Abstract
Glutamate racemase activity in Bacillus anthracis is of significant interest with respect to chemotherapeutic drug design, because l-glutamate stereoisomerization to d-glutamate is predicted to be closely associated with peptidoglycan and capsule biosynthesis, which are important for growth and virulence, respectively. In contrast to most bacteria, which harbor a single glutamate racemase gene, the genomic sequence of B. anthracis predicts two genes encoding glutamate racemases, racE1 and racE2. To evaluate whether racE1 and racE2 encode functional glutamate racemases, we cloned and expressed racE1 and racE2 in Escherichia coli. Size exclusion chromatography of the two purified recombinant proteins suggested differences in their quaternary structures, as RacE1 eluted primarily as a monomer, while RacE2 demonstrated characteristics of a higher-order species. Analysis of purified recombinant RacE1 and RacE2 revealed that the two proteins catalyze the reversible stereoisomerization of l-glutamate and d-glutamate with similar, but not identical, steady-state kinetic properties. Analysis of the pH dependence of l-glutamate stereoisomerization suggested that RacE1 and RacE2 both possess two titratable active site residues important for catalysis. Moreover, directed mutagenesis of predicted active site residues resulted in complete attenuation of the enzymatic activities of both RacE1 and RacE2. Homology modeling of RacE1 and RacE2 revealed potential differences within the active site pocket that might affect the design of inhibitory pharmacophores. These results suggest that racE1 and racE2 encode functional glutamate racemases with similar, but not identical, active site features.
Inhalational anthrax is a complex human disease that begins with deposition of Bacillus anthracis spores into lungs, followed by infiltration and proliferation of vegetative bacteria in the blood and several organs, ultimately leading to death of the host (23, 25, 36, 42). Current antibiotic treatments are effective during early stages of infection; however, the emergence of engineered (55) and naturally occurring (4) antibiotic-resistant strains underscores the need to identify new antibacterial targets (7). Moreover, due to the potential deployment of B. anthracis as a bioweapon, the development and stockpiling of new anthrax countermeasures have been prioritized by the United States government (6).
Inhibition of cell wall synthesis remains an effective approach for preventing bacterial growth (53, 54). For most eubacteria, an important constituent of the peptidoglycan cell wall is d-glutamate (38, 39, 41, 44, 45). d-Glutamate is not typically available in the environment but instead is generated by the enzyme glutamate racemase (E.C. 5.1.1.3), which catalyzes the reversible stereoinversion of l-glutamate (5, 14, 31). Insights into the cofactor-independent amino acid racemases have begun to emerge from biochemical studies of enzymes isolated from several organisms, including Bacillus subtilis, Bacillus pumilus, Bacillus sphaericus, Escherichia coli, Lactobacillus fermentum, Lactobacillus brevis, Aquifex pyrophilus, Staphylococcus haemolyticus, Brevibacterium lactofermentum, and Mycobacterium tuberculosis (1, 2, 5, 10, 14, 18, 19, 21, 28, 29, 33, 35, 40, 47, 58, 61), as well as the recently described crystal structure of RacE-d-glutamate from B. subtilis (43). Several studies have identified glutamate racemase as an essential gene in B. subtilis and E. coli, which has led to the prediction that glutamate racemase activity is important for peptidoglycan biosynthesis in these organisms (14, 31). Because glutamate racemases are not found in mammals, these enzymes have emerged as excellent targets for the design of a new class of antibacterial agents (3, 12, 22, 28, 43, 59).
Analogous to the peptidoglycan of other eubacteria, d-glutamate is predicted to be an important constituent of B. anthracis (48). However, in B. anthracis d-glutamate is also the sole component of the poly-γ-d-glutamic acid (PDGA) capsule (24), an important virulence factor that is required for dissemination in a murine model of inhalational anthrax (16) and is presumed to be required for disease in humans as well (16, 36, 46). Therefore, in addition to its proposed role in cell wall biosynthesis, glutamate racemase is also predicted to be the major source of d-glutamate for PDGA capsule synthesis in B. anthracis (9). In contrast to most bacteria that possess only one glutamate racemase gene (5, 10, 14, 18, 19, 26, 29, 33, 35, 40, 61), the B. anthracis genome contains two genes (BAS0806 and BAS4379) predicted to encode glutamate racemases, which are designated racE1 and racE2. Recently, inactivation of racE2 was reported to cause a severe growth defect in B. anthracis, while inactivation of racE1 only moderately inhibited growth (52). However, the underlying reasons for these growth phenotypes, especially for the racE2 mutant, were not identified and thus cannot be attributed at this time to defects in peptidoglycan synthesis resulting from insufficient d-glutamate availability (52). Further complicating the interpretation of the phenotypes reported for the racE1 and racE2 mutants is uncertainty concerning whether either of these two genes encodes functional enzymes with glutamate racemase activity. Thus, validation of RacE1 or RacE2 as a potential drug target awaits elucidation of the enzymatic and biochemical properties of these proteins.
Here, we characterized and compared recombinant wild-type and mutant forms of RacE1 and RacE2 from B. anthracis. The racE1 and racE2 genes were cloned from B. anthracis Sterne 7702 and expressed as recombinant proteins in E. coli. These studies revealed that in cell-free assays, B. anthracis RacE1 and RacE2 both catalyze stereoinversion of l-glutamate to d-glutamate. However, the two enzymes differ in ways that could influence future inhibitor development.
MATERIALS AND METHODS
Materials.
B. anthracis Sterne 7702 (56) was obtained from Theresa M. Koehler (Houston, TX). E. coli XL1-Blue was obtained from Stratagene (La Jolla, CA). E. coli T7 lysogen BL21(DE3) was obtained from Novagen (Madison, WI). A DNeasy tissue kit, a DNA PCR purification kit, and a DNA gel extraction kit were acquired from QIAGEN (Valencia, CA). DNA oligonucleotides were synthesized at Integrated DNA Technologies Incorporated (Coralville, IA). Picomaxx DNA polymerase for PCR amplification and a QuikChange site-directed mutagenesis kit were acquired from Stratagene (La Jolla, CA). XhoI, SalI, and BamHI restriction endonucleases were obtained from New England Biolabs (Ipswich, MA). Ni-nitrilotriacetic acid (NTA) His·Bind metal affinity resin and pET-15b were purchased from Novagen (Madison, WI). DNA sequencing was performed at the Roy J. Carver Biotechnology Center (Urbana, IL). A plasmid miniprep kit and gel filtration standards were obtained from Bio-Rad (Hercules, CA). Components for Luria-Bertani and Bacto brain heart infusion medium were purchased from BD Diagnostics (Franklin Lakes, NJ). Isopropyl-β-d-thiogalactoside (IPTG) was obtained from Fisher Scientific (Fair Lawn, NJ). l-Glutamate, d-glutamate, ampicillin, dithiothreitol (DTT), and tris(2-carboxyethyl)phosphine (TCEP) were obtained from Sigma Aldrich (St. Louis, MO). Amicon centrifugal filter devices with a molecular weight cutoff of 10,000 were obtained from Millipore (Billerica, MA).
Multiple-sequence alignment.
To compare the primary amino acid sequences of B. subtilis RacE and B. anthracis RacE1 and RacE2, a multiple-sequence alignment was generated. Amino acid sequences for B. anthracis Sterne RacE1 (BAS0806), B. anthracis Sterne RacE2 (BAS4379), and B. subtilis RacE (BSU2835) were aligned using ClustalW (http://align.genome.jp/). The multiple-sequence alignment file was then entered into the ESPript V2.2 alignment program (http://mail.bic.nus.edu.sg/ESPript/cgi-bin/ESPript.cgi). Secondary structural elements and solvent accessibility indices were imported from the crystal structure of B. subtilis 168 RacE (Protein Data Bank accession no. 1ZUW) (43).
Cloning of racE1 and racE2 and preparation of expression strains.
To obtain genomic DNA, B. anthracis Sterne 7702 was cultivated at 37°C with aeration on a rotary shaker in brain heart infusion medium (3.7% Bacto brain heart infusion, Millipore deionized water, 0.5% glycerol shaker). DNA was isolated from mid-log-phase cultures and was purified using the DNeasy tissue kit. B. anthracis Sterne racE1 (BAS0806) and B. anthracis Sterne racE2 (BAS4379) were PCR amplified using primers corresponding to the 5′ and 3′ ends of each gene (Table 1). These primers were engineered such that 5′ XhoI (racE1) or SalI (racE2) and 3′ BamHI (racE1 and racE2) restriction sites were incorporated. Each PCR product was purified using the PCR purification kit. The purified amplicons were incubated with XhoI (racE1) or SalI (racE2) and BamHI to generate directional annealing sites. The amplicons were then ligated with pET-15b to replace the XhoI-BamHI fragment within the polylinker region. The ligation mixtures were introduced into E. coli XL1-Blue by electroporation. The integrity of each gene from individual clones was confirmed by DNA sequencing. pET-15b-racE1 and pET-15b-racE2 were isolated using the plasmid miniprep kit and introduced by electroporation into E. coli T7 lysogen BL21(DE3).
TABLE 1.
Cloning of B. anthracis glutamate racemase genes, using B. anthracis Sterne 7702(pXO1+/pXO2−)
| Gene | Accession no.a | Primer
|
Predicted mol wtc | First residue of coding sequencec | Last residue of coding sequencec | |
|---|---|---|---|---|---|---|
| Direction | Sequence (5′→3′)b | |||||
| racE1 | BAS0806 | Forward | 5′-GCTGCTCGAGTCTGTATGTCATAAAC-3′ | 32,532 | Met-1 | Asn-298 |
| Reverse | 5′-AGTGGATCCCTAATTACAGATGCGA-3′ | |||||
| racE2 | BAS4379 | Forward | 5′-GTGATGTCGACAAGTTGAATAGAGCAATCGGTG-3′ | 32,316 | Met-1 | Glu-291 |
| Reverse | 5′-GCTAGGATCCTTATTCTTTTTCTAAATGAATATG-3′ | |||||
DNA sequences for racE1 and racE2 were obtained from The Institute for Genomic Research Comprehensive Microbial Resource (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi).
Oligonucleotide primers were engineered such that 5′ XhoI (racE1) or SalI (racE2) and 3′ BamHI (racE1 and racE2) restriction sites were incorporated. Primers were synthesized by Integrated DNA Technologies (Coralville, IA).
Coding sequences and molecular weights were predicted based upon additional residues contributed by the pET-15b vector N-terminal polyhistidine tag (Novagen).
Expression of racE1 or racE2.
E. coli BL21(DE3) transformed with either pET-15b-racE1 or pET-15b-racE2 was grown overnight in Luria-Bertani broth (5 ml) supplemented with ampicillin (100 μg/ml) at 37°C and with aeration. After 12 h, the starter cultures were back-diluted into fresh Luria-Bertani broth (500 ml) supplemented with ampicillin (100 μg/ml) at 37°C and grown with aeration until the optical density at 600 nm reached 0.5. Expression of racE1 or racE2 was induced by addition of IPTG (0.1 mM). The cultures were grown for an additional 4 h and then harvested by centrifugation at 5,000 × g for 15 min at 4°C. The cell pellets were resuspended in 20 ml Ni-NTA binding buffer (50 mM phosphate, 300 mM NaCl, 10 mM imidazole, 0.5 mM TCEP; pH 8.0) and disrupted by sonication (three 20-s cycles at 23 kHz and 20 W) using a model 100 Sonic Dismembrator from Fisher Scientific (Fair Lawn, NJ). The cell lysate was then clarified by centrifugation at 30,000 × g for 30 min and applied to 2 ml (bed volume) of Ni-NTA His·Bind metal affinity resin. After 1 h of gentle rotation at 4°C, the bound protein was washed with two 10-ml volumes of Ni-NTA wash buffer (50 mM phosphate, 20 mM imidazole, 300 mM NaCl, 0.5 mM TCEP; pH 8.0) and then eluted with two 6-ml volumes of Ni-NTA elution buffer (50 mM phosphate, 250 mM imidazole, 300 mM NaCl, 0.5 mM TCEP; pH 8.0). The column eluant was then exchanged into storage buffer (50 mM Tris, 100 mM NaCl, 0.2 mM DTT; pH 8.0) utilizing an Amicon centrifugal filter device.
Protein concentrations were quantified by absorbance spectroscopy using the method described by Gill and von Hippel (20). Briefly, the extinction coefficient at 280 nm was calculated based upon the amino acid composition of the polyhistidine-tagged recombinant protein. The resulting extinction coefficients (17,420 M−1 cm−1 for RacE1 and 21,430 M−1 cm−1 for RacE2) were then used to determine the concentration of the appropriate protein solution utilizing a Multiskan Spectrum UV spectrophotometer from Thermo Electron Company (Waltham, MA). The protein stocks were stored at −20°C in storage buffer (50 mM Tris, 100 mM NaCl, 0.2 mM DTT; pH 8.0) with 20% glycerol at a final concentration of 10 mg/ml for up to 2 months.
Preliminary experiments revealed no detectable differences in the kinetic properties of RacE1 or RacE2 in the presence or in the absence of the amino-terminal polyhistidine fusion peptides (data not shown). Based on these data, purified proteins with amino-terminal polyhistidine fusion peptides were utilized for all of the experiments described in this study.
CD spectroscopy of purified RacE1 and RacE2.
Circular dichroism (CD) spectra were collected for RacE1 and RacE2 in the far-UV range utilizing a J-720 CD spectropolarimeter from JASCO (Easton, MD). A cylindrical cuvette with a total volume of 350 μl and a path length of 0.1 cm was used for each assay. The CD spectra of RacE1 (2.7 μM) and RacE2 (2.4 μM) in optically clear borate buffer (50 mM potassium borate, pH 8.0) were recorded from 190 to 260 nm at a scan rate of 50 nm/s with a 1-nm wavelength step and with five accumulations.
Data acquisition was coordinated using the JASCO Spectra Manager v1.54A software. Raw data files were uploaded onto the DICHROWEB online server (http://www.cryst.bbk.ac.uk/cdweb/html/home.html) and analyzed using the CDSSTR algorithm with reference set 4, which is optimized for the analysis of data recorded in the range from 190 to 240 nm (34).
Size exclusion chromatography.
Size exclusion chromatography was conducted using an AKTA Purifier 900 fast protein liquid chromatography (FPLC) system equipped with a Superdex 200 10/300 GL size exclusion column and a UV detector, all obtained from Amersham Pharmacia Biotech (Little Chalfont, United Kingdom). RacE1 (5 mg/ml; 100 μl), RacE2 (5 mg/ml; 100 μl), or a gel filtration standard mixture was injected onto the column preequilbrated with a potassium borate buffer (50 mM boric acid, 100 mM KCl, 0.2 mM DTT; pH 8.0) liquid phase at a flow rate of 0.5 ml/min. Standard curves were generated by plotting the log of the molecular weights (provided by the supplier) of the gel filtration standards versus retention times. Experimental retention times were used to calculate the apparent molecular weights of RacE1 and RacE2 from the standard curve.
Racemization assays.
Enzyme-catalyzed stereoisomerization of d- or l-glutamate was assayed using a J-720 CD spectropolarimeter from JASCO (Easton, MD). A thermostat-equipped cylindrical cuvette with a capacity of 700 μl and a path length of 1 cm was used for each assay. d- or l-glutamate (5 mM) in optically clear potassium borate buffer (50 mM boric acid, 100 mM KCl, 0.2 mM DTT; pH 8.0) was incubated at 25°C in the absence or presence of RacE1 or RacE2 at a concentration of 0.08, 0.33, or 1.3 μM. d- or l-glutamate stereoisomerization was monitored by recording the CD signal at 217 nm. Data acquisition was performed using the JASCO Spectra Manager v1.54A software, and a nonlinear curve fit was applied using GraphPad Prism V4.03 from GraphPad Software (San Diego, CA).
Determination of steady-state kinetic parameters.
Racemase assays were carried out as described above, with the exception that the d-glutamate concentration was varied from 0.2 to 10 mM while the concentration of l-glutamate was varied from 5 to 200 mM. In addition, a 700-μl cuvette with a 1-cm path length was used for reactions with substrate concentrations less than 5 mM, and a 350-μl cuvette with a 0.1-cm path length was used for reactions with higher substrate concentrations. Reactions were initiated by addition of RacE1 (0.78 μM) or RacE2 (0.78 μM), and the levels of d- or l-glutamate were monitored by recording the CD signal at 215 nm for d- or l-glutamate concentrations from 0.2 to 10 mM and at 225 nm for l-glutamate concentrations from 30 to 200 mM at 25°C. Data acquisition was performed using the JASCO Spectra Manager v1.54A software, and a nonlinear curve fit was applied using GraphPad Prism V4.03 from GraphPad Software (San Diego, CA).
pH rate profile.
The stereoisomerization of glutamate in the l→d direction by RacE1 and RacE2 in buffers having various pH values was assayed using a J-720 CD spectropolarimeter from JASCO (Easton, MD). A cylindrical cuvette with a total volume of 350 μl and a path length of 0.1 cm was used for each assay. Seven different buffers spanning a pH range from 6.5 to 9.5 with increments of 0.5 pH unit were prepared. To maintain a well-buffered system, the following phosphate and borate buffer formulations were utilized: for pH 6.5, 7.0, and 7.5, 50 mM potassium phosphate, 100 mM KCl, 200 mM l-glutamate, 0.2 mM DTT; and for pH 8.0, 8.5, 9.0, and 9.5, 50 mM boric acid, 100 mM KCl, 200 mM l-glutamate, 0.2 mM DTT. Each buffer was prepared with 200 mM l-glutamate, which yielded fully saturating conditions for RacE1 and nearly saturating conditions (83% saturation) for RacE2, so that the initial rate data would report true kcat values. Reactions were initiated by addition of RacE1 (0.78 μM) or RacE2 (0.78 μM), and the levels of l-glutamate were monitored by recording the CD signal at 225 nm at 25°C. Data acquisition was performed using the JASCO Spectra Manager v1.54A software, and a user-defined curve fit was applied using GraphPad Prism V4.03 from GraphPad Software (San Diego, CA).
Homology models.
The homology models for RacE1 and RacE2 were constructed using The Chemical Computing Group's Molecular Operating Environment (MOE) 2006.08. The template for both models was the B. subtilis RacE-d-glutamate structure (Protein Data Bank accession no. 1ZUW), which was aligned with the sequences for RacE1 and RacE2 using the Blosum62 substitution matrix. Ten intermediate homology models resulting from permutational selection of different loop candidates and side chain rotamers were built for RacE1 and RacE2. The intermediate model which scored best according to a packing evaluation function was chosen as the final model. Each of the intermediate models was subjected to a degree of energy minimization using the force field MMFF94x, with a distance-dependent dielectric (i.e., it simulated the polar environment of water).
Docking of compound 69 to RacE1 and RacE2.
A conformational database was generated for (2R,4R)-2-amino-4-(2-benzo[b]thienyl)methyl pentanedioic acid (compound 69) (12) by using a stochastic conformational search, as implemented in MOE 2006.08 (Chemical Computing Group, Inc.). This program employs a variation of the method of Ferguson and Raber (17), in which bonds are randomly rotated, rather than using perturbation of Cartesian coordinates. The force field was MMFF94x. Minimization was performed for each conformation up to a root mean square gradient of 0.001. Any two conformers were considered identical if their optimal heavy atom root mean square superposition distance was less than a tolerance value of 0.1 Å. All conformations with an energy greater than 7 kcal/mol were excluded from the database. This yielded a conformational database of 17 unique conformations of compound 69, which were used in the docking procedure. The docking of compound 69 into RacE1 and RacE2 was performed with the Dock function in MOE 2006.08, using the Alpha Triangle placement method, and the London dG Scoring method for free energy estimation.
Site-directed mutagenesis.
Mutagenesis was performed using the QuikChange mutagenesis kit from Stratagene (La Jolla, CA). First, complementary mutagenic primers (Table 2) were engineered with the desired mutation in the center of the primer and 10 to 15 bases of correct sequence on either side. Reaction mixtures were prepared as described in the QuikChange protocol with pET-15b-racE1 or pET-15b-racE2 as the plasmid template for generation of the RacE1 and RacE2 mutants. After cycling of the reaction mixture 18 times in a thermal cycler, the resulting mixture was digested with DpnI, and the resulting DNA was transformed into supercompetent E. coli XL1-Blue cells. The resulting mutant plasmids were isolated, and the entire gene was sequenced to ensure that the appropriate mutations were introduced, while the rest of the gene sequences remained unchanged.
TABLE 2.
Primer sequences used for mutagenesis
| Gene | Desired mutationa | Primerb | Primer sequence (5′→3′)c |
|---|---|---|---|
| racE1 | Cys77Ala | racE1C77AFor | 5′-GGCTCTAGTTGTAGCAGCGAATACTGCTGCAGCTGC-3′ |
| racE1C77ARev | 5′-GCAGCTGCAGCAGTATTCGCTGCTACAACTAGAGCC-3′ | ||
| racE1 | Cys188Ala | racE1C188AFor | 5′-GATACGTTAATTCTTGGGGCGACGCATTATCCACTTTTAGAG-3′ |
| racE1C188ARev | 5′-CTCTAAAAGTGGATAATGCGTCGCCCCAAGAATTAACGTATC-3′ | ||
| racE2 | Cys74Ala | racE2C74AFor | 5′-CAAAATGTTAGTTATTGCAGCGAATACAGCAACTGCAGTTGTATTAG-3′ |
| racE2C74ARev | 5′-CTAATACAACTGCAGTTGCTGTATTCGCTGCAATAACTAACATTTTG-3′ | ||
| racE2 | Cys185Ala | racE2C185AFor | 5′-GATACACTTATTTTAGGTGCGACACATTATCCGATTTTAGGTCC-3′ |
| racE2C185ARev | 5′-GGACCTAAAATCGGATAATGTGTCGCACCTAAAATAAGTGTATC-3′ |
Residues were selected for conservative mutagenesis due to their close proximity to d-glutamate in the homology models generated from the B. subtilis RacE crystal structure (43).
Primers were designed in accordance with the QuikChange site-directed mutagenesis protocol by Stratagene (La Jolla, CA) and were synthesized by Integrated DNA Technologies (Coralville, IA).
Underlined sequences indicate engineered codon mutations.
Protease sensitivity assays.
Wild-type RacE1 (0.13 mM), RacE1 C77A (0.13 mM), and RacE1 C188A (0.13 mM) were incubated in a Tris buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM DTT; pH 8.0) with various concentrations of chymotrypsin (0, 53, 213, and 640 μg/ml) at 4°C. Wild-type RacE2 (0.13 mM), RacE2 C74A (0.13 mM), and RacE2 C185A (0.13 mM) were incubated in a Tris buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM DTT; pH 8.0) with various concentrations of chymotrypsin (0, 10, 40, and 160 μg/ml) at 4°C. After 1 h, the reactions were stopped by addition of an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer (4% SDS, 100 mM Tris, 0.4 mg bromophenol blue/ml, 0.2 M DTT, 20% glycerol). The samples were boiled for 3 min and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (16% acrylamide; 70 V for 25 min and then 200 V for an additional 100 min). The gels were soaked in fixing solution (25% isopropanol, 10% acetic acid) for 15 min and then placed in Coomassie blue stain (10% acetic acid, 0.06 mg/ml Coomassie brilliant blue G-250). After 10 h, the gels were rinsed twice with H2O, preserved by soaking in water with 3% glycerol for 10 h, and dried between gel drying films.
RESULTS
Identification and cloning of B. anthracis racE1 and racE2.
The racE1 (BAS0806) and racE2 (BAS4379) genes predicted to encode glutamate racemases in B. anthracis were identified by sequence homology to racE (BSU2835) in B. subtilis. racE1 and racE2 were PCR amplified from chromosomal DNA purified from B. anthracis Sterne 7702 (Table 1) and cloned into an expression vector for recombinant expression in E. coli. racE1 and racE2 are predicted to encode 32,532- and 32,316-Da proteins, respectively. RacE1 and RacE2 share 51% sequence identity and 67% sequence similarity (Table 1 and Fig. 1), and the lengths of their amino and carboxyl termini differ; RacE1 possesses amino-terminal (MSV) and carboxyl-terminal (NARICN) amino acid extensions that are absent in RacE2 (Fig. 1).
FIG. 1.
B. anthracis RacE1 and RacE2 possess significant sequence homology to B. subtilis RacE. Primary amino acid sequences for B. anthracis RacE1 and RacE2 and B. subtilis RacE were aligned utilizing ESPript V.2.2 (http://espript.ibcp.fr/ESPript/ESPript/). Secondary structural elements and solvent accessibility indices were imported from the crystal structure of B. subtilis 168 RacE (Protein Data Bank accession no. 1ZUW) (43). For solvent accessibility indices, the darker shaded regions indicate sections in the folded proteins that are exposed to solvent.
Expression and purification of B. anthracis RacE1 and RacE2.
Both RacE1 and RacE2 were expressed as soluble, recombinant proteins in E. coli, each with an amino-terminal hexahistidine fusion peptide to facilitate purification via nickel-chelate affinity chromatography. In this single chromatography step, both RacE1 and RacE2 were purified to greater than 98% purity, as estimated by SDS-PAGE analysis (Fig. 2A). Preliminary experiments to compare properties of the recombinant proteins before and after the amino-terminal polyhistidine fusion peptide was removed by thrombin cleavage indicated that the presence of the amino-terminal polyhistidine fusion peptide had no detectable effects on the kinetic properties of either RacE1 or RacE2 (data not shown).
FIG. 2.
RacE1 and RacE2 are both functional enzymes in a cell-free system. (A) Purification of recombinant RacE1 and RacE2. The RacE1 and RacE2 clarified lysates and fractions from nickel-chelate affinity chromatography were analyzed by 12% SDS-PAGE, followed by Coomassie brilliant blue G-250 staining. (B) RacE1 and RacE2 secondary structure. CD spectra in the far-UV region (190 to 260 nm) were recorded for RacE1 and RacE2 (2.7 and 2.4 μM, respectively, both in 50 mM potassium borate buffer; pH 8.0). (C) Gel filtration chromatography. The sizes of purified RacE1 and RacE2 were determined by size exclusion FPLC. The molecular weights (MW) of RacE1 and RacE2 were calculated from the retention times of the peak absorbance by comparison with calibration standards having known molecular weights. (D) Racemization of glutamate in the l→d direction. RacE1 and RacE2 were assessed for the capacity to convert l-glutamate to the corresponding d enantiomer by using CD to directly observe the loss of l-glutamate as it was converted to d-glutamate. l-Glutamate (5 mM) was incubated in the absence or presence of RacE1 or RacE2 (1.3, 0.33, or 0.08 μM), and the CD signal was recorded for 2.25 h. (E) Racemization of glutamate in the d→l direction. RacE1 and RacE2 were assessed for the capacity to catalyze the forward (l-glutamate → d-glutamate) and reverse (d-glutamate → l-glutamate) reactions by CD spectroscopy. l-Glutamate (5 mM) or d-glutamate (5 mM) was incubated in the presence of RacE1 (0.31 μM) or RacE2 (0.31 μM), and the CD signal was recorded for 2.25 h. For panels A to E, at least three separate experiments were performed. For each independent experiment, we used RacE1 or RacE2 from one of three independent enzyme preparations, as well as assay reagents from one of three independent preparations. For panels A to E, representative data from a single experiment are shown. In panel C, the molecular weights are reported as the means ± standard deviations from three independent experiments. mAU, milliabsorbance units.
RacE1 and RacE2 have similar secondary structures.
To compare the secondary structural compositions of recombinant RacE1 and RacE2, CD spectra in the far-UV region (190 to 260 nm) were collected for RacE1 or RacE2. These experiments revealed that recombinant RacE1 and RacE2 both yielded CD spectra indicating the presence of α-helix, β-sheet, and β-turn secondary structural elements. Moreover, comparison of the CD spectra revealed that the relative percentages of α-helix, β-sheet, and β-turn secondary structural elements were nearly identical for RacE1 and RacE2 (Fig. 2B and Table 3). These data suggest that when expressed as recombinant proteins, RacE1 and RacE2 had similar overall secondary structural properties.
TABLE 3.
Analysis of CD spectra using DICHROWEBa
| Protein | % α-helix | % β-sheet | % β-turn | % Unordered |
|---|---|---|---|---|
| RacE1 | 38 ± 3 | 15 ± 2 | 20 ± 1 | 28 ± 1 |
| RacE2 | 37 ± 3 | 16 ± 2 | 20 ± 1 | 28 ± 1 |
CD spectra were recorded in the far-UV range utilizing a J-720 CD spectropolarimeter. The spectra were recorded from 190 to 260 nm at a scan rate of 50 nm/s and a 1-nm wavelength step with five accumulations. Each reaction was performed with three independent enzyme preparations, and the data are means ± standard deviations of the means. The spectra were uploaded onto the DICHROWEB online server and analyzed as described in Materials and Methods.
RacE1 and RacE2 differ in their solution quaternary structures.
To compare the quaternary structures of RacE1 and RacE2 and to rule out the possibility that the purified proteins exist as aggregates, the apparent molecular weights of purified RacE1 and RacE2 were determined by size exclusion FPLC. These experiments revealed that both RacE1 and RacE2 eluted well after the exclusion volume of the column, indicating that neither of these proteins exists as large aggregates in solution. However, RacE2 had a shorter retention time than RacE1, suggesting that in solution RacE2 exists in a higher-molecular-weight form than RacE1 (Fig. 2C). The molecular weights of RacE1 and RacE2 were calculated from the retention times for the peak absorbance compared with the retention times of calibration standards having known molecular weights. The apparent molecular weight of RacE1 was calculated to be 32.3 × 103 ± 4.6 × 103, which is the predicted molecular weight of monomeric RacE1 (32.3 × 103). In contrast, the apparent molecular weight of RacE2 was calculated to be 51.5 × 103 ± 5.2 × 103, which is lower than the value expected for the dimeric protein (64.6 × 103), suggesting that in solution RacE2 may be polydisperse, existing as both monomers and higher-order complexes. These results suggest that although RacE1 and RacE2 share significant sequence similarity, these proteins have different quaternary structural properties.
Purified RacE1 and RacE2 are both functional in cell-free assays.
To evaluate whether racE1 and racE2 encode functional glutamate racemase enzymes, RacE1 and RacE2 were assessed to determine their capacities to convert l-glutamate to the corresponding d enantiomer by using CD to directly observe the loss of l-glutamate as it was converted to d-glutamate. These experiments revealed that in the absence of RacE1 or RacE2, stereoisomerization of l-glutamate was not detectable (Fig. 2D). In contrast, consumption of l-glutamate was readily observed in the presence of either RacE1 or RacE2. Moreover, the rate of l-glutamate consumption increased as a function of RacE1 or RacE2 concentration. These data indicated that under cell-free and highly defined conditions, RacE1 and RacE2 both catalyzed the conversion of l-glutamate to d-glutamate in a concentration-dependent fashion. These results also established, for the first time, that despite reported differences in phenotypes of the null mutants (52), racE1 and racE2 both encode functional enzymes that have glutamate racemase activity.
RacE1 and RacE2 both catalyze the reverse reaction: conversion of d-glutamate to l-glutamate.
In addition to catalyzing the conversion of l-glutamate to the corresponding d enantiomer, glutamate racemases also catalyze the reverse reaction, conversion of d-glutamate to the corresponding l enantiomer. To evaluate whether RacE1 and RacE2 share this canonical property of the glutamate racemase family, we used CD to directly measure the stereoisomerization of d-glutamate to the corresponding l enantiomer. The experiments revealed a time-dependent increase in the CD signal corresponding to the loss of d-glutamate in the presence of either RacE1 or RacE2 (Fig. 2E). Under the conditions of the reaction, the initial rate of stereoisomerization of d-glutamate was higher for RacE2 than for RacE1. In comparison, the rates of stereoisomerization of l-glutamate were similar for RacE2 and RacE1. Finally, these experiments revealed that for both RacE1 and RacE2, the rate of d-glutamate conversion is lower than the rate of l-glutamate conversion. These results indicated that RacE1 and RacE2 share one of the canonical properties of glutamate racemases, which is the capacity to catalyze the stereoisomerization of either glutamate enantiomer.
Steady-state kinetic analysis of RacE1 and RacE2.
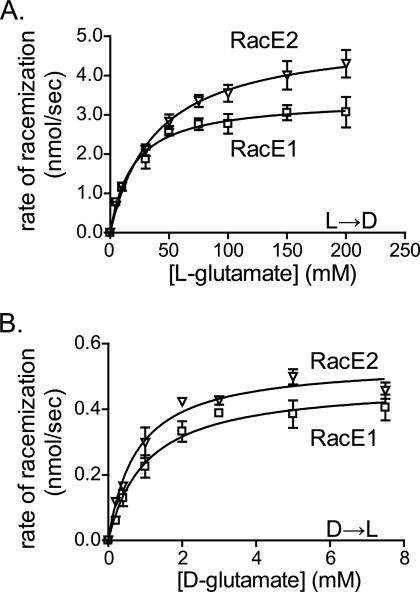
To compare the catalytic properties of RacE1 and RacE2 in more detail, we analyzed the steady-state kinetic parameters of the two enzymes in the presence of d- or l-glutamate. RacE1 or RacE2 was incubated in a potassium borate buffer in the presence of various concentrations of d-glutamate or l-glutamate, and the change in magnitude of the CD signal was monitored. Initial rate data were obtained for RacE1 and RacE2 for each of the substrate concentrations, and plots of the rate of glutamate racemization (nmol/s) versus substrate concentration (mM) were generated for RacE1 and RacE2 in the presence of l- and d-glutamate (Fig. 3). Steady-state kinetic parameters for RacE1 and RacE2 in the presence of l- or d-glutamate were obtained by applying a nonlinear curve fit to the data (Table 4). In the l→d direction, differences in the individual kinetic parameters for RacE1 and RacE2 were statistically significant (for RacE1 kcat = 12 ± 0.6 s−1 and for RacE2 kcat = 18 ± 0.6 s−1 [P = 0.0003]; for RacE1 Km = 19 ± 4 mM and for RacE2 Km = 38 ± 6 mM [P = 0.01]). In the d→l direction, differences in the individual kinetic parameters for RacE1 and RacE2 were not statistically significant (for RacE1 kcat = 1.8 ± 0.1 s−1 and for RacE2 kcat = 2.0 ± 0.1 s−1 [P = 0.07]; for RacE1 Km = 1.0 ± 0.2 mM and for RacE2 Km = 0.77 ± 0.1 mM [P = 0.1]). These findings are in contrast to those for B. subtilis, in which there are approximately 100-fold differences in the catalytic efficiencies of RacE and YrpC (1, 2). Overall, these data indicated that RacE1 and RacE2 have similar, but not identical, steady-state kinetic properties under cell-free conditions.
FIG. 3.
Steady-state kinetic analysis of RacE1 and RacE2. RacE1 (0.78 μM) or RacE2 (0.78 μM) was incubated in a potassium borate buffer (50 mM boric acid, 100 mM KCl, 0.2 mM DTT; pH 8.0) in the presence of various concentrations of d-glutamate (0.1 to 10 mM) (A) or l-glutamate (5 to 200 mM) (B), and the change in magnitude of the CD signal was monitored. (A) Steady-state kinetic parameters for the racemization of glutamate in the l→d direction by RacE1 and RacE2. The data are expressed as initial rate of racemization for RacE1 or RacE2 as a function of the l-glutamate concentration. Steady-state kinetic parameters for RacE1 and RacE2 in the presence of l-glutamate were obtained by applying a nonlinear curve fit to the data. (B) Steady-state kinetic parameters for the racemization of glutamate in the d→l direction by RacE1 and RacE2. The data are expressed as the initial rate of racemization for RacE1 and RacE2 as a function of the d-glutamate concentration. Steady-state kinetic parameters for RacE1 or RacE2 in the presence of d-glutamate were obtained by applying a nonlinear least-squares regression utilizing GraphPad Prisim V4.03. For all studies, at least three independent experiments were performed. For each independent experiment, we used RacE1 or RacE2 from one of three independent enzyme preparations, as well as assay reagents from one of three independent preparations. The symbols indicate the means of the data from three independent experiments, and the error bars indicate the standard deviations of the means.
TABLE 4.
Kinetic parameters for RacE1 and RacE2
| Protein |
l→d racemizationa
|
d→l racemizationa
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1)b | Km (mM)c | kcat/Km (103 M−1 s−1)d | kcat (s−1)e | Km (mM)f | kcat/Km (103 M−1 s−1)g | |
| RacE1 | 12 ± 0.6 | 19 ± 4 | 0.64 ± 0.2 | 1.8 ± 0.1 | 1.0 ± 0.2 | 1.7 ± 0.2 |
| RacE2 | 18 ± 0.6 | 38 ± 6 | 0.48 ± 0.2 | 2.0 ± 0.1 | 0.77 ± 0.1 | 2.6 ± 0.2 |
The racemization of both d- and l-glutamate by RacE1 and RacE2 was analyzed as described in Materials and Methods. Each reaction was performed with three independent enzyme preparations, and the data are means ± standard deviations of the means. The kinetic parameters for RacE1 and RacE2 were assessed for statistically significant differences by the Student t test.
P = 0.0003.
P = 0.01.
P = 0.4.
P = 0.07.
P = 0.1.
P = 0.005.
pH dependence of RacE1 and RacE2 catalysis.
To probe active site features of RacE1 and RacE2 that might contribute to catalysis, we investigated the pH dependence of RacE1- and RacE2-catalyzed glutamate racemase activity. These experiments revealed that the rates of stereoisomerization in the l→d direction catalyzed by RacE1 or RacE2 are both pH dependent, with pH optima of 8.2 and 8.1, respectively. The kcat-versus-pH data fit a bell-shaped curve (equation 1) representative of an enzyme utilizing two ionizable side chains for catalysis (Fig. 4):
 |
(1) |
where kcatapp is the measured kcat, kcatmax is the true kcat, and pKa1 and pKa2 are the pKas of two ionizable groups important for enzyme activity. The pKa values for the two ionizable groups were calculated to be 6.3 ± 0.1 and 9.7 ± 0.1 for RacE1 and 6.1 ± 0.1 and 9.7 ± 0.1 for RacE2. These data suggest that the racemization reactions catalyzed by RacE1 and RacE2 are both dependent upon the ionization state of two catalytic residues. Moreover, the pKa values for the two residues are strikingly similar for the two enzymes, suggesting that RacE1 and RacE2 utilize similar residues for catalysis.
FIG. 4.
pH dependence of RacE1 and RacE2 catalysis. RacE1 and RacE2 were assayed in buffers having pH values ranging from 6.5 to 9.5 with increments of 0.5 pH unit. Each buffer was prepared with a concentration of glutamate (200 mM) that was fully saturating for RacE1 and nearly saturating (83% saturating) for RacE2, so that the initial rate data would report true kcat values. Initial rate data were then obtained for RacE1 (0.78 μM) and RacE2 (0.78 μM) in each of the different buffer formulations by measuring the change in magnitude of the CD signal over time. The data are expressed as the turnover number (kcat) as a function of the reaction pH. The symbols indicate the means of the data from three independent experiments. For each independent experiment, we used RacE1 or RacE2 from one of three independent enzyme preparations, as well as assay reagents from one of three independent preparations. The error bars indicate standard deviations of the means.
Site-directed mutagenesis reveals active site residues important for catalysis in both RacE1 and RacE2.
To further compare active site features of RacE1 and RacE2, we constructed three-dimensional homology models for both B. anthracis RacE1 and RacE2, based upon the available crystal structure for B. subtilis RacE (43). RacE shares 53 and 59% sequence identity with RacE1 and RacE2, respectively. The three-dimensional homology models generated for B. anthracis RacE1 and RacE2 based on the B. subtilis RacE-d-glutamate template exhibited Cα root mean square deviation (RMSD) values of 7.5 Å and 2.6, respectively, indicating that RacE2 may share significant structural similarities with B. subtilis RacE, while RacE1 may possess a more divergent structural arrangement. The homology models for both RacE1 and RacE2 revealed that two cysteine residues (C77 and C188 for RacE1 and C74 and C185 for RacE2) are predicted to be in close proximity to the predicted location of the d-glutamate substrate (Fig. 5A), suggesting that these two cysteine residues may be important for the enzymatic activities of RacE1 or RacE2.
FIG. 5.
Site-directed mutagenesis reveals active site residues important for catalysis in both RacE1 and RacE2. (A) Three-dimensional homology models. Homology models for RacE1 and RacE2 were constructed using the Chemical Computing Group's MOE 2006.08. The template for both models was the B. subtilis glutamate racemase-d-glutamate structure (Protein Data Bank accession no. 1ZUW), which was aligned with the sequences for B. anthracis RacE1 (BAS0806) and RacE2 (BAS4379) using the Blosum62 substitution matrix. (B) Demonstration of residues important for catalysis. The two putative catalytic cysteine residues in RacE1 (Cys77 and Cys188) and RacE2 (Cys74 and Cys185) were independently changed to alanine by site-directed mutagenesis and assessed for the capacity to racemize glutamate in the l→d direction. The four mutant enzymes (0.31 μM) or two wild-type enzymes (0.31 μM) were independently incubated in the presence of l-glutamate (5 mM). The differential absorption of CD light by glutamate was constantly monitored for 2.25 h utilizing a J-720 CD spectropolarimeter. (C) Chymotrypsin sensitivity patterns. Chymotrypsin protease sensitivity patterns were generated for the wild-type and mutant forms of RacE1 and RacE2. Wild-type RacE1 (0.13 mM), RacE1 C77A (0.13 mM), and RacE1 C188A (0.13 mM) were incubated in a Tris buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM DTT; pH 8.0) with various concentrations of chymotrypsin (lane A, 0 μg/ml; lane B, 53 μg/ml; lane C, 213 μg/ml; lane D, 640 μg/ml) at 4°C. Wild-type RacE2 (0.13 mM), RacE2 C74A (0.13 mM), and RacE2 C185A (0.13 mM) were incubated in a Tris buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM DTT; pH 8.0) with various concentrations of chymotrypsin (lane A, 0 μg/ml; lane B, 10 μg/ml; lane C, 40 μg/ml; lane D, 160 μg/ml) at 4°C. After incubation for 1 h, the reactions were stopped by addition of SDS sample buffer, and the samples were electrophoresed on a 16% SDS-polyacrylamide gel and stained with Coomassie brilliant blue G-250. The experiments in panels B and C were performed three separate times. For each independent experiment, we used wild-type or mutant forms of RacE1 or RacE2 from one of three independent enzyme preparations, as well as assay reagents from one of three independent preparations. Representative data from a single experiment are shown. WT, wild type.
To determine whether the predicted RacE1 and RacE2 active site residues are important for catalysis, RacE1 C77 and C188 and RacE2 C74 and C185 were independently changed to alanine by site-directed mutagenesis. These experiments revealed that for RacE1, an alanine substitution at C77 or C188 attenuated detectable racemase activity by at least 100-fold (Fig. 5B). Likewise, for RacE2, an alanine substitution at C74 or C185 attenuated detectable activity by at least 100-fold. The lower limit of detectable activity was obtained by diluting RacE1 or RacE2 until racemase activity was no longer detectable (data not shown). To ensure that the alanine substitution had not resulted in gross structural changes, we compared protease sensitivity patterns of wild-type and mutant forms of RacE1 and RacE2. These experiments revealed similar chymotrypsin sensitivity patterns for full-length RacE1 and RacE1 C77A or RacE1 C188A, as well as for full-length RacE2 and RacE2 C74A or RacE2 C185A (Fig. 5C), indicating a lack of gross structural differences between wild-type and mutant forms of the full-length proteins. Taken together, these data indicate that C77 and C188 are important for the racemization activities of RacE1 and C74 and C185 are important for the racemization activities of RacE2 and suggest that RacE1 and RacE2 may possess several similar active site features. In addition, these results suggest that the homology models that we generated for RacE1 and RacE2 may be useful for generating future predictions about the structure-function relationships underlying the activities of these enzymes.
DISCUSSION
While most bacteria possess a single glutamate racemase gene, B. anthracis has two genes, racE1 and racE2, each predicted to encode a glutamate racemase. Only several other low-G+C-content gram-positive bacteria, including B. subtilis, are predicted or have been shown experimentally to encode two glutamate racemases (30). The presence of two putative glutamate racemases in B. anthracis is potentially of great importance from a chemotherapeutic standpoint because such functional redundancy could mandate that both enzymes be effectively inhibited to successfully prevent the growth of bacilli during infection. On the one hand, if both enzymes possess similar enzymatic properties and active site features, a single compound might be sufficient to inhibit both enzymes. On the other hand, the RacE1 and RacE2 active sites may be sufficiently divergent that a single compound might be ineffective at preventing d-glutamate production by B. anthracis.
A recent study reported a significantly more severe growth defect for a B. anthracis racE2 deletion mutant than for a racE1 deletion mutant (52), suggesting that there are fundamental differences between these two genes. However, it was not demonstrated that the growth defects were due specifically to reduced stereoisomerization of l-glutamate to d-glutamate by the mutant bacilli. Moreover, at the time of this study, neither racE1 nor racE2 had been demonstrated to encode functional glutamate racemases. The overall objective of the current study was therefore twofold: to establish if either racE1 or racE2 encodes a functional glutamate racemase and, if so, to explore whether there are fundamental differences in the biochemical properties between RacE1 and RacE2, which could provide insights into the growth phenotype differences between the racE1 and racE2 mutant strains (52). Notably, the two glutamate racemase isoenzymes in B. subtilis (RacE and YrpC) were demonstrated to have markedly different catalytic properties, with RacE exhibiting 100-fold-higher catalytic efficiency than YrpC (1, 2). In addition, racE is an essential gene, while yrpC is dispensable for growth in rich medium, supporting the idea that in B. subtilis only one of two glutamate racemases is necessary for converting l-glutamate to the d-glutamate required for rapid proliferation (31).
Our data indicated that racE1 and racE2 both encode functional glutamate racemases. In addition, we demonstrated that in a highly defined, cell-free system, RacE1 and RacE2 both catalyze the stereoisomerization of glutamate. In contrast to the disparate properties demonstrated for B. subtilis RacE and YrpC (1, 2), steady-state kinetic analysis identified B. anthracis as the first organism harboring genes encoding two glutamate racemases that in cell-free assays have similar, although not identical, catalytic properties. Analysis of the pH dependence of l-glutamate racemization suggested that RacE1 and RacE2 each possess two active site residues with titratable side chains having nearly identical pKa values. Furthermore, based on homology models that we generated for RacE1 and RacE2, we used directed mutagenesis to demonstrate the importance of two cysteine residues predicted to be in the active sites of both enzymes. Taken together, these results suggested that RacE1 and RacE2 may share similar active site features. However, a more detailed comparison of the predicted active site geometries of the RacE1 and RacE2 homology models (based on the B. subtilis RacE-d-glutamate crystal structure) suggested that several conserved active site residues may be positioned differently relative to bound d-glutamate (Fig. 6A). For example, the distances from the nitrogen atom of d-glutamate to the analogous oxygen atoms of two conserved active site side chains varied for RacE1 (3.1 Å for T189 and 3.0 Å for S15) and RacE2 (3.4 Å for T186 and 2.6 Å for S12). Furthermore, the proximities of the α-carbon of d-glutamate to the analogous sulfur atoms of RacE1 C77 (3.1 Å) and RacE2 C74 (3.4 Å) were different.
FIG. 6.
Three-dimensional homology models reveal differences in RacE1 and RacE2 fine active site features. Three-dimensional homology models for RacE1 and RacE2 were constructed using the Chemical Computing Group's MOE 2006.08. The template for both models was the B. subtilis RacE-d-glutamate structure (Protein Data Bank accession no. 1ZUW), which was aligned with the sequences for B. anthracis RacE1 (BAS0806) and RacE2 (BAS4379) using the Blosum62 substitution matrix. (A) RacE1 and RacE2 exhibit differences in the spatial arrangement of active site residues. Residues predicted to be important for catalysis in B. anthracis RacE1 (green) and RacE2 (purple) and B. subtilis RacE (gray) were aligned. The RMSD of the superposition of Cα atoms between B. anthracis RacE1 and B. subtilis RacE is 7.5 Å, while the RMSD between B. anthracis RacE2 and B. subtilis RacE is 2.6 Å. (B and C) Differences in inhibitor docking to the active sites of RacE1 and RacE2. The glutamate racemase inhibitor compound 69 (12) is shown docked within the active site of B. anthracis RacE1 (B) and overlaid with the active site of B. anthracis RacE2 (C). Residues that are present at the entrance to the hydrophobic binding pocket of B. anthracis RacE1 (green) and RacE2 (purple) are shown. Compound 69 was unable to dock within the active site of RacE2, presumably due to the larger side chain of RacE2 V149, which aligns with A152 in RacE1.
Differences in nonconserved residues surrounding the RacE1 and RacE2 active sites may also impart altered sensitivity to inhibitors. Compound 69 is a potent inhibitor of Streptococcus pneumoniae RacE (50% inhibitory concentration, 0.036 μg/ml), yet its inhibitory properties are highly attenuated for Staphylococcus aureus and Moraxella catarrhalis RacE (12). It has been suggested that the replacement of an alanine at position 149 with a valine may be responsible for these altered pharmacodynamic properties in S. pneumoniae and S. aureus (43). Interestingly, B. anthracis RacE1 possesses an alanine (Ala-152) and RacE2 possesses a valine (Val-149) at the same positions in the predicted structural models, which suggests that these two enzymes may exhibit altered sensitivity to inhibition by compound 69. To explore this possibility further, we computationally generated a library of conformers of compound 69 and scored the conformer library for high-affinity binding in the active sites of the RacE1 and RacE2 homology models (see Materials and Methods). Compound 69 bound tightly (−6.17 kcal/mol) within the active site of RacE1 and adopted a structure analogous to that of the bound substrate (Fig. 6B). However, entrance of compound 69 into the active site of RacE2 was prevented by the presence of Val-149 (Fig. 6C), and no favorable active site docking conformations could be identified. These results suggest that fitting of a pharmacophore into the catalytic pockets of RacE1 and RacE2 may have different requirements, underscoring the potential importance of considering the active sites of both enzymes during the pharmacophore design process.
The results of these studies suggest that differences in the racE1 and racE2 phenotypes reported earlier are unlikely to be due solely to differences between the intrinsic catalytic efficiencies of the two enzymes (52). However, we cannot rule out the possibility that the catalytic properties of RacE1 and RacE2 may be substantially different within the bacterium. Several other possible levels of regulation of racE1 and racE2 inside the organism may contribute to the observed phenotypic differences in the respective deletion mutants. Transcript levels of virulence factors including the PDGA capsule are regulated by the global transcriptional regulator atxA (8, 11, 32, 60). The increased production of PDGA capsule that occurs in response to CO2 induction may lead to a larger demand for cytoplasmic levels of d-glutamate; thus, it is plausible to speculate that racE1 and racE2 could be differentially regulated at the transcriptional level. In addition, the relative importance or roles of RacE1 and RacE2 may be associated with differential localization of the two proteins. Differential intracellular localization of proteins has been shown to be important and tightly coupled with the life cycle of B. subtilis (49-51). Putative localization signals were not computationally identified for RacE1 or RacE2 (data not shown). An alternative possibility is that the enzymatic activities of RacE1 and RacE2 are regulated at the posttranslational level. Indeed, the enzymatic activity of glutamate racemase from E. coli is regulated by a peptidoglycan precursor (15, 27), a function that raises the possibility that a metabolite or peptidoglycan precursor could regulate the enzymatic activity of RacE1 or RacE2. Finally, our experiments revealed that RacE1 and RacE2 had distinct properties in solution when they were analyzed by gel filtration. The source of the unexpected apparent molecular weight for RacE2 in solution (51.5 × 103 ± 5.2 × 103) (Fig. 2C) is currently unknown but could be one or more heretofore unrecognized factors, including polydispersity, the binding of cofactors, posttranslational modifications, etc. Moreover, the significance of the apparent quaternary structural differences between RacE1 and RacE2 in solution is unclear, as is whether these differences are associated with regulation of enzymatic activities within the cell. Notably, within the family of glutamate racemases, quaternary structure differences apparently exist, as enzymes from B. subtilis (YrpC) (1), B. pumilus (33), L. fermentum (13, 19), and Pediococcus pentosaceus (37) have been reported to be monomers, while a dimeric form has been reported for A. pyrophilus (29) and there are conflicting reports about the quaternary structure of the E. coli enzyme (15, 62). Furthermore, RacE from B. subtilis has been reported to exist in equilibrium between a monomer and a dimer (57). Studies are currently under way in our laboratory to further explore the cellular roles of RacE1 and RacE2 and to elucidate the basis of regulation of their cellular activities.
In summary, we have demonstrated that racE1 and racE2 encode functional glutamate racemases, suggesting that the enzymatic activities of both enzymes should be targeted for inhibition. Although we have demonstrated experimentally that RacE1 and RacE2 possess similar, but not identical, enzymatic properties and catalytic residues, several differences in active site features are predicted for RacE1 and RacE2, suggesting that both active sites need to be considered when drugs for effective inhibition of glutamate racemase activity in B. anthracis are designed.
Acknowledgments
This work was supported by NIH-NIAID award U54-AI057156 to the Western Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research (S.R.B.; principal investigator, D. Walker).
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Ashiuchi, M., K. Soda, and H. Misono. 1999. Characterization of yrpC gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci. Biotechnol. Biochem. 63:792-798. [DOI] [PubMed] [Google Scholar]
- 2.Ashiuchi, M., K. Tani, K. Soda, and H. Misono. 1998. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-gamma-glutamate. J. Biochem. (Tokyo) 123:1156-1163. [DOI] [PubMed] [Google Scholar]
- 3.Ashiuchi, M., T. Yoshimura, N. Esaki, H. Ueno, and K. Soda. 1993. Inactivation of glutamate racemase of Pediococcus pentosaceus with l-serine O-sulfate. Biosci. Biotechnol. Biochem. 57:1978-1979. [Google Scholar]
- 4.Athamna, A., M. Athamna, N. Abu-Rashed, B. Medlej, D. J. Bast, and E. Rubinstein. 2004. Selection of Bacillus anthracis isolates resistant to antibiotics. J. Antimicrob. Chemother. 54:424-428. [DOI] [PubMed] [Google Scholar]
- 5.Baltz, R. H., J. A. Hoskins, P. J. Solenberg, and P. J. Treadway. September 1999. Method for knockout mutagenesis in Streptococcus pneumoniae. U.S. patent 5,981,281.
- 6.Borio, L. L., and G. K. Gronvall. 2005. Anthrax countermeasures: current status and future needs. Biosecur. Bioterror. 3:102-112. [DOI] [PubMed] [Google Scholar]
- 7.Bossi, P., D. Garin, A. Guihot, F. Gay, J. M. Crance, T. Debord, B. Autran, and F. Bricaire. 2006. Bioterrorism: management of major biological agents. Cell. Mol. Life Sci. 63:2196-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Candela, T., and A. Fouet. 2006. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 60:1091-1098. [DOI] [PubMed] [Google Scholar]
- 10.Choi, S. Y., N. Esaki, T. Yoshimura, and K. Soda. 1991. Overproduction of glutamate racemase of Pediococcus pentosaceus in Escherichia coli clone cells and its purification. Protein Expr. Purif. 2:90-93. [DOI] [PubMed] [Google Scholar]
- 11.Dai, Z., J. C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 12.de Dios, A., L. Prieto, J. A. Martin, A. Rubio, J. Ezquerra, M. Tebbe, B. Lopez de Uralde, J. Martin, A. Sanchez, D. L. LeTourneau, J. E. McGee, C. Boylan, T. R. Parr, Jr., and M. C. Smith. 2002. 4-Substituted d-glutamic acid analogues: the first potent inhibitors of glutamate racemase (MurI) enzyme with antibacterial activity. J. Med. Chem. 45:4559-4570. [DOI] [PubMed] [Google Scholar]
- 13.Diven, W. F. 1969. Studies on amino acid racemases. II. Purification and properties of the glutamate racemase from Lactobacillus fermenti. Biochim. Biophys. Acta 191:702-706. [DOI] [PubMed] [Google Scholar]
- 14.Doublet, P., J. van Heijenoort, J. P. Bohin, and D. Mengin-Lecreulx. 1993. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J. Bacteriol. 175:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublet, P., J. van Heijenoort, and D. Mengin-Lecreulx. 1994. The glutamate racemase activity from Escherichia coli is regulated by peptidoglycan precursor UDP-N-acetylmuramoyl-l-alanine. Biochemistry 33:5285-5290. [DOI] [PubMed] [Google Scholar]
- 16.Drysdale, M., S. Heninger, J. Hutt, Y. Chen, C. R. Lyons, and T. M. Koehler. 2005. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J. 24:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, D. M., and J. Raber. 1989. A new approach to probing conformational space with molecular mechanics: random incremental pulse search. J. Am. Chem. Soc. 111:4371-4378. [Google Scholar]
- 18.Fotheringham, I. G., S. A. Bledig, and P. P. Taylor. 1998. Characterization of the genes encoding d-amino acid transaminase and glutamate racemase, two d-glutamate biosynthetic enzymes of Bacillus sphaericus ATCC 10208. J. Bacteriol. 180:4319-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo, K. A., and J. R. Knowles. 1993. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry 32:3981-3990. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 21.Glaser, L. 1960. Glutamic acid racemase from Lactobacillus arabinosus. J. Biol. Chem. 235:2095-2098. [PubMed] [Google Scholar]
- 22.Glavas, S., and M. E. Tanner. 1997. The inhibition of glutamate racemase by d- N-hydroxyglutamate. Bioorg. Med. Chem. Lett. 7:2265-2270. [Google Scholar]
- 23.Guidi-Rontani, C., M. Weber-Levy, E. Labruyere, and M. Mock. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31:9-17. [DOI] [PubMed] [Google Scholar]
- 24.Hanby, W. E., and H. N. Rydon. 1946. The capsular substance of Bacillus anthracis. Biochem. J. 40:297-309. [PubMed] [Google Scholar]
- 25.Hanna, P. 1998. Anthrax pathogenesis and host response. Curr. Top. Microbiol. Immunol. 225:13-35. [DOI] [PubMed] [Google Scholar]
- 26.Harth, G., S. Maslesa-Galic, M. V. Tullius, and M. A. Horwitz. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58:1157-1172. [DOI] [PubMed] [Google Scholar]
- 27.Ho, H. T., P. J. Falk, K. M. Ervin, B. S. Krishnan, L. F. Discotto, T. J. Dougherty, and M. J. Pucci. 1995. UDP-N-acetylmuramyl-l-alanine functions as an activator in the regulation of the Escherichia coli glutamate racemase activity. Biochemistry 34:2464-2470. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, K. Y., C. S. Cho, S. S. Kim, H. C. Sung, Y. G. Yu, and Y. Cho. 1999. Structure and mechanism of glutamate racemase from Aquifex pyrophilus. Nat. Struct. Biol. 6:422-426. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. S., I. G. Choi, S. H. Kim, and Y. G. Yu. 1999. Molecular cloning, expression, and characterization of a thermostable glutamate racemase from a hyperthermophilic bacterium, Aquifex pyrophilus. Extremophiles 3:175-183. [DOI] [PubMed] [Google Scholar]
- 30.Kimura, K., L. S. Tran, and Y. Itoh. 2004. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 150:2911-2920. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, K., S. D. Ehrich, A. Albertini, et. al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L., T. Yoshimura, K. Endo, N. Esaki, and K. Soda. 1997. Cloning and expression of the glutamate racemase gene of Bacillus pumilus. J. Biochem. (Tokyo) 121:1155-1161. [DOI] [PubMed] [Google Scholar]
- 34.Lobley, A., L. Whitmore, and B. A. Wallace. 2002. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18:211-212. [DOI] [PubMed] [Google Scholar]
- 35.Malathi, K. C., M. Wachi, and K. Nagai. 1999. Isolation of the murI gene from Brevibacterium lactofermentum ATCC 13869 encoding d-glutamate racemase. FEMS Microbiol. Lett. 175:193-196. [DOI] [PubMed] [Google Scholar]
- 36.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima, N., K. Tanizawa, H. Tanaka, and K. Soda. 1986. Cloning and expression in Escherichia coli of the glutamate racemase gene from Pediococcus pentosaceus. Agric. Biol. Chem. 50:2823-2830. [Google Scholar]
- 38.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, J. T. 1996. The murein sqcculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 40.Pucci, M. J., J. A. Thanassi, H. T. Ho, P. J. Falk, and T. J. Dougherty. 1995. Staphylococcus haemolyticus contains two d-glutamic acid biosynthetic activities, a glutamate racemase and a d-amino acid transaminase. J. Bacteriol. 177:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes. Chapman & Hall, London, United Kingdom.
- 42.Ross, J. M. 1955. On the histopathology of experimental anthrax in the guinea-pig. Br. J. Exp. Pathol. 36:336-339. [PMC free article] [PubMed] [Google Scholar]
- 43.Ruzheinikov, S. N., M. A. Taal, S. E. Sedelnikova, P. J. Baker, and D. W. Rice. 2005. Substrate-induced conformational changes in Bacillus subtilis glutamate racemase and their implications for drug discovery. Structure 13:1707-1713. [DOI] [PubMed] [Google Scholar]
- 44.Salton, M. R. J. 1994. The bacterial cell envelope, p. 1-22. In J. M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall. Elsevier Science Publishing Co., Amsterdam, The Netherlands.
- 45.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scorpio, A., D. J. Chabot, W. A. Day, D. K. O'Brien, N. J. Vietri, Y. Itoh, M. Mohamadzadeh, and A. M. Friedlander. 2007. Poly-gamma-glutamate capsule-degrading enzyme treatment enhances phagocytosis and killing of encapsulated Bacillus anthracis. Antimicrob. Agents Chemother. 51:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sengupta, S., M. Shah, and V. Nagaraja. 2006. Glutamate racemase from Mycobacterium tuberculosis inhibits DNA gyrase by affecting its DNA-binding. Nucleic Acids Res. 34:5567-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Severin, A., K. Tabei, and A. Tomasz. 2004. The structure of the cell wall peptidoglycan of Bacillus cereus RSVF1, a strain closely related to Bacillus anthracis. Microb. Drug Resist. 10:77-82. [DOI] [PubMed] [Google Scholar]
- 49.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 50.Shapiro, L., and R. Losick. 1997. Protein localization and cell fate in bacteria. Science 276:712-718. [DOI] [PubMed] [Google Scholar]
- 51.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 52.Shatalin, K. Y., and A. A. Neyfakh. 2005. Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol. Lett. 245:315-319. [DOI] [PubMed] [Google Scholar]
- 53.Silver, L. L. 2006. Does the cell wall of bacteria remain a viable source of targets for novel antibiotics? Biochem. Pharmacol. 71:996-1005. [DOI] [PubMed] [Google Scholar]
- 54.Silver, L. L. 2003. Novel inhibitors of bacterial cell wall synthesis. Curr. Opin. Microbiol. 6:431-438. [DOI] [PubMed] [Google Scholar]
- 55.Stepanov, A. V., L. I. Marinin, A. P. Pomerantsev, and N. A. Staritsin. 1996. Development of novel vaccines against anthrax in man. J. Biotechnol. 44:155-160. [DOI] [PubMed] [Google Scholar]
- 56.Sterne, M. 1946. Avirulent anthrax vaccine. Onderstepoort J. Vet. Sci. Anim. Ind. 21:41-43. [PubMed] [Google Scholar]
- 57.Taal, M. A., S. E. Sedelnikova, S. N. Ruzheinikov, P. J. Baker, and D. W. Rice. 2004. Expression, purification and preliminary X-ray analysis of crystals of Bacillus subtilis glutamate racemase. Acta Crystallogr. D Biol. Crystallogr. 60:2031-2034. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, M., Y. Kato, and S. Kinoshita. 1961. Glutamic acid racemase from Lactobacillus fermenti. Purification and properties. Biochem. Biophys. Res. Commun. 4:114-117. [DOI] [PubMed] [Google Scholar]
- 59.Tanner, M. E., and S. Miao. 1994. The synthesis and stability of aziridinoglutamate, an irreversible inhibitor of glutamate racemase. Tetrahedron Lett. 35:4073-4076. [Google Scholar]
- 60.Uchida, I., J. M. Hornung, C. B. Thorne, K. R. Klimpel, and S. H. Leppla. 1993. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. J. Bacteriol. 175:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yagasaki, M., K. Iwata, S. Ishino, M. Azuma, and A. Ozaki. 1995. Cloning, purification, and properties of a cofactor-independent glutamate racemase from Lactobacillus brevis ATCC 8287. Biosci. Biotechnol. Biochem. 59:610-614. [DOI] [PubMed] [Google Scholar]
- 62.Yoshimura, T., M. Ashiuchi, N. Esaki, C. Kobatake, S. Y. Choi, and K. Soda. 1993. Expression of glr (murI, dga) gene encoding glutamate racemase in Escherichia coli. J. Biol. Chem. 268:24242-24246. [PubMed] [Google Scholar]