were grown in rich medium containing 3.2% tryptone, 2% yeast extract, and 1% NaCl. Strains harboring resistance determinants were grown in the presence of the appropriate antibiotic(s): ampicillin (Amp) (100 μg/ml), kanamycin (Kan) (50 μg/ml), and/or chloramphenicol (Cm) (34 μg/ml). Unless otherwise stated, all reagents and chemicals were obtained from Sigma-Aldrich Canada Ltd. (Oakville, ON).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic | Reference or source |
|---|---|---|
| Strains | ||
| E. coli BL21[λDE] CodonPlus | F−ompT hsdSB (rB− mB−) dcm met gal(λDE3)endA Hte[argU ileY leuW Tetr] | Stratagene |
| E. coli DH5α | K-12 φ80d lacZΔM15 endAI hsdR17 (rK− mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF+)U169 F− | Invitrogen |
| E. coli W3110 | F−trpA23 xyl glySL | 16 |
| E. coli MHD79 | sltY mltA mltB mltC mltD emtA (Cmr Kanr Tetr) | 15 |
| E. coli MC1061 | araD139 Δ(ara-leu)7696 Δ(lac)174 galU galK hsdR2(rK− mK−) mcrB1 rpsL (Strr) | 4 |
| Pseudomonas aeruginosa PAO1 | Serotype O5; A+ B+ | 13 |
| Plasmids | ||
| pET30a(+) | IPTG-inducible expression vector; Kanr | Novagen |
| pET32-EK/LIC | IPTG-inducible expression vector; Ampr | |
| pET30a(+)-Ampr | pET30a(+) derivative containing bla on a DraIII/AlwNI fragment; Ampr | This study |
| pACJW2 | pET30a(+) derivative containing P. aeruginosa pbpA on an NdeI/HindIII fragment; Kanr | 23 |
| pACBL17 | pACJW2 derivative encoding the N-terminal n-PB module of P. aeruginosa PBP 2 of Met1to Ser250; Kanr | This study |
| pACBL18 | pET30a(+) derivative containing E. coli pbpA on an NdeI/HindIII fragment; Kanr | This study |
| pACBL19 | pACBL18 derivative encoding site-specific replacement of Ser330→Ala on E. coli pbpA; Kanr | This study |
| pACBL24 | pACBL18 derivative encoding the N-terminal n-PB module of E. coli PBP 2 of Met1 to Thr252; Kanr | This study |
| pACBL26 | pET30a(+)-Ampr derivative containing mutant E. coli pbpA on an NdeI/HindIII fragment encoding site-specific replacement of Ser330→Ala; Ampr | This study |
| pACBL28 | pET30a(+)-Ampr derivative containing mutant E. coli pbpA on an NdeI/HindIII fragment encoding N-terminal n-PB module of Met1 to Ser253; Ampr | This study |
| pACBL29 | pET30a(+)-Ampr derivative containing E. coli pbpA on an NdeI/HindIII fragment; Ampr | This study |
| pACCA11 | pET30a(+)-Ampr derivative containing P. aeruginosa pbpA on an NdeI/HindIII fragment; Ampr | This study |
| pACDC38 | pACJW2 derivative encoding site-specific replacement of Ser327→Ala on P. aeruginosa pbpA; Kanr | 23 |
| pACKD16 | pET30a(+) derivative containing pbpA on an NdeI/HindIII fragment encoding an N-terminal truncation of PBP 2 of Pro2 to Arg38; Kanr | 23 |
| pACSR1 | pET30a(+) derivative containing P. aeruginosa ftsI on an NdeI/XhoI fragment encoding PBP 3; Kanr | This study |
| pUCH6-wzz1 | wzz1 cloned via XbaI-PstI from pETH6-wzz1 into pUCP27 (N-terminal His6 tag) | 6 |
Construction of expression plasmids.
Unless otherwise stated, all PCRs used to amplify DNA were performed using the polymerase Expand Long from Roche Molecular Biochemicals (Laval, Quebec, Canada), following the manufacturer's instructions. Chromosomal DNA from P. aeruginosa PAO1 and E. coli W3110 was prepared as a template from overnight cultures, using DNAzol (Invitrogen Canada Inc., Burlington, ON), according to the manufacturer's protocol. Restriction enzymes and T4 DNA ligase for cloning were obtained from New England Biolabs (Pickering, Ontario, Canada). Successfully cloned constructs were verified by sequencing over the entire nucleotide sequence of the genes.
The oligonucleotide primers used in the amplification of DNA and construction of vectors pBLAC17, pBLAC18, and pBLAC24 are presented in Table 2. After amplification using the appropriate genomic DNA as template, PCR products were cloned into the expression vector pET30a(+) via the NdeI and HindIII restriction sites. These constructs were designed to generate each protein as a fusion with a C-terminal hexa-His tag.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | 5′-to-3′ sequence (restriction site) | Description | Construct |
|---|---|---|---|
| PBP2-NTerm1 | CTTTAAGAAGGAGATATACATATGCCGCAG (NdeI) | Amplification of n-PB of P. aeruginosa PBP 2 | pACBL17 |
| PBP2-NTerm2 | CCCGAAGCTTGCTCAGGACGATGTC (HindIII) | Amplification of n-PB of P. aeruginosa PBP 2 | pACBL17 |
| EcPBP2-5 | CCCGAAAACGCAGCATATGAAACTACAGAACTC (NdeI) | Amplification of E. coli pbpA | pACBL18 |
| EcPBP2-2 | CCGTCAAAGCTTATGGTCCTCCGCTG (HindIII) | Amplification of E. coli pbpA | pACBL18 |
| EcPBP2-SA-SDM1 | TCCTCCCGCGGCTACAGTTAAAC | SDM of Ser→330Ala on E. coli pbpA | pACBL19 |
| EcPBP2-SA-SDM2 | GTTTAACTGTAGCCGCGGGAGGA | SDM of Ser→330Ala on E. coli pbpA | pACBL19 |
| EcPBP2-1 | GAAAACGCAGCATATGAAACTACAGAACTC (NdeI) | Amplification of n-PB of E. coli PBP 2 | pACBL24 |
| EcPBP2nPB-1 | GAGTTTGAGAAGCTTCGTCAGGTAAATATC (HindIII) | Amplification of n-PB of E. coli PBP 2 | pACBL24 |
| PaPBP3-1 | GGTGGCCATATGAAACTGAATTATTTCCAGG (NdeI) | Amplification of P. aeruginosa ftsI (pbpB) | pACSR1 |
| PaPBP3-2 | GGCCCTCGAGGCCACGCCCTCC (XhoI) | Amplification of P. aeruginosa ftsI (pbpB) | pACSR1 |
Site-specific replacement of Ser330 to an Ala residue in E. coli PBP2 was performed using the QuickChange site-Directed mutagenesis kit (Stratagene, La Jolla, CA), following the manufacturer's recommended method. pBLAC18 was used as the template for the PCR with the mutagenic primers EcPBP2-SA-SDM1 and EcPBP2-SA-SDM2 (Table 2). The resulting construct was named pBLAC19.
For transformations of E. coli MHD79, which possess an Kan resistance marker, a variant of the pET30a(+) vector from Novagen had to be engineered to replace its Kan resistance marker with Amp. The Kan resistance cassette from pET30a(+) was removed by restriction endonuclease digestion using DraIII and AlwNI, and the bla gene from pET32-Ek/LIC was excised using the same restriction enzymes. This bla gene was then ligated into the digested pET30a(+), resulting in pET30a(+)-Ampr. pET30a(+)-Ampr was used as a parent vector to harbor the E. coli and P. aeruginosa pbpA variants. Each of the desired pbpA genes was excised from its respective pET30a(+) construct using NdeI and HindII. The recovered genes were then ligated into pET30a(+)-Ampr, which had been predigested with the same restriction enzymes, generating plasmids pACCA11, pBLAC26, pBLAC28, and pBLAC29.
Isolated genomic DNA from P. aeruginosa PAO1 was also used to PCR amplify ftsI, encoding full-length PBP 3 (24). The oligonucleotide primers used are presented in Table 2, and NdeI/XhoI restriction sites were used to facilitate the cloning into the pET30a(+) vector to provide pACSR1, designed to generate PBP 3 with a C-terminal hexa-His tag.
Transformations and protein overproduction.
E. coli BL21(λDE3) CodonPlus (pLysS) was used as the host for routine overproduction studies of the E. coli and P. aeruginosa PBP 2s and their derivatives. Freshly transformed cells were grown overnight in LB or enriched medium containing the appropriate antibiotics at 37°C with aeration (200 rpm). Overexpression of the cloned genes was induced with the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Roche).
For protein production in E. coli MHD79 and MC1061, it was necessary to integrate the λDE3 prophage into the chromosomes of these strains such that the lysogenized host could be used to express genes cloned into the pET vectors under the control of the inducible T7 promoter. The λDE3 lysogenization kit (Novagen) was used for this site-specific integration according to the manufacturer's instructions, and λDE3 lysogens were verified using the Tester Phage.
PBP analyses.
Detection of expressed PBP 2s and their derivatives took advantage of the fusion of the C-terminal His6 tag and a commercially available mouse anti-His antibody (“His Probe H-3”; Santa Cruz Biotechnology, Santa Cruz, CA). Western immunoblotting was performed as previously described (23). The PBPs were also analyzed for penicillin-binding activity by the SDS-PAGE-based assay (36) using fluorescent BOCILLIN FL (Molecular Probes Inc., Eugene, OR) and biotinylated ampicillin as previously described (23).
Growth curves and viable counts.
The growth characteristics of E. coli BL21 transformed with the different plasmids were determined by incubating a 1/50 dilution of overnight cultures in 50 ml LB containing the appropriate antibiotic at 37°C with shaking (200 rpm). Growth of the cultures, in triplicate, was monitored by determining both the optical density at 600 nm and viable counts for at least 6 h. For viable counts, samples of the growing cultures were serially diluted in sterile saline (10−2, 10−3, 10−4, 10−6, and 10−8), and 100-μl aliquots were plated on LB plates containing 5% agar (Difco). After 16 h of incubation at 37°C, the colonies were counted and viable counts determined.
Preparation of cells for microscopy.
Samples of cells were recovered from cultures at different stages of their growth and heat fixed to a glass slide. Cells were then stained with safranine for 15 s, rinsed with water, and then observed by phase-contrast microscopy using Nomarski optics on a Leica microscope with differential interference contrast.
For scanning electron microscopy (SEM), cell pellets were washed and resuspended in 0.07 M Sorensen's phosphate buffer, pH 6.8, placed on a 13-mm carbon planchette (Canemoc & Marivac, Lakefield, Quebec, Canada), and fixed with 2% glutaraldehyde in the same buffer for 1 h. The samples were then rinsed in several changes of buffer, dehydrated through a series of ethanol washes, critical point dried using carbon dioxide, and sputter coated with 20 nm of gold/palladium in a Hummer VII sputter coater (Anatech Corp., Alexandria, VA). To visualize the bacteria, filters were scanned using a Hitachi S-570 SEM (Tokyo, Japan) and images were collected directly from the SEM using Quartz PCI software (Quartz Imaging Corp., Vancouver, Canada).
Analysis of peptidoglycan cross-linking.
Cells for analysis were harvested by centrifugation at 8,000 × g for 15 min and washed twice in 10 mM sodium phosphate (pH 7.0). Insoluble peptidoglycan was extracted from whole cells using the boiling SDS procedure and purified as previously described (5). Analysis of peptidoglycan cross-linking associated with the different E. coli transformants was performed using the high-performance liquid chromatography-based method of Glauner (10) following its digestion with 20 U/ml mutanolysin at 37°C for 16 h. Any residual insoluble material was removed by centrifugation (12,000 × g, 10 min, room temperature), and the soluble muropeptides in the supernatants were reduced with sodium borohydride in 0.5 M sodium borate buffer, pH 9, before being applied to a 4.6-by-250-mm Gemini C18 (5 μ) analytical column (Phenomenex Inc., Torrance, CA). Detection of eluting muropeptides was achieved by monitoring the A205 value.
Other analytical techniques.
Agarose gel electrophoresis was performed using 1% agarose (OmniPur, Darmstadt, Germany), while SDS-PAGE (21) was performed using 12.5% (wt/vol) acrylamide (Bio-Rad Laboratories [Canada] Ltd., Mississauga, Ontario, Canada). DNA sequencing was performed by the Guelph Molecular Supercenter, University of Guelph. Protein concentrations were determined using a commercial bicinchoninic acid assay (Pierce, Rockford, IL) with bovine serum albumin (0.05 to 2 mg/ml) as the standard.
RESULTS AND DISCUSSION
Overproduction of PBP 3 and of PBP 2 and its derivatives.
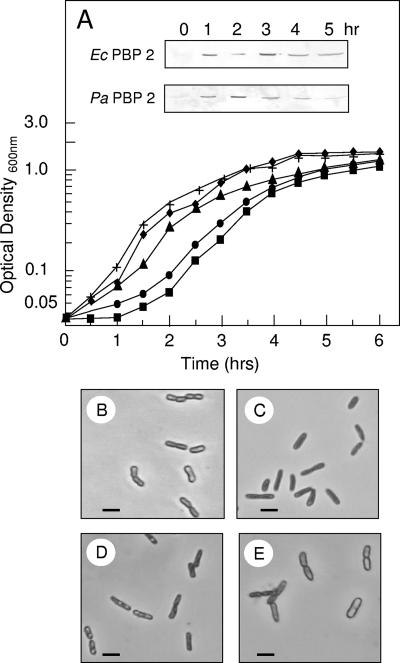
Cells of E. coli BL21(λDE3) CodonPlus harboring pACBL18 and pACJW2 were grown in LB medium at 37°C and induced with 1 mM IPTG for the overproduction of wild-type E. coli PBP 2-His6 and P. aeruginosa PBP 2-His6, respectively (Fig. 1). This overproduction was confirmed by SDS-PAGE and Western blot analysis using a monoclonal anti-His-tag antibody (Fig. 2). The level of overproduction continued to increase with time for 4 h after induction before beginning to level off. Approximately 0.06 mg of purified PBP 2 could be obtained from 1 liter of these cultures (data not shown). Their functional activity was confirmed by PBP assays using both BOCILLIN FL and biotin-ampicillin (data not shown).
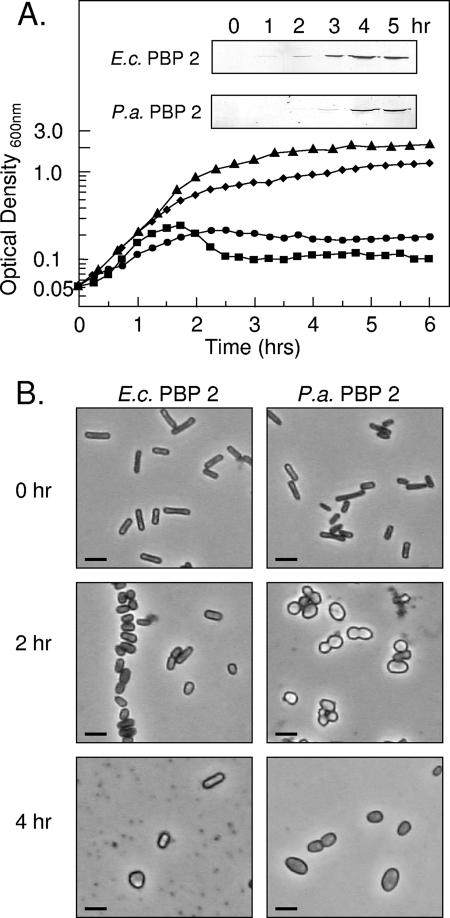
FIG. 2.
Effect of overproduction of E. coli (E.c.) or P. aeruginosa (P.a.) PBP 2 on growth and morphology of E. coli BL21. (A) Growth curves of E. coli BL21 incubated in LB at 37°C following IPTG induction of (▴) no plasmid, WT control; (⧫) pET30a(+) with no insert; (•) pACBL18 encoding wild-type E.c. PBP 2; or (▪) pACJW2 encoding wild-type P.a. PBP 2. Inserts: Associated Western immunoblots assaying overproduction of respective PBPs with time. (B) Phase-contrast micrographs of representative cells sampled at the times indicated from the respective cultures overproducing the two wild-type PBPs. Cells were prepared for microscopy as described in Materials and Methods. Bar, 2 μm.
To facilitate the production of P. aeruginosa PBP 2 in sufficient quantities for purification and subsequent characterization, we previously engineered a soluble derivative that lacks its 38-amino-acid N-terminal membrane anchor (Fig. 1). This truncated derivative of PBP 2, encoded on pACKD16, is overproduced by E. coli BL21 in significantly greater quantities than the wild-type form, and yields of approximately 1 mg of purified protein can be obtained per liter of culture (23). Like full-length PBP 2, this truncated derivative still retained its ability to bind penicillins.
It was shown previously that replacement of Ser330 with Ala results in the inactivation of the transpeptidase module of E. coli PBP 2 (38). Likewise, replacement of the homologous Ser327 residue in P. aeruginosa PBP 2 precluded penicillin-binding capacity, thereby identifying this Ser residue as the catalytic nucleophile of the PB module (23). Analogous to the situation with wild-type cells, IPTG induction of E. coli BL21 harboring plasmids pACBL19 and pACDC38 led to the overproduction of E. coli Ser330→Ala PBP 2 and P. aeruginosa Ser327→Ala PBP 2, respectively, in modest amounts (Fig. 3A) compared to the N-terminal truncated, soluble forms of the proteins (23) as detected by Western immunoblotting analysis. Both PBP 2 derivatives were confirmed to lack the ability to bind penicillins covalently (data not shown).
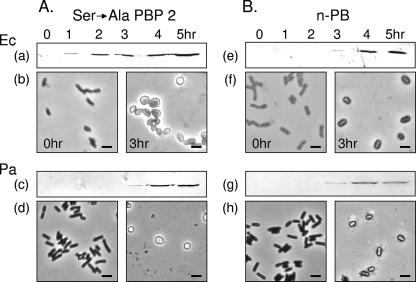
FIG. 3.
Effect of overproduction of inactive derivatives of E. coli (Ec) (top) or P. aeruginosa (Pa) (bottom) PBP 2 on cellular morphology of E. coli BL21. Cells harboring the respective plasmids for the overproduction of (A) the catalytic Ser→Ala mutant PBPs or (B) the isolated n-PB modules were induced with 1 mM IPTG and grown in LB at 37°C for the times indicated. (a, e, c, and g) Western immunoblot analysis of PBP 2 production during the 5-h incubation of induced cells. (b, d, f, and h) Phase-contrast micrographs of representative cells sampled at 0 and 3 h post-IPTG induction.
P. aeruginosa PBP 3 shares 29% identity (45% similarity) and 25% identity (44% similarity) with P. aeruginosa and E. coli PBP 2, respectively. It is predicted to have the same topology as PBP 2 with an N-terminal membrane anchor linked to n-PB and PB periplasmic modules via a noncleavable signal sequence. The ftsI (also named pbpB) gene encoding P. aeruginosa PBP 3 (24) was cloned and overexpressed in E. coli BL21. PBP assays confirmed the protein product exhibited PBP activity (data not shown).
Effect of overproduction of PBP 2 and its derivatives on growth.
Growth of E. coli BL21 overproducing the different variants of PBP 2 in LB broth at 37°C was monitored turbidometrically. When E. coli cells harboring pACBL18 were induced with IPTG to overproduce wild-type E. coli PBP 2, growth continued for about 2 h before reaching an apparent stationary phase (Fig. 2). Viable counts confirmed the cessation of growth and furthermore indicated the cells entered a death phase (data not shown). In contrast, control cells of E. coli BL21 alone or those harboring the empty pET30a(+) vector continued to grow after addition of IPTG for 4 h before achieving a maximal density of greater than 1.2 optical density units. The effect of wild-type P. aeruginosa PBP 2 overproduction in E. coli was more dramatic, as growth ceased 1.5 h postinduction, followed by an apparent death phase, which also was confirmed by viable counts.
The detrimental effect of PBP 2 overproduction was found to be dependent upon the phase of growth at which cultures were induced. Induction of E. coli BL21 harboring pACKD16 (P. aeruginosa PBP 2) at early exponential phase had the greatest effect on cell growth. In contrast, addition of IPTG at mid-exponential phase or later had significantly less effect. Hence, all subsequent experimentation involved IPTG induction of cell cultures during their early exponential phase of growth.
To investigate the possibility that the detrimental effect of PBP 2 overproduction was caused by the presence of increased transpeptidase activity associated with the PB module of the protein, peptidoglycan from these cells was isolated and analyzed for cross-linking by the method of Glauner (10). These analyses gave 20.7% cross-linking for transformants overproducing P. aeruginosa PBP 2, which was virtually the same as the 19.4% value obtained for wild-type E. coli BL21. That growth cessation and cell death were not the result of any extraneous transpeptidase activity was confirmed by examining the growth of E. coli harboring pACBL19 or pACDC38 overproducing inactive E. coli Ser330→Ala or P. aeruginosa Ser327→Ala PBP 2, respectively. Indeed, overproduction of the catalytically inactive derivatives of the two PBP 2s from the respective plasmids also inhibited growth in a manner identical to that observed with the wild-type enzymes. In contrast, cells overproducing the N-terminal truncated, soluble derivative of P. aeruginosa PBP 2, which is retained in their cytoplasm (23), remained viable and grew at the same rate as control E. coli cells. Thus, the death and lysis effect of PBP 2 appeared to correlate with the overproduction of the membrane-associated forms of the PBPs.
This lethal effect was found to be specific to the overproduction of PBP 2 based on growth studies with control E. coli BL21 harboring pUCH6-Wzz1 coding for P. aeruginosa Wzz1, a nonrelated membrane-anchored protein associated with lipopolysaccharide biosynthesis (6). As with the soluble form of PBP 2, these cells overproducing Wzz1 grew at the same rate and to the same density as the control cells (data not shown). Hence, these data suggested that the inhibitory and eventually lethal effect of overproducing the membrane-associated form of PBP 2 was not simply a consequence of saturating the cytoplasmic membrane of E. coli BL21 with too much protein but apparently was specific to its unique property(ies).
Effect of overproduction of PBP 2 on E. coli morphology.
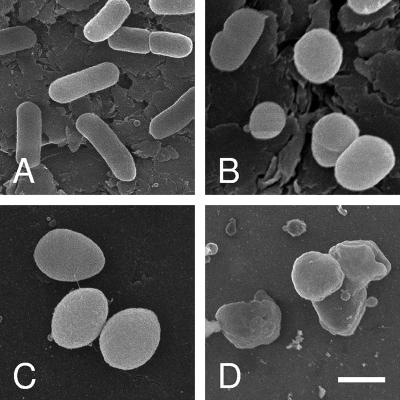
E. coli BL21 bacteria overproducing the different forms of PBP 2 were harvested at different time points during their culture and examined by both phase-contrast microscopy and SEM. Overproduction of wild-type E. coli PBP 2 caused morphological changes after 2 h of induction with IPTG, the same time that growth appeared to slow down and then cease (Fig. 2B). Initially, this change involved transformation to stubby or shortened rods, which became enlarged spheres by 4 h of incubation. Based on SEM analysis (Fig. 4), the average cell volumes were estimated to increase two- to threefold, from approximately 1.5 μm3 to 3.8 to 4.5 μm3. At this time, evidence of cell lysis was apparent, which continued with prolonged incubation. E. coli overproducing wild-type P. aeruginosa PBP 2 followed a similar pattern of morphological changes but, as expected from its growth pattern described above, at a faster pace. In this case, transformation to stubby rods occurred within 45 min to 1 h postinduction, with enlarged spheres forming by 2 to 2.5 h. Lysis of these cells typically began by 2 h of incubation and continued rapidly over the next 2 h, which was clearly observed by SEM (Fig. 4). In contrast, controls, E. coli BL21 harboring pET30a(+), remained typical rod-shaped cells throughout the induction time course.
FIG. 4.
Effect of PBP 2 overproduction on cellular morphology of E. coli BL21. Cells harboring (A) pET30a(+) with no insert (control), (B) pACBL8 (E. coli PBP 2), or (C and D) pACJW2 (P. aeruginosa PBP 2) were induced with IPTG and cultured in LB. Following 3 h (B and C) or 5 h (A and D) of incubation at 37°C, cells were harvested and prepared for SEM as described in Materials and Methods. Bar, 1 μm.
Cells overproducing the engineered inactive forms of PBP 2 also became spherical, and like the pattern observed with wild-type PBP 2, this transformation process followed their pattern of growth, with the first evidence of morphological change coinciding with the onset of the premature stationary phase (Fig. 3A). Again, evidence of cell lysis was clearly observed upon prolonged incubation, as the cells entered their apparent death phase. As predicted from its growth pattern, E. coli overproducing the soluble, cytoplasm-bound form of PBP 2 did not undergo any morphological change but instead remained as short rods throughout the course of incubation. Likewise, there was no evidence of morphological changes associated with the overproduction of Wzz1 or PBP 3 in E. coli, thus confirming previous observations made by others (24). Taken together, these findings indicate another unique consequence of overproducing PBP 2 in E. coli, that of morphological changes presumably resulting from altered peptidoglycan metabolism.
Effect of overproduction of PBP 2 N-terminal module on E. coli.
The results described above indicated that the morphological changes and cell lysis caused by the overproduction of PBP 2 did not require associated transpeptidase activity. This suggested that the entire PB module may not be required and that the property of the protein responsible for the observed effects may be confined to its n-PB module. Little was previously known about the n-PB module of PBP 2, but it is generally believed that this module of the class B HMW PBPs (viz PBPs 2 and 3 of E. coli and P. aeruginosa) directs the proper folding and stability of the entire protein. Studies with E. coli PBP 3 indicated that the n-PB module involving Gly57 to Glu258 functions as a noncleaved, pseudointramolecular chaperone, while the Met1-to-Glu56 amino-terminal module serves as a noncleaved signal peptide which spans from the cytosol through the cytoplasmic membrane to the periplasm (11).
To investigate the role, if any, of the n-PB module in conferring the morphological changes described above, constructs were engineered coding for the first 253 or 250 amino acid residues of the E. coli or P. aeruginosa PBP 2, respectively, fused to C-terminal hexa-His tags. E. coli BL21 was transformed with the respective plasmids pACBL24 and pACBL17, and expression studies confirmed the overproduction of these truncated PBP 2s upon IPTG induction (Fig. 3B). The observed Mr of the modules was approximately 30,000, which is close to the expected values of 30,500 and 30,300, respectively. Penicillin-binding assays were not performed with these proteins, but a preliminary zymogram assay using insoluble peptidoglycan as a substrate revealed that while lacking lytic activity, both the full-length P. aeruginosa PBP 2 and its isolated n-PB domain bind to peptidoglycan (data not shown). Previous studies have shown that proteins binding to peptidoglycan without catalyzing its lysis can be observed as “false positives” in a zymogram when peptidoglycan is used as a substrate, appearing as faint bands in the SDS-PAGE gel counterstained with methylene blue (2).
Cultures of E. coli BL21 transformed with pACBL17 and pACBL24 and induced with IPTG were grown to late exponential phase before cells were harvested for microscopic examination. Overproduction of either of the n-PB modules, as with the full-length PBP 2s, indeed led to a morphological change in E. coli, as the cells became a mixture of shortened rods and enlarged spheres (Fig. 3B).
Overproduction of PBP 2 and derivatives in multiple lytic transglycosylase mutant strain.
We investigated the possibility that the morphological changes and/or autolysis accompanying the overproduction of membrane-bound PBP 2 and its inactive derivatives was an indirect effect caused by uncontrolled lytic transglycosylase activity. Thus, the PBP 2 genes were overexpressed in E. coli MHD79, a mutant strain lacking six lytic transglycosylases (MltA, MltB, MltC, MltD, Slt70, and EmtA) (15). Because the MHD79 strain carries a number of resistant markers, including Kan, the Kan resistance cassette of the pET30a(+) plasmids harboring the two respective PBP 2s had to be replaced. This was accomplished by transferring the Amp resistance cassette from pET32EK/LIC to provide pACBL29 and pACCA11, encoding the genes for E. coli and P. aeruginosa PBP 2, respectively.
Transformation of E. coli MHD79 with these respective plasmids led to the overproduction of PBP 2, which could be readily detected by Western blot analysis using the anti-His-tag antibody (Fig. 5A). The growth rate of these cells was only marginally slower than that of control cells possessing the modified pET30a(+) plasmid without an insert, which was likely due to protein overproduction, and lysis was abolished. Consistent with this minimal difference in growth, no change in cellular morphology was observed with the overproduction of the PBPs in this background, as cells continued to grow normally as short rods (Fig. 5B). Given that PBPs are sensitive to β-lactams, it was necessary to establish that this lack of morphological and lysis effects was not due to direct “inhibition” of PBP 2 by the Amp used to maintain the pACBL29 and pACCA11 vectors in E. coli MHD79. These plasmids were thus used to transform E. coli BL21 and were induced for overexpression. The same effects of morphology change and lysis were observed as when using the Kan resistance marker of the original pACBL18 and pACJW2 vectors (data not shown). Finally, the parent strain of MHD79 was not BL21 but instead MC1061. Hence, to control for any differences in genetic backgrounds, we performed an additional control experiment in which E. coli MC1061 was transformed with plasmids pACBL29 and pACCA1. Overproduction of the E. coli and P. aeruginosa PBP 2s in this background produced the same effects as that observed with E. coli BL21 (data not shown). These data thus suggest that the morphological changes and lysis observed in wild-type cells overproducing PBP 2 are not caused directly by the presence of this protein but instead are caused by the activity of one or more lytic transglycosylases that may associate with it.
FIG. 5.
Effect of overproduction of E. coli (Ec) or P. aeruginosa (Pa) PBP 2 on growth and morphology of E. coli mutant strain MHD79 lacking six lytic transglycosylases. (A) Growth curves of cells grown in LB at 37°C following addition of 1 mM IPTG. +, E. coli MHD79 (control); the DE3 lysogenized MHD79 strain harboring (▴) no plasmid (control), (⧫) pET30a(+) with no insert (control), (•) pACBL29 encoding wild-type E. coli PBP 2, or (▪) pACCA11 encoding WT P. aeruginosa PBP 2 is shown. Inserts, associated Western immunoblots assaying time course of overproduction of respective PBPs from pACBL29 and pACCA11. (B) Phase-contrast micrographs of representative cells from 3-h cultures of E. coli MHD79 harboring (B) no plasmid, (C) pET30a(+) with no insert, (D) pACBL29 (E. coli PBP 2), or (E) pACCA11 (P. aeruginosa PBP 2). Bar, 2 μm.
Conclusions.
The current model for the growth of the peptidoglycan sacculus invokes two opposing enzyme systems of lytic and synthetic activity. This enzymatic balance model (44) thus involves a balance between the creation of nicks in the peptidoglycan structure and insertion of new material at these sites. There is now experimental evidence, albeit limited, that the lytic transglycosylases and PBPs that catalyze these activities combine into biosynthetic complexes (reviewed in references 17 and 34). The following summarizes what is currently known regarding E. coli PBP 2 specifically: (i) it is normally produced in limited quantities compared to the other peptidoglycan biosynthetic enzymes, as low as 58 molecules per cell depending upon growth conditions (9); (ii) it functions at both the cylindrical regions and septation sites of the cell, but it is responsible for extending the length of the rod-shaped cell (7); (iii) it has to associate with wild-type RodA to function appropriately (8, 19); (iv) together with RodA, it is guided and positioned at the cylindrical cell wall by the actin-like MreB cables that underlie the cytoplasmic membrane (20); and (v) preliminary evidence indicates it also associates with MltA and Slt70 (42, 43). Little is known at the molecular level as to how these complexes form and are organized, but it has been suggested the multimodular class B PBPs interact via their n-PB modules with other components within their complexes (41). The results of this study would appear to provide some experimental evidence for this proposal, since associations with the isolated n-PB module would account for the morphological changes and eventual lysis observed when it is overproduced in E. coli. Thus, overproduced wild-type PBP 2s and their individual n-PB modules but not the N-terminal truncated form of PBP 2 would be transferred to the outer leaflet of the cytoplasmic membrane, where they would be deposited in a diffuse fashion around the cylindrical part of the cell wall (7). It is likely that this high concentration of transferred PBP 2 would eventually saturate and overwhelm available sites on the underlying MreB cables, allowing for the total random distribution of the proteins around the cell circumference. Since equivalent amounts of RodA and the bifunctional PBP 1a or 1b (up to approximately 220 molecules per cell (9) would not be available to complement the excessive amounts of PBP 2 or its isolated n-PB module, fully functional biosynthetic systems would not form. However, the n-PB module, either alone or as part of full-length PBP 2, may serve to recruit one or more lytic transglycosylases present in the periplasm (SltB1 or Slt70) and/or on the inner leaflet of the outer membrane (MltA), thereby positioning them proximal to the peptidoglycan sacculus. The initial activity of the lytic transglycosylase(s) associated with these randomly placed complexes would then act to weaken the peptidoglycan sacculus. Assuming that the glycan strands of peptidoglycan are indeed oriented as “hoops” around the girth of the rod-shaped cell (46), the internal turgor pressure exerting against the weakened sacculus would result in the collapse of its rod shape to the more innately stable morphology of the observed spheres. With time, the continued lytic activity would eventually lead to bacteriolysis.
While the proposal presented above is highly speculative and other explanations may exist, it would also account for the lysis observed when inactive variants of PBP 1b, which is known to complex with PBP 3 and MltA among other proteins (see reference 1 and references therein), are overproduced in E. coli (28). Again, the excessive amounts of PBP 1b would not permit the formation of complete biosynthetic complexes but nonetheless would attract lytic transglycosylase(s) to eventually cause the autolysis. Recently Bertsche et al. (1) demonstrated that a truncated derivative of PBP 3 comprising only the N-terminal 56 amino acids was capable of interacting with PBP 1B in vitro, thus providing further evidence for the binding function of the n-PB module of the class B PBPs. Moreover, this truncated PBP 3 derivative was shown to possess the structural determinants required to target the protein to the cell division site in vivo (34). Finally, we have preliminary data based on affinity chromatography experiments that demonstrate a direct interaction between P. aeruginosa PBP 2 and the lytic translglycosylase SltB1 (22).
As discussed above, we are proposing that the n-PB module of PBP 2 plays a role in the nucleation of enzyme complexes for the elongation of the peptidoglycan sacculus in E. coli and by extension P. aeruginosa. In E. coli, these complexes are thought to involve both a soluble and a membrane-bound lytic transglycosylase (Slt70 and MltA, respectively) and a class A PBP that function together with PBP 2 within the periplasm to extend the sacculus (recently reviewed in references 3 and 35). The transmembrane helix of PBP 2 associates with the integral membrane proteins MreD, RodA, and MreC, and collectively they complex with the MreB cables underlying the cytoplasmic membrane. Considerably less is known about the precise arrangement of the periplasmic components, but a dedicated “scaffolding” protein, MipA, has been found in E. coli which appears to complex specifically with PBP 1b and MltA (42). Hence, it is conceivable that the n-PB module of PBP 2, which extends from the periplasmic face of the cytoplasmic membrane, functions in concert with MipA to recruit and organize the other components of the complex, including lytic transglycosylase B. By extension of this postulate, the n-PB module of PBP 3 may function similarly with MipA to help organize the “divisome,” a large complex of a variety of enzymes and proteins involved in the process of cell division, including those comprising the Z-ring (reviewed in reference 14). Clearly, however, further detailed investigation will be required to validate this proposed function for the n-PB module of PBPs 2 and 3 and advance our understanding of their function in the greater architecture of these protein networks.
Acknowledgments
We thank W. Vollmer, Universitat Tubingen, for providing the multiple lytic transglycosylase mutant E. coli MHD79 and both C. Daniels and J. Lam, Department of Molecular and Cellular Biology, University of Guelph, for providing plasmid pUCH6-wzz1. We also thank Sandra Ruiloba and Kathy Daniels for assistance in cloning P. aeruginosa ftsI, coding for PBP 3, Monika Gorzelak for technical help, and Sandy Smith, Department of Food Science, University of Guelph, for her expert technical assistance with SEM.
These studies were supported by an operating grant to A.J.C. from the Canadian Institutes of Health Research (MOP-49623), and a postgraduate scholarship (PGS B) to B.A.L. from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Bertsche, U., T. Kast, B. Wolf, C. Fraipont, M. E. G. Aarsman, K. Kannenberg, M. von Reschenberg, M. Nguyen-Distèche, T. den Blaauwen, J.-V. Höltje, and W. Vollmer. 2006. Interaction between two murein (peptidoglycan) synthases, PBP 3 and PBP 1B, in Escherichia coli. Mol. Microbiol. 61:675-690. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, N. T., and A. J. Clarke. 2002. Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry 41:1001-1013. [DOI] [PubMed] [Google Scholar]
- 3.Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601-610. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, A. J. 1993. Extent of peptidoglycan O acetylation in the tribe Proteeae. J. Bacteriol. 175:4550-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, C., C. Griffiths, B. Cowles, and J. S. Lam. 2002. Pseudomonas aeruginosa O-antigen chain length is determined prior to ligation to lipid A core. Environ. Microbiol. 4:883-897. [DOI] [PubMed] [Google Scholar]
- 7.Den Blaauwen, T., M. E. Aarsman, N. O. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 8.de Pedro, M. A., W. D. Donachie, J.-V. Holtje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougherty, T. J., K. Kennedy, R. E. Kessler, and M. J. Pucci. 1996. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 178:6110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glauner, B. 1988. Separation and quantification of muropeptides with high performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 11.Goffin, C., C. Fraipont, J. Ayala, M. Terrak, M. Nguyen-Disteche, and J.-M. Ghuysen. 1996. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J. Bacteriol. 178:5402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 15.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J.-V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höltje, J.-V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 18.Höltje, J. V., D. Mirelman, N. Sharon, and U. Schwarz. 1975. Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 124:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, and B. G. Spratt. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 261:7024-7031. [PubMed] [Google Scholar]
- 20.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Legaree, B. A., and A. J. Clarke. 2006. Using affinity chromatography to investigate protein-protein interactions involved in peptidoglycan synthesis in Pseudomonas aeruginosa, abstr. 20. Fifty-sixth Annu. Meet. Can. Soc. Microbiol., London, Ontario, Canada.
- 23.Legaree, B. A., K. E. Daniels, J. T. Weadge, D. Cockburn, and A. J. Clarke. 2007. Function of penicillin-binding protein 2 in viability and morphology of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 59:411-424. [DOI] [PubMed] [Google Scholar]
- 24.Liao, X., and R. E. W. Hancock. 1995. Cloning and characterization of the Pseudomonas aeruginosa pbpB gene encoding penicillin-binding protein 3. Antimicrob. Agents Chemother. 39:1871-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, X., and R. E. W. Hancock. 1997. Identification of a penicillin-binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase-dependent expression. J. Bacteriol. 179:1490-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macheboeuf, P., C. Contreras-Martel, V. Job, O. Dideberg, and A. Dressen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673-691. [DOI] [PubMed] [Google Scholar]
- 27.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Fraipont, J.-M. Ghuysen, and M. Nguyen-Disteche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37:1019-1031. [DOI] [PubMed] [Google Scholar]
- 28.Meisel, U., J.-V. Höltje, and W. Vollmer. 2003. Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli. J. Bacteriol. 185:5342-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro, F., A. Robin, R. D'Ari, and D. Joseleau-Petit. 1998. Analysis of the effect of ppGpp on the ftsQAZ operon in Escherichia coli. Mol. Microbiol. 29:815-823. [DOI] [PubMed] [Google Scholar]
- 30.Neu, H. C. 1983. Penicillin-binding proteins and the role of amdinocillin in causing bacterial cell death. Am. J. Med. 75:9-20. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen-Disteche, M., C. Fraipont, N. Buddelmeijer, and N. Nanninga. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci. 54:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi, J., M. Matsuhashi, and S. Matsuhashi. 1979. Comparative studies of penicillin-binding proteins in Pseudomonas aeruginosa and Escherichia coli. Eur. J. Biochem. 100:41-49. [DOI] [PubMed] [Google Scholar]
- 33.Piette, A., C. Fraipont, T. den Blaauween, M. E. G. Aarsman, S. Pastoret, and M. Nguyen-Distèche. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 186:6110-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheffers, D.-J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69:585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, J., G. Xie, P. K. Elf, K. D. Young, and R. A. Jensen. 1998. Comparative analysis of Pseudomonas aeruginosa penicillin-binding protein 7 in the context of its membership in the family of low-molecular-mass PBPs. Microbiology 144:975-983. [DOI] [PubMed] [Google Scholar]
- 36.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoker, N. G., J. K. Broome-Smith, A. Edelman, and B. G. Spratt. 1983. Organization and subcloning of the dacA-rodA-pbpA cluster of cell shape genes in Escherichia coli. J. Bacteriol. 155:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takasuga, A., H. Adachi, F. Ishino, M. Matsuhashi, T. Ohta, and H. Matsuzawa. 1988. Identification of the penicillin-binding active site of penicillin-binding protein 2 of Escherichia coli. J. Biochem. (Tokyo) 104:822-826. [DOI] [PubMed] [Google Scholar]
- 39.Vinella, D., M. Cashel, and R. D'Ari. 2000. Selected amplification of the cell division genes ftsQ-ftsA-ftsZ in Escherichia coli. Genetics 156:1483-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinella, D., R. D'Ari, and P. Bouloc. 1992. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 11:1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer, W., and J.-V. Höltje. 2001. Morphogenesis of Escherichia coli. Curr. Opin. Microbiol. 4:625-633. [DOI] [PubMed] [Google Scholar]
- 42.Vollmer, W., M. von Rechenberg, and J.-V. Höltje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 43.von Rechenberg, M., A. Ursinus, and J.-V. Höltje. 1996. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb. Drug Resist. 2:155-157. [DOI] [PubMed] [Google Scholar]
- 44.Weidel, W., and H. Pelzer. 1964. Bagshaped macromolecules—a new outlook on bacterial cell walls. Adv. Enzymol. 26:193-232. [DOI] [PubMed] [Google Scholar]
- 45.Wissel, M. C., and D. S. Weiss. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, K. D. 2006. Too many strictures on structure. Trends Microbiol. 14:156-157. [DOI] [PubMed] [Google Scholar]
- 47.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]