Abstract
The virR operon, located on the virulence plasmid of the intracellular pathogen Rhodococcus equi, contains five genes, two of which (virR and orf8) encode transcriptional regulators. The first gene of the operon (virR), encoding a LysR-type transcriptional regulator, is transcribed at a constitutive low level, whereas the four downstream genes are induced by low pH and high growth temperature. Differential regulation of the virR operon genes could not be explained by differential mRNA stability, as there were no major differences in mRNA half-lives of the transcripts representing each of the five genes within the virR operon. Transcription of virR is driven by the PvirR promoter, with a transcription start site 53 bp upstream of the virR initiation codon. The four genes downstream of virR are transcribed from PvirR and from a second promoter, Porf5, located 585 bp downstream of the virR initiation codon. VirR binds to a site overlapping the initiation codon of virR, resulting in negative autoregulation of the virR gene, explaining its low constitutive transcription level. The Porf5 promoter is induced by high temperature and low pH, thus explaining the observed differential gene expression of the virR operon. VirR has a positive effect on Porf5 activity, whereas the response regulator encoded by orf8 is not involved in regulating transcription of the virR operon. The PvirR promoter is strikingly similar to those recognized by the principal sigma factors of Streptomyces and Mycobacterium, whereas the Porf5 promoter does not share sequence similarity with PvirR. This suggests that Porf5 is recognized by an alternative sigma factor.
The mycolic acid-containing actinomycete Rhodococcus equi is an intracellular pathogen of macrophages and the causative agent of foal pneumonia. Although foals are the primary host of this pathogen, other animals, including pigs, goats, and cattle, are sporadically infected. In addition to these animal hosts, R. equi infections are increasingly encountered in immunocompromised humans, in particular in those diagnosed with AIDS (20, 24). Virulence relies on the ability of R. equi to replicate inside macrophages (13), a process which is dependent on the pathogen interfering with endosomal maturation following phagocytosis and preventing acidification of the vacuole in which it resides (9, 37, 38). Eventually, intracellular proliferation of the pathogen leads to necrosis of the macrophage, accompanied by massive damage to lung tissue characterized by cavitation and granuloma formation (18, 20, 24).
All R. equi strains isolated from foals harbor an 81-kb plasmid that has been shown to be essential for intracellular replication and virulence (10, 35, 36). This virulence plasmid contains a region of 27.5 kb that is characterized by a lower G+C content than the remainder of the virulence plasmid and the R. equi genome, indicating that it probably was acquired via lateral gene transfer (33). It was proposed that this region represents a pathogenicity island, a hypothesis that was lent credence by the fact that the transcription of genes located within it is upregulated following phagocytosis of R. equi (25, 33). Among these is a group of genes encoding a family of small proteins, one of which, virulence-associated protein A (VapA), has been shown to be a virulence factor (14).
Transcriptional regulation of pathogenicity island genes is subject to at least six different environmental parameters, including temperature and pH (3, 25, 32, 34). In contrast to this apparent regulatory complexity, only two genes encoding transcriptional regulators, located in a five-cistron operon (the virR operon, containing virR, orf5, vapH, orf7, and orf8), have been identified in the pathogenicity island (28, 33). It therefore is likely that regulators encoded by the R. equi genome play an important role in controlling the expression of pathogenicity island genes. This may be accomplished, at least in part, through controlling the expression levels of the two transcriptional regulators within the pathogenicity island. The virR gene encodes a LysR-type transcriptional regulator (LTTR) that is required for transcription of the vapA gene (28). The orf8 gene encodes a response regulator; interestingly, a cognate sensor kinase protein is not encoded on the virulence plasmid (33). Disruption of either gene attenuates R. equi, demonstrating their importance in controlling virulence (26). A recent report describing a low level of constitutive transcription of the virR gene while the transcript levels of the four downstream genes responded to changes in growth conditions (25) appeared to contradict our previous finding that the five genes constitute an operon (28). Considering the importance of the virR operon in controlling the virulence of R. equi, this apparent paradox was examined further. This paper reports that the observed difference in regulation of the virR operon genes is not due to differential mRNA stability of the virR operon transcript but to the presence of a regulated promoter within the virR gene driving transcription of orf5, vapH, orf7, and orf8.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or oligonucleotide | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| E. coli BL21(DE3)pLysE | F−ompT hsdSB(rB− mB−) dcm gal λ(DE3) (pLysE) Cmr | 31 |
| R. equi ATCC 33701 | Virulent strain with 81-kb virulence plasmid p33701 | American Type Culture Collection |
| R. equi ATCC 33701 (P−) | Avirulent strain; virulence plasmid cured | 35 |
| R.equi orf8 mutant | orf8 deletion | 26 |
| Plasmids | ||
| pBluescript II KS(+) | Apr αlacZ′ | Stratagene |
| pREV5 | Aprr oriV(pMF1) oriV(ColE1) | 19 |
| pBlueRegP1 | pBluescript KS with 6,680-bp fragment containing orf3 to orf8 from the R. equi ATCC 33701 virulence plasmid | 28 |
| pET3bvirRhis | pET3b with 931-bp fragment containing virR with a six-His tag at the 3′ end | 28 |
| pJOE814.2 | Ampr ColE1 ori xylE reporter gene | 23 |
| pREV6 | pREV5 with t tag located within two terminators derived from pJOE814.2 | This study |
| pORF3PEX | pREV6 with a 2,705-bp DNA fragment containing promoters PvirR and Porf5, virR, and orf5 | This study |
| pVirRT | pREV6 with a 2,183-bp DNA fragment containing Porf5, virR, and orf5 | This study |
| p004 | pREV6 with a 1,840-bp DNA fragment containing virR and orf5 | This study |
| pVirRT-LysR | pVirRT with virR located outside the transcriptional terminators | This study |
| pInterVir | pBluescript KS with a 459-bp DNA fragment containing the virR-orf3 intergenic region | This study |
| pCodingVir | pBluescript KS with an 846-bp DNA fragment containing 690 bp of the virR coding region | This study |
| pREV531 | pREV5 with a 1.5-kb DNA fragment containing orf3 and virR′ derived from pBlueRegP1 | This study |
| pREV5341 | pREV5 with a 3.1-kb DNA fragment containing orf3, virR, and orf5′ derived from pBlueRegP1 | This study |
Media and growth conditions.
Escherichia coli DH5α (Bethesda Research Laboratories) and E. coli BL21(DE3)pLysE (Novagen) were used for general cloning procedures and for expression of virR-his, respectively. Bacterial strains were grown in Luria-Bertani (LB) broth (29). Growth of R. equi at 37°C and pH 6.5 (inducing conditions) was used to induce virulence plasmid gene expression, whereas these genes were transcribed at low levels following growth at 30°C and pH 8.0 (noninducing conditions). Where appropriate, the following supplements were added: kanamycin, 50 μg ml−1 (E. coli) or 200 μg ml−1 (R. equi); ampicillin, 50 μg ml−1; apramycin, 30 μg ml−1 (E. coli) or 80 μg ml−1 (R. equi); chloramphenicol, 30 μg ml−1; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 20 μg ml−1; isopropyl-β-d-thiogalactopyranoside (IPTG), 0.1 mM. For solid medium, agar was added at 1.5% (wt/vol).
DNA manipulations.
Chromosomal DNA was isolated as described previously (28). Plasmid DNA was isolated via the alkaline lysis method of Birnboim and Doly (5) or by using a Wizard Plus SV miniprep kit as described by the manufacturer (Promega). DNA fragments were isolated from agarose gels by using a Genelute DNA purification kit as described by the manufacturer (Sigma-Aldrich). PCR was carried out using Taq DNA polymerase (Promega) or Deep Vent DNA polymerase (New England Biolabs) as described by the manufacturer. Other DNA manipulations were done in accordance with standard protocols (29).
Plasmid construction.
A region located on plasmid pJOE814.2 containing two transcriptional terminators and a xylE gene was amplified with Deep Vent DNA polymerase, using primers XylF and XylR. This 1,424-bp fragment was cloned into DraI-digested pREV5 to make pREV81. The xylE gene was subsequently excised by digesting pREV81 with ClaI, followed by religation of the 4,948-bp fragment, resulting in pREV6.
A series of DNA fragments serially shortened from the 5′ end was constructed by PCR with the oligonucleotide BCMRev in conjunction with either ORF3PEX2, VirRT1, or 004F, using pBlueRegP1 as a template. These fragments were ligated into the unique EcoRV restriction site of pREV6 to yield pORF3PEX, pVirRT, and p004, respectively. An intact virR gene was amplified from pBlueRegP1 by using oligonucleotides 005R and Orf3BlnI. The resulting 1,717-bp product was digested with BlnI, and the 1,296-bp fragment was cloned into SpeI-digested pVirRT.
pInterVir was constructed by amplifying a 459-bp fragment containing the orf3-virR intergenic region from pBlueRegP1, using oligonucleotides GM-Lys-F and GM-Lys-R; this fragment was subsequently ligated into EcoRV-digested pBluescript II KS. pCodingVir was constructed by amplifying an 846-bp fragment containing the virR coding region, using oligonucleotides VirRT1 and −70−50REV; this fragment was subsequently ligated into EcoRV-digested pBluescript II KS. The sequences of the oligonucleotides used in the construction of these plasmids are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′ to 3′) | Purpose | Reference |
|---|---|---|---|
| XylF | TATATAACTAGTGCCTCTTCGCTATT | Plasmid construction | This study |
| XylR | TATAGGCCGGCCCGATTCATTAATGCA | Plasmid construction | This study |
| BCMRev | GTTATCTAGGGGCAGGCGACAG | Plasmid construction | This study |
| VIRRT1 | CGCATTGAACGACAGGTTG | Plasmid construction | This study |
| ORF3PEX2 | TGACGAGCACCAATGTTTTC | Plasmid construction | This study |
| GM-Lys-R | CGGCTGCCGCAATATGAC | Plasmid construction | This study |
| GM-Lys-F | CACCCCAACCTGTCGTTC | Plasmid construction | This study |
| −70−50 REV | GACCACCTGCGATGCGTGAT | Plasmid construction | This study |
| GyrCOOH1 | GTCGAGCAGGGTCAAGTGTA | Quantification of gyrB | This study |
| GyrCOOH25′ | AGCTCCTTGGCGTTCATCT | Quantification of gyrB | This study |
| 16SrRNAF200 | ACGAAGCGAGAGTGACGGTA | Quantification of 16S rRNA | 21 |
| 16SrRNAR200 | ACTCAAGTCTGCCCGTATCG | Quantification of 16S rRNA | 21 |
| 003F | GTTTCGTCTTCCACCGTCTT | Quantification of orf3 | This study |
| 003R | AGCCTTATCGTCGCAACTGT | Quantification of orf3 | This study |
| 004F | CGGACGAGTTCGACTGGTAT | Quantification of virR | This study |
| 004R | CAAAGACGATTTGGGGTACG | Quantification of virR | This study |
| 005F | CTCTTCCTGATCGGAGTTGC | Quantification of orf5 | This study |
| 005R | GAGTCGCAGACGAGGTAAGC | Quantification of orf5 | This study |
| 006F | AGGGTTATGCAGGTGGATTG | Quantification of vapH | This study |
| 006R | TACCGATTACGGAGCTCACC | Quantification of vapH | This study |
| 007NF | ATGCACTCCCTGAAAACTATC | Quantification of orf7 | This study |
| 007NR | GGTGGGCTGGATTGACGCGCA | Quantification of orf7 | This study |
| 008F | GAACAACTGGGAATGGTGGT | Quantification of orf8 | This study |
| 008R | GTTCGCCGTTTCTAGACGAA | Quantification of orf8 | This study |
| D4-VIR5022 | GATGTGCAGGGCGTCAGC | Primer extension | This study |
| VIR5022 | GATGTGCAGGGCGTCAGC | Sequencing | This study |
| D4-2IntPex5564 | GGTACGACGCCAGCAGCCGC | Primer extension | This study |
| 2IntPex5564 | GGTACGACGCCAGCAGCCGC | Sequencing | This study |
Electroporation of R. equi.
Plasmids were introduced into R. equi strains by electroporation as described previously (19).
RNA isolation and real-time RT-PCR.
RNAs were isolated from R. equi as described previously (28). Reverse transcriptase (RT) reactions using random primers (Promega) were performed with 1 U Improm II RT following the manufacturer's recommendations, with 100 ng of total RNA as the template in a final volume of 20 μl. The product was subsequently amplified using a QuantiTect SYBR green real-time kit following the manufacturer's instructions (QIAGEN). Reaction mixtures were subjected to 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s in a LightCycler instrument (Roche) with a temperature transition rate of 20°C/s. Melting curve analysis was performed at 50 to 95°C (temperature transition rate, 0.2°C/s), with stepwise fluorescence detection following amplification. Cycle threshold (CT) values were obtained and used to calculate the number of RNA copies per μg of total RNA, using a standard curve of known amounts of DNA target with r2 coefficients of >0.997 in the range of 5 × 103 to 5 × 108 molecules per reaction. gyrB mRNA was used as a housekeeping gene to compare the amounts of RNA in each reaction. The data reported in this paper represent the results of three independent experiments in which each sample was analyzed in duplicate. The sequences of oligonucleotides used for real-time PCR are listed in Table 2.
Fluorescent primer extension and DNA sequencing.
The D4-labeled oligonucleotides D4-VIR5022 and D4-2IntPex5564 (Table 2), complementary to sequences 62 to 80 bp and 624 to 643 bp, respectively, downstream of the initiation codon of virR, were used in primer extension reactions to determine the transcriptional start sites of the virR operon. Total RNA (2 μg) and 1 μM of D4-VIR5022 were incubated at 70°C for 5 min, followed by reverse transcription at 42°C for 60 min, using 5 U of Superscript III RT in a volume of 20 μl as recommended by the manufacturer (Invitrogen). After treatment of the sample with 20 μg of RNase A at 37°C for 30 min, cDNA was precipitated and dissolved in 12 μl of nuclease-free water. The primer extension product (0.5 ng) was combined with 0.5 μl of DNA size standard kit 600 (Beckman Coulter) and 40 μl of CEQ sample loading solution (Beckman Coulter) and analyzed with a CEQ 8000 fragment analysis system on a CEQ 8000 DNA sequencer (Beckman Coulter). In addition, dideoxy sequencing reactions using either VIR5022 or 2IntPex5564 (Table 2) and 60 ng of NheI-digested pORF3PEX (Table 1) were performed using a CEQ DCTS kit as described by the manufacturer (Beckman Coulter). The D4-labeled primer extension products (50 pg) were added to the sample prior to analyzing the sequence on a CEQ 8000 DNA sequencer to identify transcriptional start sites.
mRNA half-life determination.
RNAs were isolated from R. equi in the mid-logarithmic phase of growth (optical density at 600 nm = 0.5) at selected intervals following inhibition of transcription by the addition of 200 μg ml−1 rifampin (Sigma). The number of mRNA copies was determined using real-time RT-PCR, followed by linear regression to determine the half-life. 16S rRNA was used for internal normalization in this study.
EMSA.
Expression and purification of virR-his were done as previously described (28). A DNA fragment containing the virR promoter region, pInterVir, was digested with EcoRI and HindIII and labeled with [α-32P]dATP, using Klenow DNA polymerase as described previously (28). The labeled fragment was subsequently purified using a QIAquick PCR purification kit according to the manufacturer's instructions (QIAGEN). Radiolabeled DNA fragments (2 ng) were incubated with purified VirR-His at 30°C for 30 min in electrophoretic mobility shift assay (EMSA) binding buffer, 20 μg of bovine serum albumin, and 1 μg of poly(dI-dC) DNA (Amersham Biosciences) in a volume of 20 μl. The samples were separated by electrophoresis in a prerun 5% nondenaturing polyacrylamide gel containing TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA) and run at 4°C and 10 V cm−1. Following drying, the gel was analyzed by autoradiography.
DNA restriction protection assay.
A 459-bp fragment containing the virR promoter region was amplified using the oligonucleotides GM-Lys-F and GM-Lys-R, with pInterVir as the template. To obtain a negative control DNA region, an 846-bp fragment was amplified using the oligonucleotides VirRT1 and −70−50REV. DNA fragments (500 ng) were incubated with purified VirR-His at 30°C for 30 min in EMSA binding buffer. HincII (10 units) was added to each sample, which was further incubated at 30°C for 30 min. The samples were separated by electrophoresis and visualized using SYBR green (Sigma) per the manufacturer's instructions.
RESULTS
Transcription of the virR operon is controlled by temperature and pH.
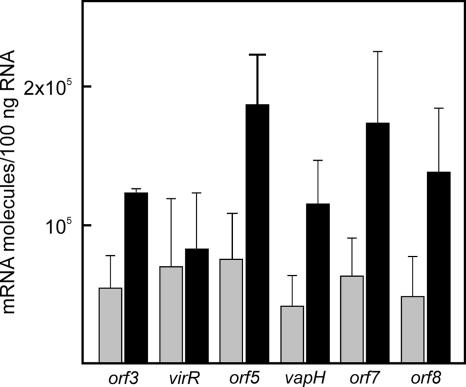
Using a microarray to study the transcription levels of pathogenicity island genes, Ren and Prescott (25) showed that the transcription of four genes (orf5, vapH, orf7, and orf8) downstream of virR, but not that of virR itself, is upregulated when cells are grown at pH 6.5 and 37°C compared to that at pH 8.0 and 30°C. Since this observation appeared to contradict our previous findings that these genes are organized in an operon (28), mRNAs from R. equi grown under these two conditions were isolated and quantified using real-time RT-PCR (Fig. 1). The transcription levels of the four genes downstream of virR were significantly increased (threefold) when cells were grown at pH 6.5 and 37°C compared to those for cells grown at pH 8.0 and 30°C. In contrast, there was no significant difference in transcription levels of virR, the first gene of the operon, thus confirming the earlier microarray data (25).
FIG. 1.
Regulation of virR operon gene transcription by temperature and pH. mRNAs were isolated from Rhodococcus equi cells grown under noninducing (30°C, pH 8.0) or inducing (37°C, pH 6.5) growth conditions, followed by absolute quantification of the mRNA molecules using real-time RT-PCR. Transcription levels are indicated for each gene of the virR operon (virR, orf5, vapH, orf6, orf7, and orf8) and the divergently transcribed orf3 gene following growth under noninducing (gray bars) and inducing (black bars) conditions.
mRNA stability of the virR operon transcript.
Although virR, orf5, vapH, orf7, and orf8 are organized in an operon (28), their transcription is not coordinately regulated (25). A possible explanation for this apparent paradox may be that mRNAs derived from orf5, vapH, orf7, and orf8 are more stable than that of virR, allowing the former four transcripts to accumulate. To examine this possibility, the mRNA half-lives of these genes were determined following inhibition of transcription by the addition of rifampin (Table 3). The half-lives of the transcripts representing each of the five genes within the virR operon varied from 1.3 to 2.3 min. Since the virR transcript had a half-life of 1.8 min, differential mRNA stability of the virR operon transcript does not account for the observed differences in transcript levels of virR and its four downstream genes.
TABLE 3.
Transcript half-lives of genes within the virR operon
| Gene | Half-life (min) |
|---|---|
| virR | 1.8 ± 0.2 |
| orf5 | 1.3 ± 0.3 |
| vapH | 2.3 ± 0.3 |
| orf7 | 2.2 ± 0.1 |
| orf8 | 1.3 ± 0.1 |
The virR operon contains an internal promoter regulated by pH and temperature.
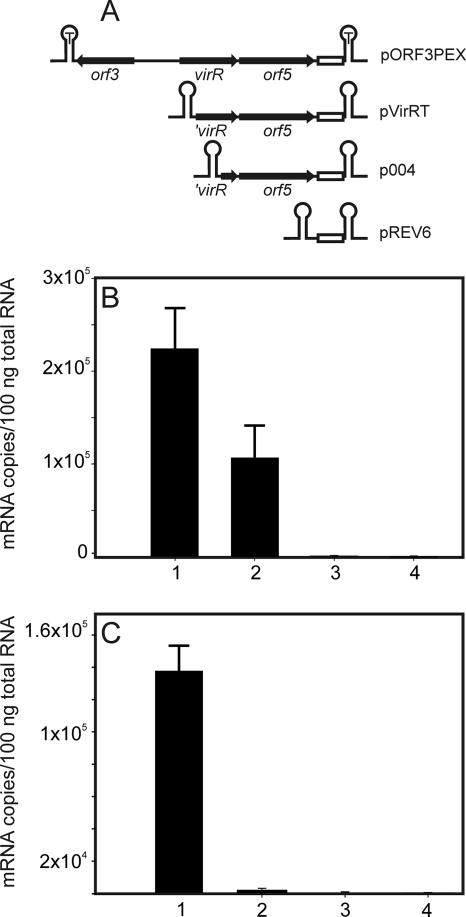
An alternative explanation for the observed differential regulation of the virR operon genes is the presence of a promoter located downstream of the virR operon promoter (PvirR) driving transcription of the orf5, vapH, orf7, and orf8 genes. Transcription levels were analyzed using the promoter probe vector pREV6, which contains a small DNA fragment (t tag) located between two transcriptional terminators which is not present in R. equi (Fig. 2A). Since there is no promoter, transcription of the t tag does not occur (Fig. 2B). Introduction of a DNA fragment containing the orf3-virR intergenic region as well as orf5 into pREV6 resulted in transcription of the t tag, demonstrating the presence of an active promoter. Deletion of the orf3-virR intergenic region, including the 5′ end of virR (1 to 111 bp), reduced but did not abolish transcription of the t tag. Transcription of the t tag was completely abolished when a larger 5′ fragment (1 to 404 bp) of virR was deleted, demonstrating that a promoter (Porf5) or sequences required for its activity are located between bp 111 and 404 of the virR gene (Fig. 2B).
FIG. 2.
Determination of PvirR and Porf5 promoter activities by real-time RT-PCR. (A) Schematic representation of inserts in the promoter probe vector pREV6. The hairpin structure marked with a “T” represents the transcriptional terminators. Genes and their direction of transcription are indicated by black arrows. The open white box represents the t tag. The transcription level of the t tag was measured by real-time RT-PCR. (B) mRNA transcript levels of the t tag in R. equi cells harboring pORF3PEX, containing both PvirR and Porf5 (1); pVirRT, containing Porf5 (2); p004 (3); or pREV6 (4). (C) mRNA transcript levels of the t tag in R. equi cells harboring pVirRT containing only Porf5 grown at 37°C and pH 6.5 (1) or at 30°C and pH 8.0 (2) and in R. equi cells harboring pREV6 grown at 37°C and pH 6.5 (3) or 30°C and pH 8.0 (4).
To examine whether the internal promoter Porf5 is regulated by temperature and pH, the transcription levels of the t tag were examined following growth of R. equi(pVirRT) under either inducing (pH 6.5, 37°C) or noninducing (pH 8.0, 30°C) growth conditions. Transcription of the t tag increased 67-fold under inducing compared to noninducing conditions, whereas transcription of the t tag did not occur from pREV6 (Fig. 2C).
Mapping of the transcriptional start sites of the virR operon.
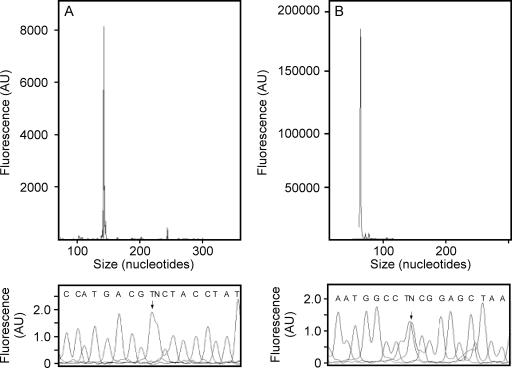
To determine the transcriptional start site of the virR gene, a primer extension reaction using the fluorescently labeled oligonucleotide D4-VIR5022, complementary to the virR gene, was carried out. Using mRNA isolated from R. equi grown under inducing conditions as the template, a 133-bp DNA fragment was observed (Fig. 3A). The transcriptional start site of PvirR was subsequently determined to be an adenine located 53 bp upstream of the virR initiation codon (Fig. 3A and 4D). Interestingly, the −10 sequence (nucleotides −12 to −7 [TAGCAT]) of the PvirR promoter is strikingly similar to that of the consensus σhrdB promoter (TAGART), which is recognized by the principal sigma factor of Streptomyces coelicolor (7, 15). However, there is no clear similarity in the −35 region to the consensus σhrdB promoter (TTGACA). Using the same approach, the internal transcriptional start site of Porf5 was mapped to a guanidine 585 bp downstream of the virR initiation codon (Fig. 3B and 4D). The PvirR and Porf5 promoters do not share any obvious sequence similarity in either the −10 or −35 area.
FIG. 3.
Determination of transcriptional start sites of the virR operon of R. equi. Fluorescent primer extension was carried out with a Cy5-labeled primer and 5 μg of total cellular RNA extracted from R. equi cells grown under inducing conditions (37°C and pH 6.5). The upper panels show the D4-labeled primer extension products combined with DNA size standards and analyzed with a CEQ 8000 fragment analysis system. The lower panels show dideoxy sequencing reactions spiked with the D4-labeled primer extension products. The arrows indicate the transcriptional start sites where the D4-labeled cDNA and sequencing products overlapped. (A) Determination of the PvirR transcriptional start site, using D4-VIR5022. (B) Determination of the Porf5 transcriptional start site, using D4-2IntPex5564. AU, arbitrary units.
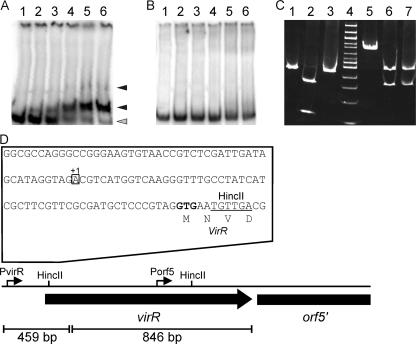
FIG. 4.
Analysis of VirR DNA binding. Various concentrations of VirR were incubated with 2 ng of radiolabeled DNA containing (A) the PvirR promoter region in a 459-bp DNA fragment or (B) the Porf5 promoter region in an 846-bp DNA fragment. The amount of protein added to each lane was as follows: lanes 1, radiolabeled DNA fragment only; lanes 2, 50 ng VirR-His; lanes 3, 100 ng VirR-His; lanes 4, 200 ng VirR-His; lanes 5, 300 ng VirR-His; and lanes 6, 400 ng VirR-His. The reaction volume was 20 μl. Protein-DNA complexes are indicated with black arrowheads. Nonbound DNA is indicated with a gray arrowhead. (C) HincII restriction protection assay. DNAs were incubated with VirR and HincII, followed by analysis of the restriction digest by gel electrophoresis. Lanes 1 to 3 contain a 459-bp DNA fragment with PvirR, and lanes 5 and 6 contain an 846-bp DNA fragment with Porf5. Lanes 1 and 5, no VirR or HincII added; lanes 2 and 6, HincII added; lanes 3 and 7, HincII and VirR added. (D) Schematic representation of the 5′ end of the virR operon. Genes and the direction of transcription are indicated by black arrows. The HincII sites used in the restriction protection assay and the positions of PvirR and Porf5 are indicated above the arrows. The nucleotide sequence of the sequence upstream of virR containing the PvirR promoter is shown. The PvirR transcriptional start site is indicated (+1).
VirR binds to the orf3-virR intergenic region.
To determine whether VirR interacts with the PvirR and Porf5 promoters, band shift experiments were carried out. Incubation of purified VirR-His with a 459-bp radiolabeled DNA fragment containing the orf3-virR intergenic region resulted in retardation of the DNA fragment. At low VirR-His concentrations, a single retarded band was visible, whereas a second retarded band became visible at higher concentrations (Fig. 4A, lane 5). In contrast, VirR-His failed to bind to a DNA fragment containing only the internal Porf5 promoter (Fig. 4B).
To further corroborate that VirR-His binds to a DNA fragment containing the PvirR promoter, a restriction enzyme protection assay was carried out. The 459-bp DNA fragment containing the orf3-virR intergenic region was incubated with VirR-His and subsequently subjected to digestion with HincII, whose recognition site is located 59 to 64 bp downstream of the PvirR transcription initiation site. While VirR-His protected this 459-bp DNA fragment from HincII digestion, it failed to prevent digestion by HincII of the adjacent 846-bp DNA fragment. These data show that the VirR binding site overlaps the HincII site downstream of PvirR within the virR coding region (Fig. 4C).
VirR is a negative regulator of the virR gene.
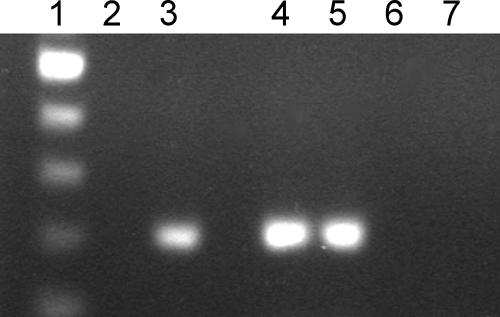
The results of the restriction enzyme protection assay show that VirR protects a HincII site located 59 to 64 bp downstream from the PvirR transcriptional start site. This strongly suggests that VirR acts as a repressor of PvirR driving the transcription of virR. To examine this possibility, plasmids with DNA fragments containing either orf3, an intact virR gene and the 5′ end of orf5 (pREV5341), or orf3 and the 5′ end of virR (pREV531) were electroporated into R. equi P−. Transcription of virR was subsequently analyzed by RT-PCR, using primers that amplified a virR fragment located upstream of Porf5, thus analyzing only the activity of PvirR. While transcription of virR was below the detection limit in strains harboring pREV5341 (Fig. 5, lane 2), it was clearly detectable in the absence of an intact virR gene (pREV531) (Fig. 5, lane 3).
FIG. 5.
Autoregulation of virR transcription. mRNAs were isolated from R. equi P− harboring either pREV531 (orf3-virR′) or pREV5341 (orf3-virR-orf5′). virR and gyrB transcripts were subsequently detected by RT-PCR as 200-bp amplification products. The oligonucleotides used for this experiment (Table 2) amplified a region upstream of Porf5. Lane 1, 100-bp marker; lane 2, R. equi(pREV5341) (virR); lane 3, R. equi(pREV531) (virR); lane 4, R. equi(pREV5341) (gyrB); lane 5, R. equi(pREV531) (gyrB); lanes 6 and 7, RT-PCR of gyrB genes of R. equi(pREV5341) and R. equi(pREV531), respectively, without RT.
VirR is a positive regulator of the orf5-orf8 cluster.
Since the orf5 and orf8 genes are transcribed from the PvirR promoter as well as the Porf5 promoter, it is likely that VirR is involved in transcription from Porf5 as well as that from PvirR. To investigate this possibility, pVirRT, containing Porf5 and the 3′ end of virR, was introduced into R. equi P−. The transcription levels of the t tag in this strain were 14-fold lower than those in the wild-type strain, indicating that the presence of virulence plasmid genes is required for wild-type transcription levels. The transcription levels were restored only partially (twofold increase) following introduction of an intact virR gene into pVirRT (but outside the transcriptional terminators of pREV6), demonstrating that VirR is required but not sufficient for full activity of the Porf5 promoter.
orf8 does not regulate transcription of the virR operon.
The orf8 gene encodes a response regulator that probably interacts with an as yet unidentified chromosomally encoded sensor kinase protein to regulate virulence plasmid gene expression. To determine whether orf8 is involved in transcriptional regulation of the virR operon, the transcription levels of orf5 were compared in the wild-type strain and an orf8 disruption mutant. The orf5 transcription level in the wild-type strain was 5.6 × 105 molecules mRNA/100 ng RNA ± 7.6 × 104 molecules mRNA/100 ng RNA, whereas this level was 4.7 × 105 molecules mRNA/100 ng RNA ± 5.6 × 104 molecules mRNA/100 ng RNA for the R. equi orf8 mutant, indicating that orf8 does not play a role in regulating transcription of the virR operon.
DISCUSSION
The virR operon contains two genes encoding transcriptional regulators which are required for the virulence of R. equi; one of these, the LTTR protein VirR, is required for expression of the virulence factor VapA (26, 28). The transcriptional organization of virR is unusual in that it is the first gene in a five-cistron operon (28); in most instances, LTTR-encoding genes are transcribed as monocistronic transcripts. In addition, an analysis of virulence plasmid transcript levels following growth under a variety of conditions showed that virR is transcribed at a low and constant level, whereas the transcription of the downstream genes is regulated (25). The aim of this paper was to analyze the transcriptional regulation of the virR operon in order to explain the observed differential regulation of virR compared to the four downstream genes.
Many LTTRs autoregulate their expression by acting as a repressor of the LTTR-encoding gene (30). The data presented here show that in this respect, virR behaves as a typical LTTR. Inactivation of virR in a virulence plasmid-free background resulted in a dramatic increase of virR transcription, which is consistent with the notion that VirR acts as a repressor of virR transcription. This is supported by the finding that VirR binds to a site that overlaps a HincII restriction site located 59 to 64 bp downstream of PvirR within the 5′ end of the virR gene, which is a typical location for a repressor binding site (8). The VirR autoregulatory circuit thus results in a constant low level of transcription of the virR operon from the PvirR promoter, independent of growth temperature and pH.
Differential mRNA stability within a polycistronic transcript has been shown to lead to vastly different mRNA levels for individual genes within an operon, resulting in differential gene expression (11, 17). This could account for the observed increase in orf5, vapH, orf7, and orf8 transcripts under inducing conditions. However, the half-life of virR mRNA (1.8 min) was the same as the average half-life of the transcripts of all five genes within the virR operon and therefore does not account for the observed differential regulation of the four genes downstream of virR in the virR operon. The observed short half-lives are typical of the majority of bacterial transcripts, as recently shown for Escherichia coli and Bacillus subtilis, where over 80% of transcripts are unstable, with half-lives of <8 min (4, 12).
Deletion analysis of the virR operon showed that in addition to the PvirR promoter, transcription of the four genes downstream of virR is also driven by a second promoter located within the virR gene. In contrast to PvirR, the activity of the Porf5 promoter is regulated by temperature and pH, resulting in a 67-fold increase in transcript levels following growth under inducing conditions compared to those obtained under noninducing growth conditions. Interestingly, the activity of the Porf5 promoter in a virulence plasmid-free R. equi strain was significantly lower than that in the presence of the virulence plasmid, suggesting that one or more components encoded by the virulence plasmid are required for full activity of Porf5. One of these components is the virR gene, as inclusion of this gene on the plasmid containing Porf5 resulted in a twofold higher activity, indicating that virR is required but not sufficient for full activity of Porf5.
The p004 plasmid contains the Porf5 promoter and its upstream region (−140 nucleotides) but did not display any promoter activity, indicating that a far upstream region required for Porf5 activity had been deleted. The requirement for far upstream sequences for promoter activity has been shown for many bacterial systems, for example, Pseudomonas aeruginosa and Mycobacterium tuberculosis (22, 27). Often these involve two-component regulatory systems interacting with either σ54- or σ70-type RNA polymerases (6, 8). However, disruption of orf8, encoding a response regulator, had no significant effect on orf5 transcription levels, showing that it is not involved in controlling transcription of the virR operon under the experimental conditions used. This strongly suggests that genome-encoded transcriptional regulators are involved in regulating the activity of the Porf5 promoter, and therefore the expression levels of the response regulator Orf8.
The −10 regions of promoters recognized by the principal sigma factors of Streptomyces coelicolor, Mycobacterium smegmatis, and M. tuberculosis (HrdB and MysA) are highly conserved (2, 7, 15). Interestingly, the −10 regions of PvirR and the previously characterized vapA promoter (28) are highly similar to those recognized by HrdB and MysA, indicating that these promoters are recognized by the main principal sigma factor of R. equi. In contrast, the −10 and −35 sequences of PvirR and Porf5 do not share any obvious sequence similarities, strongly suggesting that these two promoters are recognized by different sigma factors. The involvement of alternative sigma factors in the regulation of virulence factor expression, as proposed here, is increasingly being observed (1, 16). The R. equi genomic sequence was recently completed (http://www.sanger.ac.uk/Projects/R_equi/), and a preliminary genome analysis revealed the presence of at least 20 potential sigma factor-encoding genes (R. J. Fahey and W. G. Meijer, unpublished results). We are currently analyzing the functions of these sigma factors.
The model that emerges from these studies is that the virR operon is transcribed at a low constitutive level under noninducing conditions from the PvirR promoter, which is most likely recognized by the principal sigma factor. The noninducing growth conditions resembled those encountered during saprophytic growth of R. equi, where expression of virulence factors is not required. The constitutive low-level transcription of the virR operon is maintained through binding of VirR to a site overlapping the virR initiation codon, resulting in an autoregulatory circuit. Following a change to inducing growth conditions, which are detected by an as yet unidentified genome-encoded signal transduction pathway, Porf5 becomes active, resulting in a significant increase of orf5, vapH, orf7, and orf8, but not virR, expression. Transcription from Porf5 is dependent on an alternative sigma factor and involves far upstream sequences. The orf8 gene encodes a response regulator which is required for virulence (26) but not for Porf5 activity, whereas the roles of orf5, orf7, and vapH in virulence are not clear. Upregulation of Porf5 activity by inducing growth conditions may act as a master switch, resulting in increased expression of the response regulator Orf8, leading to subsequent full induction of the virulence genes. The validity of this model for the regulation of virulence gene expression is currently under investigation.
Acknowledgments
We thank Shinji Takai and John Prescott for making available the virulence plasmid-cured strain of R. equi and the R. equi orf8 mutant, respectively, and Gesche S. Heiss for providing us with pJOE814.2.
This work was supported by Science Foundation Ireland under grant 02/IN.1/B203. D.A.R. was supported by a grant from the Health Research Board to W.G.M.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Bashyam, M. D., and S. E. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301-308. [DOI] [PubMed] [Google Scholar]
- 2.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2002. H2O2, which causes macrophage-related stress, triggers induction of expression of virulence-associated plasmid determinants in Rhodococcus equi. Infect. Immun. 70:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening of recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, W. C., and R. G. Kranz. 1998. A bacterial ATP-dependent, enhancer binding protein that activates the housekeeping RNA polymerase. Genes Dev. 12:1884-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buttner, M. J., K. F. Chater, and M. J. Bibb. 1990. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 172:3367-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collado-Vides, J., B. Magasanik, and J. D. Gralla. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55:371-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Mora, E., M. Polidori, A. Lührmann, U. E. Schaible, and A. Haas. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635-653. [DOI] [PubMed] [Google Scholar]
- 10.Giguère, S., M. K. Hondalus, J. A. Yager, P. Darrah, D. M. Mosser, and J. F. Prescott. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect. Immun. 67:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 12.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 13.Hondalus, M. K., and D. M. Mosser. 1994. Survival and replication of Rhodococcus equi in macrophages. Infect. Immun. 62:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain, S., B. R. Bloom, and M. K. Hondalus. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. Mol. Microbiol. 50:115-128. [DOI] [PubMed] [Google Scholar]
- 15.Kang, J. G., M. Y. Hahn, A. Ishihama, and J. H. Roe. 1997. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Res. 25:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klug, G. 1993. The role of mRNA degradation in the regulated expression of bacterial photosynthesis genes. Mol. Microbiol. 9:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Lührmann, A., N. Mauder, T. Sydor, E. Fernandez-Mora, J. Schulze-Luehrmann, S. Takai, and A. Haas. 2004. Necrotic death of Rhodococcus equi-infected macrophages is regulated by virulence-associated plasmids. Infect. Immun. 72:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangan, M. W., G. A. Byrne, and W. G. Meijer. 2005. Versatile Rhodococcus equi-Escherichia coli shuttle vectors. Antonie Leeuwenhoek 87:161-167. [DOI] [PubMed] [Google Scholar]
- 20.Meijer, W. G., and J. F. Prescott. 2004. Rhodococcus equi. Vet. Res. 35:383-396. [DOI] [PubMed] [Google Scholar]
- 21.Miranda-Casoluengo, R., P. S. Duffy, E. P. O'Connell, B. J. Graham, M. W. Mangan, J. F. Prescott, and W. G. Meijer. 2005. The iron-regulated iupABC operon is required for saprophytic growth of the intracellular pathogen Rhodococcus equi at low iron concentrations. J. Bacteriol. 187:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohr, C. D., D. W. Martin, W. M. Konyecsni, J. R. Govan, S. Lory, and V. Deretic. 1990. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172:6576-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nga, D. P., J. Altenbuchner, and G. S. Heiss. 2004. NpdR, a repressor involved in 2,4,6-trinitrophenol degradation in Rhodococcus opacus HL PM-1. J. Bacteriol. 186:98-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren, J., and J. F. Prescott. 2003. Analysis of virulence plasmid gene expression of intra-macrophage and in vitro grown Rhodococcus equi ATCC 33701. Vet. Microbiol. 94:167-182. [DOI] [PubMed] [Google Scholar]
- 26.Ren, J., and J. F. Prescott. 2004. The effect of mutation on Rhodococcus equi virulence plasmid gene expression and mouse virulence. Vet. Microbiol. 103:219-230. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, E. A., A. Clark, S. McBeth, and R. L. Friedman. 2004. Molecular characterization of the eis promoter of Mycobacterium tuberculosis. J. Bacteriol. 186:5410-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, D. A., G. A. Byrne, E. P. O'Connell, C. A. Boland, and W. G. Meijer. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J. Bacteriol. 186:5576-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 31.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and H. J. W. Dubendorf. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 32.Takai, S., N. Fukunaga, K. Kamisawa, Y. Imai, Y. Sasaki, and S. Tsubaki. 1996. Expression of virulence-associated antigens of Rhodococcus equi is regulated by temperature and pH. Microbiol. Immunol. 40:591-594. [DOI] [PubMed] [Google Scholar]
- 33.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Osaki, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuka, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai, S., M. Iie, Y. Watanabe, S. Tsubaki, and T. Sekizaki. 1992. Virulence-associated 15- to 17-kilodalton antigens in Rhodococcus equi: temperature-dependent expression and location of the antigens. Infect. Immun. 60:2995-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai, S., T. Sekizaki, T. Ozawa, T. Sugawara, Y. Watanabe, and S. Tsubaki. 1991. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect. Immun. 59:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tkachuk-Saad, O., and J. Prescott. 1991. Rhodococcus equi plasmids: isolation and partial characterization. J. Clin. Microbiol. 29:2696-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyooka, K., S. Takai, and T. Kirikae. 2005. Rhodococcus equi can survive a phagolysosomal environment in macrophages by suppressing acidification of the phagolysosome. J. Med. Microbiol. 54:1007-1015. [DOI] [PubMed] [Google Scholar]
- 38.Zink, M. C., J. A. Yager, J. F. Prescott, and M. A. Fernando. 1987. Electron microscopic investigation of intracellular events after ingestion of Rhodococcus equi by foal alveolar macrophages. Vet. Microbiol. 14:295-305. [DOI] [PubMed] [Google Scholar]