Abstract
The AbsA two-component signal transduction system, comprised of the sensor kinase AbsA1 and the response regulator AbsA2, acts as a negative regulator of antibiotic production in Streptomyces coelicolor, for which the phosphorylated form of AbsA2 (AbsA2∼P) is the agent of repression. In this study, we used chromatin immunoprecipitation to show that AbsA2 binds the promoter regions of actII-ORF4, cdaR, and redZ, which encode pathway-specific activators for actinorhodin, calcium-dependent antibiotic, and undecylprodigiosin, respectively. We confirm that these interactions also occur in vitro and that the binding of AbsA2 to each gene is enhanced by phosphorylation. Induced expression of actII-ORF4 and redZ in the hyperrepressive absA1 mutant (C542) brought about pathway-specific restoration of actinorhodin and undecylprodigiosin production, respectively. Our results suggest that AbsA2∼P interacts with as many as four sites in the region that includes the actII-ORF4 promoter. These data suggest that AbsA2∼P inhibits antibiotic production by directly interfering with the expression of pathway-specific regulators of antibiotic biosynthetic gene clusters.
Streptomycetes are gram-positive, soil-dwelling bacteria that possess an abundant secondary metabolism and are the sources of numerous antibiotics, chemotherapeutic agents, immunosuppressants, and other agents in clinical use (10). The members of the Streptomyces genus have been predicted to produce >100,000 distinct secondary metabolites, of which only a fraction have been identified so far (49). One model organism for this genus, Streptomyces coelicolor, produces three chromosomally encoded, well-characterized antibacterial compounds: actinorhodin, undecylprodigiosin, and calcium-dependent antibiotic (CDA). Actinorhodin and undecylprodigiosin exhibit blue and red pigmentations, respectively, providing experimentally useful visual cues for antibiotic production.
Antibiotic biosynthesis is mediated by large contiguous gene clusters ranging in size from a few kilobases to over 100 kilobases (7). Located within these clusters are genes encoding biosynthetic enzymes, resistance determinants, and pathway-specific regulators. There are several types of pathway-specific regulators, the best-characterized being the Streptomyces antibiotic regulatory proteins (SARPs), which are transcriptional activators that contain an N-terminal winged helix-turn-helix motif (52). SARPs have the most direct impact on antibiotic production as they activate transcription of the biosynthetic genes. In S. coelicolor, the SARPs for actinorhodin and undecylprodigiosin production are ActII-ORF4 and RedD, respectively (5, 42). CdaR is a SARP homologue and proposed activator of CDA production though there is no direct evidence for this at present (36). Another pathway-specific regulator is redZ (located in the undecylprodigiosin biosynthetic gene cluster), which encodes a response regulator-like protein that activates the expression of the SARP-encoding redD as well as redD-independent biosynthetic genes involved in undecylprodigiosin production (20, 22, 23). Many secondary metabolites are produced in a growth-phase-dependent manner in both liquid and solid media. In liquid culture, the onset of antibiotic production occurs in the stationary phase, whereas on solid media, it coincides with the formation of a sporulating surface layer of cells called the aerial mycelium, a distinguishing feature of the streptomycetes (6, 7). The cues that trigger antibiotic production are unknown and are likely to be complex. Among probable candidates are nutritional (carbon, nitrogen, and phosphate levels), physiological (cyclic AMP, GTP, and ppGpp levels), and environmental factors (cell density and small diffusible signaling molecules) (6, 7, 10).
In addition to the pathway-specific regulators, which exert their primary effect on one antibiotic at a time, pleiotropic regulators that regulate the production of more than one antibiotic have been identified. Indeed, it has recently been reported that some pathway-specific regulators might also be pleiotropic, adding an unanticipated additional layer of complexity to the regulation of secondary metabolite biosynthesis (23). For example, constitutive expression of redZ not only resulted in the premature increase of redD and redD-independent transcripts but also enhanced the transcription of redD-dependent and act genes, resulting in the synthesis of undecylprodigiosin and actinorhodin at earlier times (23). In addition, the constitutive expression of cdaR resulted in an increase in the abundance of cda and act transcripts at times when they are not normally expressed, whereas the constitutive expression of actII-ORF4 resulted in an initial decrease and then an increase in the abundance of cda transcripts and a decrease in the expression of redD-independent genes at times when they are normally elevated (23).
More than 15 pleiotropic regulators of antibiotic biosynthesis and 11 pathway-specific regulators of antibiotic biosynthesis have been identified so far (see Tables 2 and 3). Interestingly, with the exception of the SARP-encoding genes, redZ and absA, the majority of genes encoding antibiotic regulators are located outside of the antibiotic biosynthetic gene clusters. Most of these regulatory genes are located in the core region, away from the chromosomal ends, where genetic instability is most prominent (48). In most instances, the mechanism of action of these regulators is unknown. Furthermore, the connectivity of these diverse mechanisms is not understood; however, it is likely that they constitute a complex regulatory network.
TABLE 2.
Promoters of pleiotropic antibiotic regulatory genes investigated in the ChIP assaya
| Pleiotropic regulator | Function/protein | Chromosomal location | Primers | Chromosomal region of primers | Reference(s) |
|---|---|---|---|---|---|
| abaA (SCO0701) | Unknown | 742242-742685 | 218/219 | 743521-743300 | 15 |
| abaB (SCO3919) | Putative LysR regulator | 4314389-4315294 | 137/138 | 4314100-4314444 | 37 |
| 139/140 | 4314353-4314580 | ||||
| 141/142 | 4314562-4314793 | ||||
| absA (SCO3225/6) | Two-component system | 3536945-3538660 | 97/98 | 3536605-3536846 | 3 |
| 95/96 | 3536818-3537050 | ||||
| 99/100 | 3537037-3537259 | ||||
| absB (SCO5572) | RNase III | 6069987-6070805 | 93/94 | 6069590-6069817 | 35 |
| 91/92 | 6069793-6070052 | ||||
| afsQ1 (SCO4907) | Response regulator | 5339525-5340202 | 220/221 | 5340427-5340179 | 24 |
| afsR (SCO4426) | SARP | 4842787-4845768 | 186/187 | 4846080-4845913 | 17 |
| 188/189 | 4845900-4845699 | ||||
| afsS (SCO4425) | Unknown | 4842382-4842573 | 222/223 | 4842862-4842654 | 47 |
| cprA (SCO6312) | γ-Butyrolactone receptor | 6972028-6972675 | 232/233 | 6972986-6972737 | 33 |
| 234/235 | 6972749-6972569 | ||||
| kbpA (SCO4422) | AfsK kinase inhibitor | 4838327-4839085 | 212/213 | 4838187-4838434 | 45 |
| mia (up from SCO0541) | Unknown | 575057-575176 | 87/88 | 575128-575488 | 11 |
| 129/130 | 575057-575176 | ||||
| ppk (SCO4145) | Polyphosphate kinase | 4559965-4562289 | 230/231 | 4562519-4562306 | 13 |
| relA (SCO1513) | GTP pyrophosphokinase | 1616607-1619150 | 228/229 | 1619339-1619044 | 9 |
| relC (SCO4648) | Ribosomal protein | 5072851-5073285 | 244/245 | 5072672-5072899 | 31 |
| scbA (SCO6266) | Putative γ-butyrolactone biosynthesis enzyme | 6891293-6892237 | 224/225 | 6891006-6891255 | 41, 43 |
| scbR (SCO6265) | γ-Butyrolactone receptor | 6890528-6891175 | 226/227 | 6891519-6891292 | 41, 43 |
No AbsA2 interaction with any of the pleiotropic antibiotic regulatory genes was observed.
TABLE 3.
Promoters of pathway-specific antibiotic regulatory genes investigated in the ChIP assay
| Pathway-specific regulator | Function/protein | Chromosomal location | Primers | Chromosomal region of primers | AbsA2 interactiona | Reference |
|---|---|---|---|---|---|---|
| actII-ORF4 (SCO5085) | SARP | 5528094-5528861 | 85/86 | 5527922-5528171 | + | 42 |
| 125/126 | 5527998-5528291 | + | ||||
| atrA (SCO4118) | TetR-like regulator | 4523502-4524395 | 172/173 | 4524671-4524428 | − | 44 |
| 174/175 | 4524418-4524132 | − | ||||
| cdaR (SCO3217) | Putative SARP | 3529272-3531188 | 105/106 | 3528594-3528893 | + | 36 |
| 107/108 | 3528856-3529080 | + | ||||
| 109/110 | 3529059-3529288 | + | ||||
| 111/112 | 3529230-3529472 | + | ||||
| cprB (SCO6071) | γ-Butyrolactone receptor | 6663344-6663991 | 206/207 | 6662972-6663270 | − | 33 |
| 208/209 | 6663259-6663496 | − | ||||
| cutRS (SCO5862/3) | Two-component system | 6419308-6419961 (cutR) | 254/255 | 6419124-6419307 | − | 12 |
| ecrA1/A2 (SCO2517/8) | Two-component system | 2712740-2713423 (ecrA1) | 246/247 | 2713644-2713427 | − | 27 |
| eshA (SCO7699) | Putative nucleotide binding protein | 8535532-8536947 | 204/205 | 8535298-8535525 | − | 25 |
| kasO (SCO6280) | SARP | 6938012-6939643 | 216/217 | 6937780-6938026 | − | 43 |
| 190/191 | 6937724-6937937 | − | ||||
| 192/193 | 6937905-6938119 | − | ||||
| lipAR (SCO0712/713) | Lipase (secreted protein)/AfsR-like regulator | 756314-757246 (lipA) | 214/215 | 757550-757253 | − | 46 |
| redD (SCO5877) | SARP | 6432566-6433618 | 131/132 | 6432353-6432597 | − | 5 |
| 133/134 | 6432594-6432826 | − | ||||
| 135/136 | 6432690-6432982 | − | ||||
| redZ (SCO5881) | Response regulator | 6438206-6438859 | 121/122 | 6439115-6438867 | + | 20 |
| 123/124 | 6438958-6438674 | + |
+, presence of interaction; −, absence of interaction.
One of the first pleiotropic regulators of secondary metabolism in S. coelicolor to be identified was the absA locus, encoding the sensor kinase AbsA1 and the response regulator AbsA2. The absA operon is embedded within the CDA biosynthetic cluster (between genes SCO3224 and SCO3227) and has strong regulatory effects on CDA production but, surprisingly, exerts strong effects on actinorhodin, undecylprodigiosin, and methylenomycin as well (2, 3, 4, 36). Certain alleles of absA1 cause nearly complete inhibition of actinorhodin, undecylprodigiosin, and CDA production, while the deletion of either absA1 (in frame) or absA2 confers the so-called Pha phenotype, for which levels of production of these antibiotics are enhanced relative to those for parent strains (4, 8). We have shown that the cytoplasmic domain of AbsA1 can phosphorylate AbsA2 and dephosphorylate phosphorylated AbsA2 (AbsA2∼P). Furthermore, mutations that impair AbsA1 kinase activity enhance antibiotic production in vivo, while those that impair AbsA2∼P phosphatase activity dramatically reduce antibiotic production (38). In sum, these data are consistent with a model in which AbsA2∼P is a repressor of antibiotic production (38).
Transcript analysis suggested that mutations in absA strongly influence the expression of the CDA biosynthetic genes, which might be independent of the predicted CDA SARP-encoding gene cdaR (36). In contrast, other analyses suggested that the levels of transcription of actII-ORF4 and redD, encoding pathway-specific activators of actinorhodin and undecylprodigiosin, respectively, were reduced in absA1 mutants in which AbsA2∼P phosphatase activity was compromised and enhanced in absA null strains, suggesting that regulation of production of these antibiotics by absA might act via these pathway-specific regulators (1). However, the direct interaction of AbsA2∼P with its targets has yet to be shown and therefore remains a most important goal. In this work, we have used a chromatin immunoprecipitation (ChIP) assay to identify targets of AbsA2 in vivo. We show that this protein interacts with the actII-ORF4, redZ, and cdaR promoter regions in vivo and in vitro and that the in vitro interactions are stimulated by AbsA2 phosphorylation. We show that, consistent with direct regulatory links between absA and the pathway-specific regulators, induced expression of the first two of these activators in a strain bearing a hyperrepressive allele of absA1 restores antibiotic production in a pathway-specific manner.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. Escherichia coli XL1-Blue was used to propagate all plasmids, whereas E. coli BL21(DE3) and ER2058 were used for protein overexpression. E. coli strains were grown at 37°C in Luria-Bertani medium. Plasmids were introduced into Streptomyces by conjugation from E. coli ET12567/pUZ8002. Antibiotic concentrations used for plasmid selection were 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 50 μg/ml apramycin. Thiostrepton was added at 50 μg/ml to induce genes under the control of the tipA promoter. S. coelicolor strains were grown in tryptone soy broth (TSB; Oxoid) and production medium (pH 7.0) (40), on solid R2YE and MS agar (26). Staphylococcus aureus (ATCC 29213) was grown on Oxoid nutrient agar (ONA) with or without 12 mM calcium nitrate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/description | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal met λ (DE3) | Novagen |
| ER2058 | F−ara-14 leuB6 fhuA2 Δ(argF-lac)U169 lacY1lon::mini-Tn10(Tetr) glnV44 galK2 rpsL20(Strr)xyl-5 mtl-5 Δ(malB) zjc::Tn5(Kanr) Δ(mcrC-mrr)HB101 | New England Biolabs |
| ET12567/pUZ8002 | dam-13::Tn9 dcm-6 hsdM; containing the nontransmissible oriT mobilizing plasmid pUZ8002 | 16 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1lac[F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| pBluescript II SK(+) | Phagemid cloning vector | Stratagene |
| pPCR-Script Amp | Phagemid cloning vector; variant ofpBluescript II SK(+) containing a SrfI site for insertion of blunt-end amplicons | Stratagene |
| pMAL-c2X | Protein overexpression vector resulting in an N-terminal MBP fusion; Tac promoter | New England Biolabs |
| pMALA2 | absA2 in pMAL-c2X | 38 |
| S. coelicolor | ||
| J1501 | hisA1 uraA1 strA1 pgl SCP1 SCP2 | 26 |
| C542 | J1501 hisA1 uraA1 strA1 pgl absA1-542 SCP1 SCP2 | 3 |
| pIJ6902 | Integrative PtipA expression vector ori pUC18 oriT RK2 intφC31 attP tsr aac(3)IV | 23 |
| pactII-ORF4 | actII-ORF4 in pIJ6902 | This work |
| pcdaR | cdaR in pIJ6902 | This work |
| predZ | redZ in pIJ6902 | This work |
Plasmids and primers.
The plasmids used in this study are listed in Table 1. Primers were purchased from Sigma-Aldrich or the Mobix Laboratory at McMaster University. PCR was performed using VentR polymerase (New England Biolabs). DNA sequencing was performed by the Mobix Laboratory.
Preparation of antiserum against denatured His6-AbsA1 and His6-AbsA2.
His6-tagged fusion proteins of AbsA2 and the cytoplasmic portion of AbsA1 were purified as previously described (38). Aliquots of 100 μg and 50 μg of each protein were resolved on 12% sodium dodecyl sulfate-polyacrylamide gels, stained with Coomassie brilliant blue, and excised. The excised proteins in the gel slices were sent to Cocalico Biologicals (Reamston, PA) for rabbit immunization.
CDA bioassay.
Pregerminated S. coelicolor J1501 spores were inoculated into 250 ml TSB medium and grown at 30°C. Every 12 h until 72 h of growth, 10 ml of culture supernatant was removed and centrifuged, and the supernatant was filtered through 0.45-μm-pore-size Acrodisc syringe filters (Pall). ONA plates with or without 12 mM Ca(NO3)2 were overlaid with an overnight culture of Staphylococcus aureus diluted 1/100 in soft nutrient agar (1:1 ONA plus Oxoid nutrient broth). Sterilized one-quarter-inch discs (catalogue no. 10328171; Schleicher & Schuell) were placed on top of the lawn of Staphylococcus aureus. Seventy-five microliters of each sample was spotted on the filter discs, and the plates were incubated first for 3 h at 4°C and then overnight at 37°C.
Preparation of S. coelicolor membrane and cytoplasmic fractions and AbsA1 Western analysis.
S. coelicolor J1501 was grown at 30°C on cellophane discs overlaid on R2YE medium and harvested every 12 h until 48 h of growth. Cells were resuspended in P (protoplast) buffer (26) containing 2 mg/ml lysozyme and 2 g of 3-mm glass beads and vortexed for 1.5 min. Cells were then incubated for 1 h 45 min in a 30°C water bath and gently inverted every 15 min. Protoplasts were filtered through cotton wool, centrifuged at 6,000 rpm for 10 min, and resuspended in lysis buffer (50 mM HEPES, 300 mM NaCl, 5 mM EDTA, pH 7.5, 1 × protease inhibitor cocktail [Roche]). Cells were lysed by three passages through a French pressure cell and centrifuged at 3,000 rpm for 10 min, and the supernatant was subjected to ultracentrifugation (100,000 × g, 60 min). Membranes were resuspended in lysis buffer, and 20% glycerol (final concentration) was added to both the membrane and cytoplasmic fractions. Samples were frozen in liquid nitrogen and stored at −80°C. Thirty micrograms of the membrane fraction and 30 μg of the cytoplasmic fraction were subjected to Western analysis. Affinity-purified anti-AbsA1 antibodies (as described in reference 7a) were used to detect the presence of AbsA1 by chemiluminescence.
ChIP.
Pregerminated S. coelicolor J1501 spores were inoculated into 250 ml TSB medium. After 27 h of growth at 30°C, cultures were cross-linked by the addition of 1% formaldehyde and 10 mM NaPO4, pH 8.0 (final concentrations), for 30 min at 30°C. To stop the reaction, 125 mM glycine (final concentration) was added, and the cultures were incubated for 5 min at room temperature with gentle swirling every 1 min. Cells were washed twice with cold phosphate-buffered saline, pH 7.4, and stored in 100-mg (wet weight) aliquots at −80°C. One hundred milligrams of cells was resuspended in 400 μl of phosphate-buffered saline buffer containing 10 mg/ml lysozyme (final concentration) and 1 × protease inhibitor cocktail (Roche). Following incubation for 20 min at 37°C, cells were subjected to sonication (10 times for 15 seconds at half power; Fisher Sonic Dismembrator model 300), which sheared the DNA to 500 to 1,000-bp fragments (verified by gel electrophoresis; data not shown). Cell debris and unlysed cells were removed by centrifugation (twice for 5 min at 14,000 rpm at 4°C). Clarified lysate that was not subjected to immunoprecipitation served as a positive control. Two hundred twenty-five microliters of clarified lysate was combined with 500 μl of immunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% Triton X-100) plus 1× protease inhibitor cocktail and precleared for 1 h at 4°C with gentle agitation with 80 μl of a 50% protein A-Sepharose slurry (Sigma-Aldrich). Seventy-three microliters of anti-AbsA2 antibodies (Cocalico Biologicals) was added to the precleared sample and incubated overnight at 4°C with gentle agitation. To isolate the immunoprecipitated complexes, 80 μl of a 50% protein A-Sepharose slurry was added and incubated for 4 h at 4°C with gentle agitation. The protein A-Sepharose beads were washed four times (15-min incubations at 4°C with gentle agitation) with 1 ml of immunoprecipitation buffer. Beads were transferred to a fresh Eppendorf tube after the first wash to prevent contamination of protein/DNA that had adsorbed to the sides of the Eppendorf tube. To elute the protein/DNA complexes, the beads were incubated overnight at 55°C in 240 μl of elution buffer (Tris-EDTA [TE], pH 7.6, 0.2 mg/ml proteinase K, 1% sodium dodecyl sulfate). One hundred thirty microliters of elution buffer was also added to the clarified lysate that was not subjected to immunoprecipitation. This positive-control sample was subsequently treated in the same manner as the experimental sample. Following overnight incubation at 55°C, the samples were incubated at 65°C for 30 min. The supernatant was recovered, and the beads were washed with 50 μl of TE, pH 7.6, which was combined with the eluate. Two microliters of 20 mg/ml glycogen mix (Sigma G-1767) was added, and the samples were extracted with an equal volume of phenol-chloroform (1:1). The DNA was precipitated with 0.1 volumes of 3 M sodium acetate, pH 4.8, and 2 volumes of 100% ethanol, washed with 70% ethanol, and air dried. The experimental sample was resuspended in 25 μl of TE, whereas the positive-control sample was resuspended in 100 μl. To determine the identity of the immunoprecipitated DNA, PCR using primer sets shown in Tables 2 and 3 was performed on 2 μl of the immunoprecipitated DNA and 100 ng of the positive-control DNA. PCR samples were resolved on a 2% agarose gel, stained with ethidium bromide, and analyzed by Polaroid film.
Construction of an absA2 overexpression vector.
absA2 was cloned into pMAL-c2X to generate pMALA2 and overexpressed as an N-terminal maltose binding protein (MBP) fusion in E. coli ER2058 as previously described (38).
EMSAs.
Promoter fragments generated by PCR amplification were purified from 12% polyacrylamide gels by the crush and soak method and 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). Approximately 0.89 ng of end-labeled DNA was incubated with AbsA2 fusion protein in 15-μl reaction mixtures in electrophoretic mobility shift assay (EMSA) buffer (10 mM Tris-HCl, pH 7.8, 2 mM dithiothreitol, 150 mM NaCl, 10% glycerol, 45 ng salmon sperm DNA) for 15 min at 30°C. The AbsA2 fusion protein was preincubated for 30 min at 30°C in phosphorylation buffer (50 mM Tris-HCl pH 7.0, 25 mM MgCl2, 0.1 M NaCl) in the presence or absence of 100 mM phosphoramidate (PA) (a kind gift from L. Kenney; synthesized according to the method used in reference 29 with the aid of K. Koteva). Reaction mixtures were resolved in an 8% (see Fig. 3) or 10% (see Fig. 4) polyacrylamide nondenaturing gel for either 2 h 45 min (see Fig. 3) or 2 h 10 min (see Fig. 4) at 70 V. Wet gels were exposed to X-Omat Blue Kodak film with an intensifying screen and incubated overnight at −80°C prior to film exposure. The primer sets used to generate the DNA probes are as follows (for primers in Tables 2 and 3, only the numbers are listed here): cdaR (230 bp), no. 109 and 110; redZ (249 bp), no. 121 and 122; actII-ORF4 (250 bp), no. 85 and 86; actII-ORF4 fragment I (90 bp), 5′ GAGGACCCTTCCGAGGAC 3′ (forward) and 5′ TGCTGGATTTTACCGAGAGG 3′ (reverse); actII-ORF4 fragment II (92 bp), no. 85 and 5′ CACAAGCGATCTCCTATTG 3′ (reverse); actII-ORF4 fragment III (90 bp), 5′ GATCAATAGGAGATCGCTTG 3′ (forward) and 5′ GTCGAGATTCTCCGTCTC 3′ (reverse); actII-ORF4 fragment IV (122 bp), 5′ GTTATTGTCGCCCCCAG 3′ (forward) and no. 86; actII-ORF4 fragment V (94 bp), 5′ CCGCTTAAATCCTCGAAG 3′ (forward) and 5′ CACAACTCCTCGATGAGCAC 3′ (reverse); actII-ORF4 fragment VI (109 bp), 5′ GTGCTCATCGAGGAGTTGTG 3′ (forward) and 5′ CTCGTCACCCGGTGCTC 3′ (reverse).
FIG. 3.
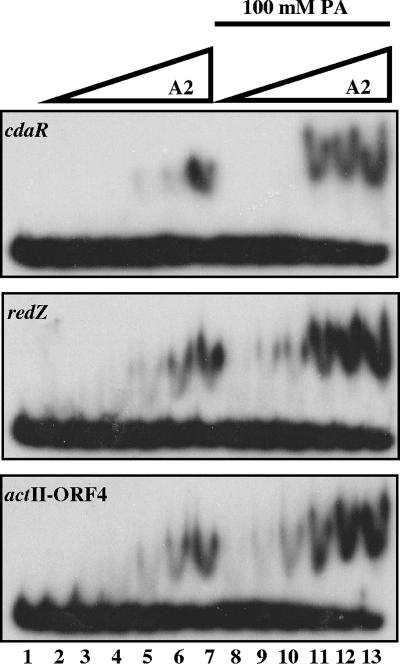
EMSAs with purified MBP-AbsA2 recombinant protein with each of the three target promoters identified by ChIP (cdaR primers 109/110, redZ primers 121/122, and actII-ORF4 primers 85/86). 32P-labeled probe (0.89 ng) was incubated at 30°C for 15 min with various concentrations of protein that had been preincubated at 30°C for 30 min with or without 100 mM PA. Reaction mixtures were resolved on a native 1.5% glycerol-containing, 8% polyacrylamide gel with 1× TBE plus 1.5% glycerol as the running buffer. Lane 1 contains probe alone. Lanes 2 and 8 contain 0.527 μM of MBP-AbsA2. Lanes 3 and 9 contain 0.675 μM of MBP-AbsA2. Lanes 4 and 10 contain 0.810 μM of MBP-AbsA2. Lanes 5 and 11 contain 0.972 μM of MBP-AbsA2. Lanes 6 and 12 contain 1.166 μM of MBP-AbsA2. Lanes 7 and 13 contain 1.4 μM of MBP-AbsA2.
FIG. 4.
Identification of the region within the actII-ORF4 promoter that is required for AbsA2 binding. (A) Schematic representation of the actII-ORF4 promoter region. (B) EMSAs with various amounts of MBP-AbsA2 protein with small overlapping fragments shown in panel A. 32P-labeled probe (0.89 ng) was incubated at 30°C for 15 min with various concentrations of protein that had been preincubated at 30°C for 30 min with or without 100 mM PA. Lanes 1 contain probe alone (0.89 ng of radiolabeled DNA). Lanes 2 and 4 contain 0.675 μM of MBP-AbsA2. Lanes 3 and 5 contain 1.4 μM of MBP-AbsA2.
Construction of actII-ORF4, cdaR, and redZ thiostrepton-inducible constructs.
The 780-bp actII-ORF4 gene was amplified by PCR with primers actII-ORF4-F (5′ GACGGGGGCGCATATGAGATTCAAC 3′, with an NdeI site at positions 11 to 16 [underlined]) and actII-ORF4-R (5′ ACCGCGTTCTAGAACCGGTGCTAC 3′, with an XbaI site at positions 8 to 13 [underlined]). The 1,917-bp cdaR gene was amplified by PCR with primers cdaR-F (5′ GTATCAAGAGCATATGGATCTTC 3′, with an NdeI site at positions 11 to 16 [underlined]) and cdaR-R (5′ GTCGCCGTTCTAGACTCAGTCGGT 3′, with an XbaI site at positions 9 to 14 [underlined]). The 657-bp redZ gene was amplified by PCR with primers redZ-F (5′ CGAAAGTCAACATATGACGACCCG 3′, with an NdeI site at positions 11 to 16 [underlined]) and redZ-R (5′ GAAGCGGCACGTCTAGACCGGGGC 3′, with an XbaI site at positions 12 to 17 [underlined]). All amplified genes were initially cloned into pPCR-Script Amp, sequenced, digested with NdeI and XbaI, and ligated to pIJ6902 digested with the same enzymes to generate pactII-ORF4, pcdaR, and predZ. The constructs were introduced into S. coelicolor strain C542 by conjugation.
RESULTS
Identification of AbsA2 target promoters.
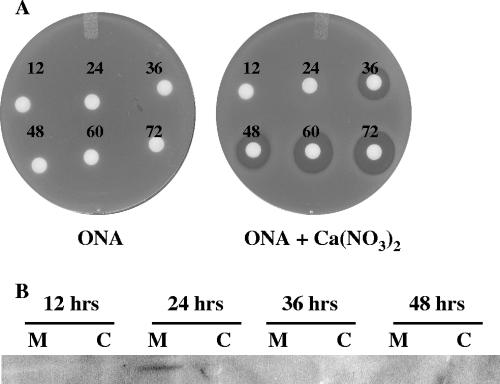
To determine if AbsA2 binding sites are present in the known antibiotic regulatory gene promoter regions, we established a ChIP assay for the AbsA2 protein using S. coelicolor J1501 that had been treated with formaldehyde to cross-link AbsA2 to its DNA targets (39). In order to do this, we first needed to establish the point in the S. coelicolor growth cycle/phase when AbsA2 was likely interacting with its target promoters. We first carried out a CDA bioassay on liquid-grown S. coelicolor. Seventy-five-microliter aliquots of culture supernatant from cells grown for 12 to 72 h were spotted on filter discs and overlaid on a lawn of Staphylococcus aureus in the presence and absence of Ca(NO3)2. Zones of inhibition that depended on the presence of calcium, indicative of the presence of CDA, were observed in 36-hour culture supernatants and from all samples from later time points (Fig. 1A). Since CDA biosynthetic gene expression is sensitive to mutations in absA (4), we interpreted this as evidence that AbsA2 might interact with its target promoters prior to 36 h of growth, thereby inhibiting CDA production. Consistent with this, Western analysis of membranes isolated from cells grown on solid R2YE medium for 12 to 72 h demonstrated that AbsA1 was present in cells grown for 24 h, and it was not detected at later time points (Fig. 1B). These data, and the fact that the absA transcript was detected in liquid-grown cultures between 18 and 54 h (4), suggested that AbsA2 might be bound to its target promoters at ∼27 h postgermination.
FIG. 1.
CDA bioassay (A) and Western analysis (B) indicate when the AbsA system may be negatively regulating antibiotic production. (A) CDA bioassay to determine when CDA is produced in S. coelicolor J1501 grown in liquid TSB medium. Seventy-five microliters of culture supernatant from cells grown for 12, 24, 36, 48, 60, and 72 h was spotted on discs placed on a lawn of Staphylococcus aureus on ONA with or without calcium nitrate and incubated first for 3 h at 4°C and then overnight at 37°C. Zones of inhibition were first observed at 36 h and indicate the presence of CDA. (B) Western analysis of membrane (M) and cytoplasmic (C) fractions of S. coelicolor grown at 30°C for 12, 24, 36, and 48 h on cellophane discs overlaid on solid R2YE medium. Anti-AbsA1 antibodies were used to detect the presence of AbsA1 in the membrane fraction of cells grown for 24 h.
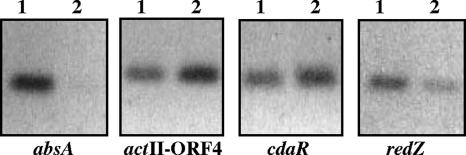
Antibodies we raised against AbsA2 could detect 50 ng of purified protein by Western analysis (data not shown). To determine whether they could immunoprecipitate the folded protein, we prepared 35S-radiolabeled AbsA2 by in vitro transcription/translation and carried out trial immunoprecipitations. We found that the antibodies could indeed immunoprecipitate AbsA2 (data not shown). We therefore cross-linked S. coelicolor cells with formaldehyde after 27 h of growth in liquid culture, isolated and fragmented the cross-linked DNA, and carried out immunoprecipitation using anti-AbsA2 antibodies (see Materials and Methods). We then carried out PCR assays using primers that flanked the intergenic regions of candidate genes that had been shown to contain their promoters or that we predicted to contain their promoters. Analysis of regulators that affect both antibiotics and morphogenesis (e.g., bld genes) was not performed, as AbsA2 has been shown to primarily affect antibiotic production. Fifteen global regulatory genes and 11 pathway-specific genes (Tables 2 and 3) were analyzed by PCR using immunoprecipitated cross-linked DNA and cross-linked DNA that had not been immunoprecipitated as a control. Several sets of primers were used for some promoters. PCR products were obtained using all of the primer sets when unimmunoprecipitated DNA was used as a control. In addition, primers directed against three genes, actII-ORF4, cdaR, and redZ, amplified fragments of the correct size when immunoprecipitated DNA was used as the template. Importantly, for all three of these regulators, several different primer sets gave a positive read-out (Fig. 2 and data not shown). In contrast, primers directed against the other 23 genes listed in Tables 2 and 3 amplified DNA fragments only in the unimmunoprecipitated pools of DNA and not in the anti-AbsA2-immunoprecipitated DNA; as an example, this scenario included the promoter region of absA itself (Fig. 2 and data not shown). The promoter regions of actII-ORF4, cdaR, and redZ had therefore been selectively enriched as a result of ChIP using anti-AbsA2 antibodies.
FIG. 2.
In vivo identification of AbsA2 target promoters by ChIP. Anti-AbsA2 antibodies were used to immunoprecipitate AbsA2/DNA complexes from cells treated with formaldehyde. PCR was performed with primers flanking putative target promoters (absA primers 97/98, actII-ORF4 primers 85/86, cdaR primers 109/110, and redZ primers 121/122). DNA used for PCR was total DNA prior to immunoprecipitation (lanes 1) and immunoprecipitated DNA (lanes 2). PCR using primers that flank the absA promoter is representative of all primer sets that did not result in amplification of the immunoprecipitated DNA. Amplification of the actII-ORF4, cdaR, and redZ promoters from the immunoprecipitated DNA indicated that these three promoters are targets of AbsA2.
In vitro interaction of AbsA2 with target promoters.
To determine whether AbsA2 could interact with the promoters of actII-ORF4, redZ, and cdaR in vitro, we carried out EMSAs. In a previous study, we purified AbsA2 both as an N-terminally His6-tagged protein and as an N-terminally MBP fusion and demonstrated that both could serve as substrates for phosphorylation and dephosphorylation by the catalytic fragment of AbsA1 (38). DNA substrates for EMSAs were prepared by PCR amplification, gel purification, and 32P end labeling of the cdaR, redZ, and actII-ORF4 promoters that were 230, 249, and 250 base pairs in size, respectively. Each fragment (0.89 ng) was then used for gel mobility shift experiments along with the two purified AbsA2 proteins (MBP-AbsA2 and His6-AbsA2).
As shown in Fig. 3, unphosphorylated MBP-AbsA2 was able to bind to all three DNA fragments, although the interactions were weak: the cdaR probe was shifted only when incubated with ≥1.4 μM MBP-AbsA2. The redZ and actII-ORF4 probes exhibited moderately stronger binding, showing reproducible shifts when 1.16 μM protein was used. It was previously demonstrated that His6-AbsA2 can autophosphorylate itself to near completion when incubated with the small molecule phosphodonor PA (32). To determine whether the interaction of MBP-AbsA2 with putative target proteins was stimulated by phosphorylation, the binding reactions were repeated in the presence of 100 mM PA. The phosphodonor clearly stimulated binding such that substantial shifts occurred at 0.97 μM MBP-AbsA2 (cdaR and redZ) or 0.81 μM MBP-AbsA2 (actII-ORF4). The experiments shown in Fig. 3 were carried out with the MBP- and His6-AbsA2 fusion proteins, and identical results were obtained except that the degree of shift of the probe was greater with the MBP fusion. Since the shifts are easier to visualize with this protein, we have shown the MBP fusion data only. Each of these shifts was competed by a 40-fold excess of unlabeled specific DNA but not by a nonspecific unlabeled control (data not shown).
DNase I footprinting experiments using these promoter fragments proved unsuccessful, so as an alternative strategy, we focused on the actII-ORF4 promoter and created a series of six small overlapping fragments (ranging in size from 90 to 122 base pairs) extending from −200 bp to +272 bp relative to the transcriptional start site (Fig. 4A). We performed EMSAs with approximately 0.89 ng of each of the small overlapping fragments and either 0.67 μM or 1.4 μM of purified MBP-AbsA2 protein in the presence or absence of PA. Phosphorylated MBP-AbsA2 at a concentration of 1.4 μM bound to four fragments within the vicinity of the actII-ORF4 promoter (fragments I, II, IV, and VI) (Fig. 4B). Fragment VI (+163 bp to +272 bp) is an unusual place for a repressor to bind and may suggest that AbsA2∼P represses transcription by DNA looping. We consistently found that, unlike the observation with the larger fragments used in Fig. 3, there was little or no shift detected with these smaller fragments when unphosphorylated MBP-AbsA2 was used. Again, each of these shifts was competed by a 40-fold excess of unlabeled specific DNA but not by a nonspecific unlabeled control (data not shown).
In vivo bypass of the negative regulatory effects of AbsA2.
To determine whether the negative regulatory effects of AbsA2∼P could be circumvented by uncoupling the expression of the putative targets in vivo, we cloned actII-ORF4, redZ, and cdaR into pIJ6902 under the control of the thiostrepton-inducible promoter PtipA and introduced them into the antibiotic-deficient strain C542 (3). In this strain, absA1 contains two point mutations (corresponding to amino acid mutations I360L and R365Q) that are proposed to inactivate AbsA1 phosphatase activity, resulting in increased phosphorylation of AbsA2 and, subsequently, increased transcriptional repression. Induction of the actII-ORF4 gene in C542 resulted in the production of a blue, secreted pigment indicative of actinorhodin production (Fig. 5). Similarly, induction of redZ in C542 resulted in the production of a red, cell-associated pigment indicative of undecylprodigiosin. These results demonstrate that the negative regulatory effects of AbsA2 could be bypassed when actII-ORF4 and redZ are overexpressed in an absA1 mutant strain deficient in antibiotic production and further support the notion that these genes are targets of AbsA2. We attempted to bypass the effect of the absA1 mutations by using a PtipA-controlled cdaR gene; however, the analysis was complicated by the fact that the standard assay strains for CDA (e.g., Staphylococcus aureus strains) were also sensitive to thiostrepton, a technical roadblock also encountered by other investigators (23).
FIG. 5.
Bypass of the absA1 mutant by actII-ORF4 and redZ. Genes were cloned into pIJ6902 under the control of the strong thiostrepton-inducible promoter. Induction of actII-ORF4 in the antibiotic-deficient strain C542 restored actinorhodin production, whereas induction of redZ restored undecylprodigiosin production. Strains were grown on R2YE containing 50 μg/ml thiostrepton for 7 days at 30°C.
DISCUSSION
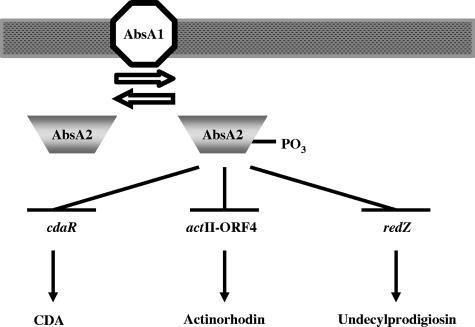
We have identified three AbsA2 target promoters: redZ, cdaR, and actII-ORF4 (Fig. 2), all of which encode pathway-specific regulators for specific antibiotic biosynthetic gene clusters. redZ encodes a response regulator-like activator that activates the expression of the SARP gene, redD, as well as several redD-independent genes (20, 22, 23). actII-ORF4 encodes a SARP that directly activates transcription of actinorhodin biosynthetic genes (19). cdaR encodes a putative SARP that activates transcription of the CDA biosynthetic genes (23, 36). All three interactions were reproduced in vitro, and the interaction of AbsA2 with these DNA targets was enhanced when the protein was phosphorylated (Fig. 3). Finally, the negative regulatory effects of absA on actinorhodin and undecylprodigiosin production could be bypassed by replacing the natural promoters of actII-ORF4 and redZ, with a thiostrepton-inducible promoter (which, naturally, is not an absA target), supporting the idea that AbsA2∼P is a negative regulator of these pathway-specific activators (Fig. 5). These data support the model shown in Fig. 6, whereby AbsA1 controls the phosphorylation state of AbsA2, which, when phosphorylated, interacts with pathway-specific activator genes of each antibiotic cluster to reduce their expression.
FIG. 6.
Schematic representation of the AbsA two-component signal transduction system in S. coelicolor. AbsA2∼P represses transcription of the cdaR, actII-ORF4, and redZ promoters, thereby preventing the production of CDA, actinorhodin, and undecylprodigiosin, respectively.
Some response regulators that bind DNA primarily as phosphoproteins can nevertheless bind their targets when unphosphorylated with reduced affinity (18, 21, 28). This is consistent with our observation of weaker interactions of unphosphorylated AbsA2 with DNA. AbsA2∼P clearly had a stronger interaction with DNA. For example, we could see evidence of binding to the redZ and actII-ORF4 promoter fragments at a protein concentration as low as 0.8 μM in the presence of PA (Fig. 3). Furthermore, AbsA2 did not bind the smaller actII-ORF4 fragments in the absence of PA (Fig. 4B).
Given the robustness of our ChIP results, we were surprised by the weakness of the AbsA2∼P/DNA interactions. One possible explanation for this is that AbsA2 possesses autophophatase activity such that the half-life of AbsA2∼P is 68.6 min (38). Given this, we imagine that during the >2-h gel running time required for our EMSA experiments, a significant fraction of the AbsA2∼P/DNA complexes would decay to less-stable AbsA2/DNA complexes and then to free protein and DNA, resulting in an underestimation of the binding strength. In contrast, in the ChIP assay, once the DNA and protein are cross-linked to one another, the complex is frozen regardless of whether the response regulator subsequently dephosphorylates. It is also equally likely that the weak interactions observed in vitro are a result of the MBP-AbsA2 fusion protein interacting with linear DNA, whereas in vivo, the native protein would interact with supercoiled DNA.
The observation that AbsA2 regulates transcription of actII-ORF4 and redZ is consistent with a previous transcriptional analysis performed in the Champness laboratory, whereby the researchers proposed that actII-ORF4 and redD were AbsA2 targets (1). Since transcription of redD is dependent on redZ (22, 23, 50), a decrease in redZ transcription will in turn result in reduced redD transcription. Therefore, although the Champness laboratory did not report redZ as a target of AbsA2, our data are in agreement. Our data do not agree, however, with a previous report suggesting that cdaR expression was not sensitive to mutations in absA (36). We point out that this might be explained by the fact that the previous study was carried out with cells grown on solid medium on which effects of additional pathways would be in play.
It is most intriguing that a regulatory mechanism that is embedded in one antibiotic biosynthetic gene cluster would have such profound effects on the production of chemically unrelated antibiotics such as undecylprodigiosin and actinorhodin. A similar situation exists in Streptomyces clavuligerus whereby ccaR, a gene embedded within the cephamycin C cluster, encodes an antibiotic regulatory protein that regulates both cephamycin C and clavulanic acid production (34). While the biological significance of the cross-regulation in S. coelicolor is unknown, it is possible to imagine a number of advantages for the organism. For example, cross-regulation between antibiotic biosynthetic clusters might help ensure that disparate biosynthetic genes are activated when the organism is most metabolically fit for secondary metabolism. Interestingly, AbsA2 did not appear to interact with the promoter of kasO (Table 3). kasO encodes the only other known SARP that is embedded within the S. coelicolor cpk (cryptic type I polyketide synthase) antibiotic cluster (43). Indeed, while our data do not suggest any additional targets, they certainly do not rule them out.
The role of the absA two-component system in antibiotic regulation may extend beyond S. coelicolor. Genes related to absA1/absA2 are found in several antibiotic biosynthetic clusters in diverse streptomycetes. In Streptomyces fradiae, an absA-like operon is present in the biosynthetic gene cluster for the lipopeptide antibiotic A54145 (30). Interestingly, A54145 belongs to the same class of antibiotics as the CDAs from S. coelicolor that are cyclic lipopeptides active against gram-positive pathogens. In addition, absA-like operons are also found in the biosynthetic gene cluster for blasticidin S, cinnamycin, and enduracidin in Streptomyces griseochromogenes, S. cinnamoneus, and S. fungicidicus, respectively (14, 51, 53). A unifying feature among the AbsA1-like sensor kinases is their predicted atypical transmembrane topologies. Most AbsA1 family members are predicted to have four or more N-terminal transmembrane domains separating very small extracellular and intracellular loops, unlike the prototypical sensor kinase topology of a large N-terminal sensory domain bordered by two transmembrane domains and an intracellular C-terminal kinase domain. The significance of this is unknown; however, it likely reflects some aspect of their signal response mechanism. Understanding this mechanism is a current goal of our work.
Acknowledgments
We are grateful to Wendy Champness for providing the C542 strain. We also thank Linda Kenney for supplying us with PA and Kalinka Koteva for assisting in making it. We thank David Andrews for supplying the S30 E. coli lysate for in vitro transcription/translation. We thank Marie Elliot for helpful suggestions and comments and Maureen Bibb and Dagmara Jakimowicz for advice on performing ChIP on Streptomyces. We also thank Kapil Tahlan for critical reading of the manuscript. Lastly, we thank the reviewers for their constructive comments.
N.L.M. was supported by an Ontario Graduate Scholarship and a CIHR doctoral research award. This research was supported by grant no. MOP-57684 from the Canadian Institutes for Health Research.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Aceti, D. J., and W. C. Champness. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180:3100-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamidis, T., and W. Champness. 1992. Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J. Bacteriol. 174:4622-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamidis, T., P. Riggle, and W. Champness. 1990. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, T. B., P. Brian, and W. C. Champness. 2001. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol. Microbiol. 39:553-566. [DOI] [PubMed] [Google Scholar]
- 5.Arias, P., M. A. Fernandez-Moreno, and F. Malpartida. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181:6958-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb, M. 1996. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. [DOI] [PubMed] [Google Scholar]
- 7.Bibb, M. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 7a.Bio-Rad. 2005. Affi-Gel 10 manual. Bio-Rad, Hercules, CA.
- 8.Brian, P., P. J. Riggle, R. A. Santos, and W. C. Champness. 1996. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J. Bacteriol. 178:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champness, W. 2000. Actinomycete development, antibiotic production, and phylogeny: questions and challenges, p. 11-31. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, DC.
- 11.Champness, W., P. Riggle, T. Adamidis, and P. Vandervere. 1992. Identification of Streptomyces coelicolor genes involved in regulation of antibiotic synthesis. Gene 115:55-60. [DOI] [PubMed] [Google Scholar]
- 12.Chang, H. M., M. Y. Chen, Y. T. Shieh, M. J. Bibb, and C. W. Chen. 1996. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol. Microbiol. 21:1075-1085. [PubMed] [Google Scholar]
- 13.Chouayekh, H., and M. J. Virolle. 2002. The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 43:919-930. [DOI] [PubMed] [Google Scholar]
- 14.Cone, M. C., X. Yin, L. L. Grochowski, M. R. Parker, and T. M. Zabriskie. 2003. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4:821-828. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Moreno, M. A., E. Martinez, L. Boto, D. A. Hopwood, and F. Malpartida. 1992. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 267:19278-19290. [PubMed] [Google Scholar]
- 16.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 17.Floriano, B., and M. Bibb. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385-396. [DOI] [PubMed] [Google Scholar]
- 18.Geng, H., S. Nakano, and M. M. Nakano. 2004. Transcriptional activation by Bacillus subtilis ResD: tandem binding to target elements and phosphorylation-dependent and -independent transcriptional activation. J. Bacteriol. 186:2028-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie, E. P., C. S. Flaxman, J. White, D. A. Hodgson, M. J. Bibb, and K. F. Chater. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144:727-738. [DOI] [PubMed] [Google Scholar]
- 21.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 22.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, J., J. Shi, V. Molle, B. Sohlberg, D. Weaver, M. J. Bibb, N. Karoonuthaisiri, C. J. Lih, C. M. Kao, M. J. Buttner, and S. N. Cohen. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276-1287. [DOI] [PubMed] [Google Scholar]
- 24.Ishizuka, H., S. Horinouchi, H. M. Kieser, D. A. Hopwood, and T. Beppu. 1992. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J. Bacteriol. 174:7585-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamoto, S., M. Watanabe, N. Saito, A. Hesketh, K. Vachalova, K. Matsubara, and K. Ochi. 2001. Molecular and functional analyses of the gene (eshA) encoding the 52-kilodalton protein of Streptomyces coelicolor A3(2) required for antibiotic production. J. Bacteriol. 183:6009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 27.Li, Y. Q., P. L. Chen, S. F. Chen, D. Wu, and J. Zheng. 2004. A pair of two-component regulatory genes ecrA1/A2 in S. coelicolor. J. Zhejiang Univ. Sci. 5:173-179. [DOI] [PubMed] [Google Scholar]
- 28.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattison, K., R. Oropeza, N. Byers, and L. J. Kenney. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315:497-511. [DOI] [PubMed] [Google Scholar]
- 30.Miao, V., R. Brost, J. Chapple, S. K., M. F. Gal, and R. H. Baltz. 2006. The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae. J. Ind. Microbiol. Biotechnol. 33:129-140. [DOI] [PubMed] [Google Scholar]
- 31.Ochi, K. 1990. A relaxed (rel) mutant of Streptomyces coelicolor A3(2) with a missing ribosomal protein lacks the ability to accumulate ppGpp, A-factor and prodigiosin. J. Gen. Microbiol. 136:2405-2412. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor, T. J., and J. R. Nodwell. 2005. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J. Mol. Biol. 351:1030-1047. [DOI] [PubMed] [Google Scholar]
- 33.Onaka, H., T. Nakagawa, and S. Horinouchi. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743-753. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Llarena, F. J., P. Liras, A. Rodriguez-Garcia, and J. F. Martin. 1997. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J. Bacteriol. 179:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price, B., T. Adamidis, R. Kong, and W. Champness. 1999. A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J. Bacteriol. 181:6142-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryding, N. J., T. B. Anderson, and W. C. Champness. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184:794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheu, A. K., E. Martinez, J. Soliveri, and F. Malpartida. 1997. abaB, a putative regulator for secondary metabolism in Streptomyces. FEMS Microbiol. Lett. 147:29-36. [DOI] [PubMed] [Google Scholar]
- 38.Sheeler, N. L., S. V. MacMillan, and J. R. Nodwell. 2005. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J. Bacteriol. 187:687-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solomon, M. J., and A. Varshavsky. 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. USA 82:6470-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taguchi, T., K. Itou, Y. Ebizuka, F. Malpartida, D. A. Hopwood, C. M. Surti, K. I. Booker-Milburn, G. R. Stephenson, and K. Ichinose. 2000. Chemical characterisation of disruptants of the Streptomyces coelicolor A3(2) actVI genes involved in actinorhodin biosynthesis. J. Antibiot. (Tokyo) 53:144-152. [DOI] [PubMed] [Google Scholar]
- 41.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 42.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 43.Takano, E., H. Kinoshita, V. Mersinias, G. Bucca, G. Hotchkiss, T. Nihira, C. P. Smith, M. Bibb, W. Wohlleben, and K. Chater. 2005. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56:465-479. [DOI] [PubMed] [Google Scholar]
- 44.Uguru, G. C., K. E. Stephens, J. A. Stead, J. E. Towle, S. Baumberg, and K. J. McDowall. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58:131-150. [DOI] [PubMed] [Google Scholar]
- 45.Umeyama, T., and S. Horinouchi. 2001. Autophosphorylation of a bacterial serine/threonine kinase, AfsK, is inhibited by KbpA, an AfsK-binding protein. J. Bacteriol. 183:5506-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdez, F., G. Gonzalez-Ceron, H. M. Kieser, and L. Servin-Gonzalez. 1999. The Streptomyces coelicolor A3(2) lipAR operon encodes an extracellular lipase and a new type of transcriptional regulator. Microbiology 145:2365-2374. [DOI] [PubMed] [Google Scholar]
- 47.Vogtli, M., P. C. Chang, and S. N. Cohen. 1994. afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol. Microbiol. 14:643-653. [DOI] [PubMed] [Google Scholar]
- 48.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 49.Watve, M. G., R. Tickoo, M. M. Jog, and B. D. Bhole. 2001. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176:386-390. [DOI] [PubMed] [Google Scholar]
- 50.White, J., and M. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widdick, D. A., H. M. Dodd, P. Barraille, J. White, T. H. Stein, K. F. Chater, M. J. Gasson, and M. J. Bibb. 2003. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc. Natl. Acad. Sci. USA 100:4316-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wietzorrek, A., M. Bibb, and R. Chakraburtty. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 53.Yin, X., and M. Zabriskie. 2006. The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus. Microbiology 152:2969-2983. [DOI] [PubMed] [Google Scholar]