Abstract
H10407 is a strain of enterotoxigenic Escherichia coli (ETEC) that utilizes CFA/I pili to adhere to surfaces of the small intestine, where it elaborates toxins that cause profuse watery diarrhea in humans. Expression of the CFA/I pilus is positively regulated at the level of transcription by CfaD, a member of the AraC/XylS family. DNase I footprinting revealed that the activator has two binding sites upstream of the pilus promoter cfaAp. One site extends from positions −23 to −56, and the other extends from positions −73 to −103 (numbering relative to the transcription start site of cfaAp). Additional CfaD binding sites were predicted within the genome of H10407 by computational analysis. Two of these sites lie upstream of a previously uncharacterized gene, cexE. In vitro DNase I footprinting confirmed that both sites are genuine binding sites, and cexEp::lacZ reporters demonstrated that CfaD is required for the expression of cexE in vivo. The amino terminus of CexE contains a secretory signal peptide that is removed during translocation across the cytoplasmic membrane through the general secretory pathway. These studies suggest that CexE may be a novel ETEC virulence factor because its expression is controlled by the virulence regulator CfaD, and its distribution is restricted to ETEC.
Enterotoxigenic Escherichia coli (ETEC) is a noninvasive pathogen that colonizes the small intestine, and it is one of the most common causes of bacterial diarrhea worldwide (37). Adherence to the intestinal epithelium is mediated by pili (15), although nonpilus adherence factors have also been reported (9, 12, 32). Currently, there are 22 known ETEC pilus serotypes, although a typical strain expresses only two or three. The expression of some of the most common pilus serotypes is dependent upon a transcriptional activator. This includes the CFA/I pilus, which is regulated by CfaD (GenBank accession no. P25393) (6, 31), and the CS1, CS2, CS3, and CS4 pili, which are regulated by Rns (accession no. P16114) (4, 5, 11). CfaD has alternatively been named CfaR (6), and 251 of its 265 residues are identical to Rns. Given their identity to each other, it is not surprising that CfaD and Rns are functionally interchangeable with each other (3, 6) and recognize the same DNA binding sites (23).
CfaD and Rns are members of the AraC/XylS family of transcriptional regulators (16). Like other family members, Rns is not sufficiently soluble for in vitro characterization. However, the limitation of protein solubility was overcome by the addition of maltose binding protein (MBP) to the amino terminus of Rns, which significantly increases the solubility of the regulator without disrupting its function (24). DNase I footprinting of MBP-Rns bound to the CS1 pilus promoter cooBp revealed two binding sites: one extending into the −35 hexamer and a second site between positions −93 and −129 (numbering relative to the transcription start site) (24). Single nucleotide substitutions within each site reduced Rns-dependent expression from cooBp in vivo, demonstrating that both sites are required for the full activation of the CS1 pilus promoter.
Rns also positively regulates its own expression by binding to a distal upstream site, from positions −243 to −210, and two sites downstream of the transcription start site (25). The upstream site and at least one downstream site are essential for positive autoregulation (25). Like most AraC/XylS family members, Rns has two helix-turn-helix motifs in its carboxy-terminal domain, and uracil interference assays have shown that it binds in two adjacent regions of the DNA major groove (24, 25). Thus, it is likely that each helix-turn-helix motif places a recognition helix in the major groove similar to MarA, another AraC/XylS family member for which there is a MarA-DNA cocrystal (28).
A recent study has shown that Rns and CfaD can also function as repressors by binding to a site immediately downstream of the transcription start site for nlpA, encoding an inner membrane lipoprotein, where they prevent the formation of RNA polymerase open complexes (3). Repression of nlpA may decrease the production of outer membrane vesicles, as was shown for mutants carrying nlpA::kan insertions (20). In this study, we sought to further our understanding of the CfaD and Rns regulons. We began by characterizing CfaD's binding sites upstream of the CFA/I pilus promoter. We then compiled this information with previously identified Rns binding sites and used bioinformatics to locate additional CfaD binding sites in the genome of ETEC strain H10407 (10). Two of these sites lie upstream of a previously uncharacterized gene, separate from the CFA/I pilus operon, that encodes a protein that is transported across the cytoplasmic membrane via the general secretory pathway. Because the expression of this protein is regulated by CfaD, we have given it the mnemonic cexE, for CfaD-dependent expression, extracytoplasmic protein.
MATERIALS AND METHODS
Strains and plasmids.
Genomic DNA from ETEC strain H10407 (O78:H11 CfaD+ CFA/I+ STIa+ STIb+ LT+) (10) was used as the template to amplify cexEp(−501 to +312) with primers aat-1for (GCTGGATCCCGAGCGGCGTATAAAA) and aat-1rev (GCGGAATTCCACTTGATTGTATGGAAT), cexEp(−421 to +312) with primers aat-4for (GCGGGATCCAGTCCAGGAAATCGAACG) and aat-1rev, and cexEp(−174 to +312) with primers aat-5for (GCGGGATCCTGTTAATCTCTATCAATACAT) and aat-1rev. Numbering of the cexE promoter is relative to its transcription start site (this study). Underlining in primer sequences indicates primer-template mismatches that add sites for restriction endonucleases. Plasmid pHKLac1 is a promoterless lacZ reporter plasmid with a pir-dependent origin of replication that can be integrated into the chromosome of E. coli at attBHK022 (3). Each cexEp PCR product was digested with BamHI and EcoRI and then ligated into the same sites of pHKLac1 to construct pCexELac1 [cexEp(−501 to +312)::lacZ], pCexELac2 [cexEp(−421 to +312)::lacZ], and pCexELac3 [cexEp(−174 to +312)::lacZ]. Alternatively, the PCR product carrying cexEp(−501 to +312) was cloned into the BamHI and EcoRI sites of pNEB193 (New England Biolabs) to construct pGPM1043. This plasmid was then subjected to oligonucleotide-directed mutagenesis to introduce point mutations within CfaD/Rns binding sites upstream of cexEp. Mutations were confirmed by DNA sequencing, and the mutagenized promoter fragments were then cloned into pHKLac1 as BamHI-EcoRI fragments. Plasmid pCexELac4 carries cexEo1-1, which has four point mutations: −44T to C, −43G to C, −42T to G, and −41T to G. Plasmid pCexELac5 carries cexEo2-1, which also has four point mutations: −482T to G, −481A to G, −479C to A, and −478G to C. With the exception of the point mutations noted above, pCexELac4 and pCexELac5 carry the same cexEp promoter fragment upstream of lacZ as pCexELac1. Each reporter plasmid was integrated into the chromosome of MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flhD5301 deoC1 ptsF25 rbsR] (7) as previously described (3) to produce strains GPM1070 (attBHK022::pCexELac1), GPM1096 (attBHK022::pCexELac2), GPM1097 (attBHK022::pCexELac3), GPM1113 (attBHK022::pCexELac4), and GPM1114 (attBHK022::pCexELac5). Single integrants were verified by colony PCR as previously described (17).
Primers aat-1for (GCTGGATCCCGAGCGGCGTATAAAA) and rrsP-XbaI-Rev (CGCTCTAGAAACATTTTACATAATGTAATCA) were used to amplify the cexE locus from human ETEC strains 27D (O126:nonmotile CfaD+ CFA/I+ STIb+) (21) and G427 (O28:H12 CfaD+ CFA/I+ STIa+ LT+) (data not shown) (isolated in the Middle East during Operation Desert Shield ca. 1990). The 1-kb PCR products were digested with BamHI and XbaI and then ligated into the same sites of pNEB193 to construct pGPM1039-27D and pGPM1039-G427. Plasmid pGPM1034 expresses CexE-His6 from a T7 RNA polymerase-dependent promoter and was constructed by amplifying cexE from H10407 with primers aat-1for (see above) and aat-XhoRev (GGACCTCGAGTTTATACCAATAAGGGGTGTCAC). The PCR product was digested with BamHI and XhoI and then ligated into the same sites of pET33b (Novagen).
Plasmid pMBPRns1 (3) was used for the expression of the fusion protein MBP-Rns in KS1000 [F′ lacIq lac+ pro+/araΔ(lac-pro) Δprc::kan eda51::Tn10 gyrA rpoB thi-1 argI(Am)] (New England Biolabs)/pRare2 (Novagen) (used to provide rare tRNAs). Plasmid pGPMRns (3) expresses Rns from lacp and is a derivative of pNEB193 (New England Biolabs). Transposon mutagenesis of pGPMRns produced pGPMRns<Tn>2, which carries rns::kan. Plasmid pNTP503 (36) expresses CfaD and is a derivative of cloning vector pBR322. Cloning vector pHSG576 (33) (GenBank accession no. D88215) has an expected copy number of ∼5 per cell due to its pSC101-derived replicon (30, 35) and was used to construct pEU2035 (13), which expresses Rns from lacp.
DNA sequencing of the ETEC cexE locus.
The genome of ETEC strain H10407 (10) is being sequenced by the Sanger Institute in collaboration with Ian Henderson and Mark Pallen. H10407 preliminary sequence data were obtained from http://www.sanger.ac.uk/Projects/E_coli_H10407/.
The cexE locus was cloned from two CFA/I+ strains of ETEC, 27D (21) and G427 (data not shown). For each clone (pGPM1039-27D and pGPM1039-G427), both strands of the cexE locus were sequenced. The 990-bp locus was found to be identical among ETEC strains 27D, G427, and H10407.
Software.
Software for the analysis of DNA sequences using Regular Expressions was designed and written by G. P. Munson. The Web server SignalP was used to evaluate the amino terminus of CexE (GenBank accession no. ABM92275) and pCoo087 (accession no. CAI79570) for potential secretory signal peptides (http://www.cbs.dtu.dk/services/SignalP/) (2).
Purification of RNA.
Strain GPM1070/pGPMRns was cultured aerobically in 10 ml of Luria-Bertani (LB) medium at 37°C. After the optical absorbance at 550 nm reached 1.0, 2 ml of a 5% (vol/vol) phenol-95% (vol/vol) ethanol solution was added to the culture, and the cells were pelleted. The cell pellet was suspended in 10 ml of RNA wash buffer (0.75% [vol/vol] NaCl, 0.8% [vol/vol] phenol, 15.8% [vol/vol] ethanol) and then centrifuged. The resulting pellet was suspended in 500 μl 0.9% NaCl and 500 μl water-saturated, nonbuffered phenol and then shaken at room temperature for 30 min. Subsequently, 50 μl of a 24:1 chloroform-isoamyl alcohol solution was added and then incubated for an additional 15 min. The solution was then chilled on ice for 5 min and centrifuged in a microcentrifuge at the maximum rpm for 5 min. RNA was ethanol precipitated from the supernatant and then suspended in 22 μl of RNase-free water.
Primer extension.
Two picomoles of 32P-end-labeled oligonucleotide (rrsP3rev [GCAGAATTCGCGGAGAGAGACCCCATAG]) was combined with 5 μg of total RNA and 0.8 mM deoxynucleoside triphosphates. The solution was heated to 65°C for 5 min and then chilled on ice for 2 min. The annealed primer was then extended with SuperScript III reverse transcriptase according to the supplier's protocol (Invitrogen). Heat-denatured aliquots were separated on DNA sequencing gels alongside dideoxy chain-terminated sequencing ladders (30).
Purification of MBP-Rns.
MBP-Rns was purified from strain KS1000/pRare2/pMBPRns1 as previously described (3). In brief, the strain was grown aerobically at 37°C in LB medium containing 0.2% (wt/vol) glucose, 30 μg/ml chloramphenicol, and 100 μg/ml ampicillin. Upon reaching mid-log phase, the culture was cooled to 30°C. Expression of MBP-Rns was induced for several hours by the addition isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 300 μM. Bacterial cells were then collected by centrifugation and concentrated >100-fold in cold lysis buffer (10 mM Tris Cl [pH 7.6, at room temperature], 200 mM NaCl, 1 mM EDTA, 0.5 mM CaCl2, 10 mM β-mercaptoethanol, 100 μg/ml DNase I). Cells were lysed by passage through a French press. Insoluble material was removed from the lysate by centrifugation. MBP-Rns was then bound to an amylose column equilibrated with buffer A (10 mM Tris Cl [pH 7.6, at room temperature], 200 mM NaCl, 1 mM EDTA, 15% [vol/vol] glycerol, and 10 mM β-mercaptoethanol). The fusion protein was eluted from the amylose column with buffer B (buffer A with 10 mM maltose).
Purification and sequencing of CexE-His6.
Strain BL21(DE3)/pGPM1034 was grown aerobically at 37°C in LB medium containing 0.2% (wt/vol) glucose and 50 μg/ml kanamycin. Expression of CexE-His6 was induced during mid-log phase by the addition of IPTG to a final concentration of 500 μM. After 4 h of induction, approximately 325 ml of spent culture medium was cooled to 4°C and then passed through a nickel-Sepharose column equilibrated with IMAC-A buffer (50 mM Tris Cl [pH 7.6, at room temperature], 300 mM NaCl, 10 mM imidazole). The column was washed with several column volumes of IMAC-A buffer, and CexE-His6 was then eluted by increasing the concentration of imidazole to 154 mM. Approximately 50 picomoles of purified protein was transferred onto a polyvinylidene fluoride membrane from a 15% sodium dodecyl sulfate-polyacrylamide gel by electroblotting in 10 mM N-cyclohexyl-3-aminopropanesulfonic acid (pH 11.0)-10% (vol/vol) methanol buffer. The immobilized protein was visualized on the membrane with Coomassie brilliant blue R-250 and submitted to the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for amino-terminal sequencing by Edman degradation.
DNase I footprinting.
Purified MBP-Rns was equilibrated at 37°C for 30 min with cexE promoter DNA uniquely labeled with 32P on the 5′ terminus of the coding or noncoding strand in footprinting buffer [10 mM Tris Cl (pH 7.6, at room temperature), 50 mM KCl, 1 mM dithiothreitol, 0.4 mM MgCl2, 0.2 mM CaCl2, 2 ng/μl poly(dI-dC), 10 μg/ml bovine serum albumin]. The samples were then treated with DNase I at a final concentration of 100 ng/μl for 1 min at 37°C. DNase I cleavage reactions were terminated by the addition of 10 volumes of DNase I stop buffer (570 mM ammonium acetate, 50 μg/ml tRNA, 80% [vol/vol] ethanol) and then precipitated on dry ice. DNA pellets were rinsed with 70% (vol/vol) ethanol and then dried. DNA samples were separated on DNA sequencing gels after heat denaturation in 4 μl of loading buffer (80% [vol/vol] formamide, 50 mM Tris-borate [pH 8.3], 1 mM EDTA, 0.1% [wt/vol] xylene cyanol, and bromophenol blue). The Maxam-Gilbert method was used to generate GA and TC sequence ladders (30).
β-Galactosidase assays.
Reporter strains GPM1070, GPM1096, GPM1097, GPM1113, and GPM1114 transformed with pGPMRns (Rns+), pGPMRns<Tn>2 (rns::kan), pNTP503 (CfaD+), or vector pBR322 were grown in LB medium with 100 μg/ml ampicillin. Reporter strains transformed with low-copy-number plasmid pEU2035 (Rns+) or vector pHSG576 were grown in LB medium with 30 μg/ml chloramphenicol. All strains were grown aerobically at 37°C. Cells were harvested during the log phase of growth, lysed, and assayed for β-galactosidase activity as previously described (22).
Accession numbers.
The sequence of the cexE locus from strain 27D has been submitted to the GenBank database under accession number EF205439. The amino acid sequence of CexE is available from GenBank under accession number ABM92275.
RESULTS
Identification of CfaD's binding sites at the CFA/I pilus promoter.
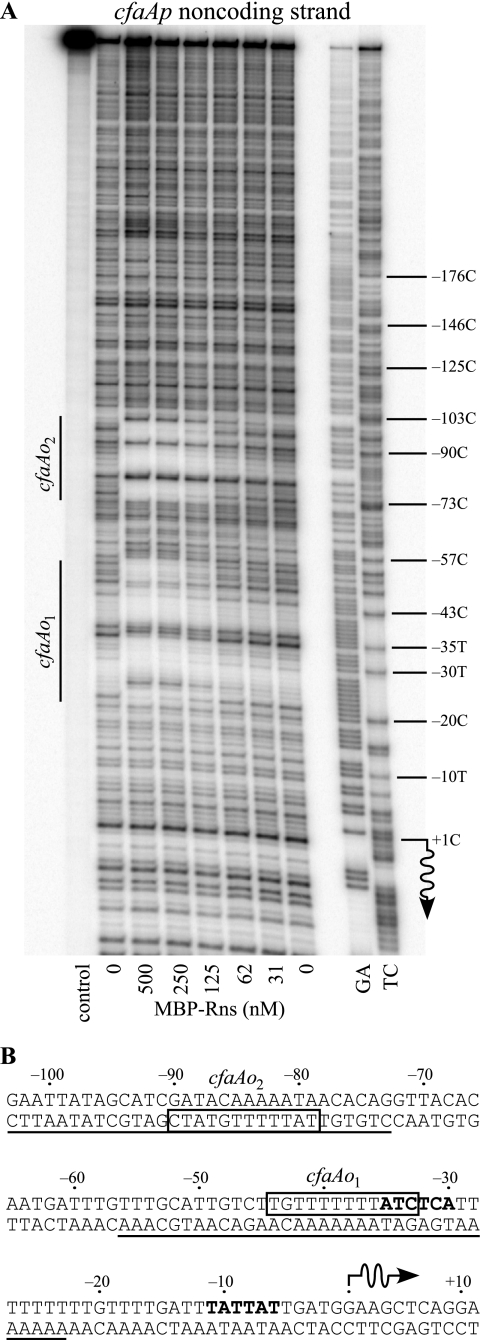
Previous studies have shown that CfaD and Rns are fully interchangeable with each other (3, 6) and recognize the same DNA binding sites (23). Therefore, for our in vitro DNA binding studies, we utilized a previously characterized MBP-Rns fusion (24, 25) to identify the CfaD/Rns binding sites on a DNA fragment carrying the CFA/I pilus promoter cfaAp from positions −469 to +91 (numbering relative to the previously reported transcription start site) (18). The DNase I footprint of MBP-Rns bound to the cfaAp fragment revealed two distinct regions of protection (Fig. 1A). We have designated the footprint encompassing positions −23 to −56 as site cfaAo1 and the footprint extending from positions −73 and −103 as site cfaAo2. DNase I is a large enzyme that overestimates DNA binding sites due to steric occlusion; however, sites cfaAo1 and cfaAo2 each contain a run of 12 nucleotides (Fig. 1B) that are similar to sequences within other CfaD/Rns binding sites (Fig. 2B). Although Rns may contact DNA beyond this central core of 12 bp, the sequence conservation within the core suggests that it contains most of the base-specific contacts for Rns. This is supported by uracil interference assays, which identified thymine C-5 methyl groups critical for Rns binding (24, 25). At the five DNA binding sites examined, thymine C-5 methyl groups beyond the central core of 12 nucleotides were not required for Rns binding, but three within the core were.
FIG. 1.
Locations of CfaD/Rns binding sites upstream of the CFA/I pilus promoter, cfaAp. Numbering is relative to the CfaD-dependent transcription start site of cfaAp, whose location is indicated by a wavy arrow. (A) DNase I protection of MBP-Rns bound to the noncoding strand of cfaAp. CfaD/Rns binding sites are indicated as cfaAo1 and cfaAo2. DNase I was not added to the DNA in the lane labeled control. Lanes labeled GA and TC contain Maxam-Gilbert sequencing ladders. (B) Sequence of cfaAp and upstream region. The underlined sequence indicates the extent of MBP-Rns DNase I footprints on the noncoding strand. Boxed sequences indicate nucleotides that are conserved among various CfaD/Rns binding sites. Probable −35 and −10 hexamers are shown in boldface type.
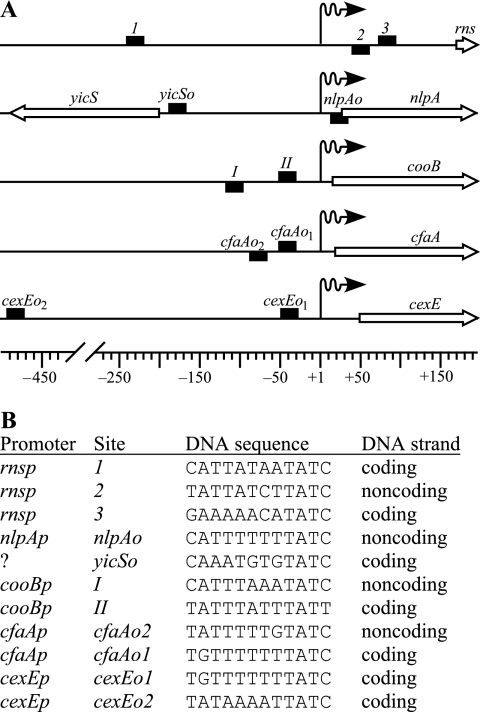
FIG. 2.
Positions and alignment of CfaD/Rns binding sites. (A) CfaD/Rns binding sites are represented as filled rectangles. The position of the rectangle above or below the line indicates whether the conserved sequence occurs on the coding or noncoding strand, respectively. Numbering is relative to the transcription start sites, which are represented by wavy arrows. ETEC promoters rnsp, cooBp (CS1 pilus), cfaAp (CFA/I pilus), and cexEp are activated by Rns or CfaD. In contrast, nlpA is common to both ETEC and K-12 strains and its promoter, nlpAp, is repressed by Rns or CfaD. It is not known if the occupancy of binding site yicSo affects expression from yicS. (B) Optimal alignment of the conserved core sequence of each CfaD/Rns binding site. Although CfaD and Rns may contact DNA beyond the nucleotides shown, the greatest sequence conservation occurs within this core of 12 bp.
We note a sequence discrepancy within site cfaAo2 between our DNA sequencing results and the CFA/I sequence file in GenBank (accession no. M55661). In M55661, nucleotides 741 to 750 are reported as being “GATACCAAAT,” but we have found the sequence to be “GATACAAAAAT” (corrections and additions are underlined). These changes have been incorporated into the sequence shown in Fig. 1B and 2B.
Prediction and verification of additional CfaD binding sites.
A recent study has shown that CfaD also represses the transcription of nlpA, which encodes an inner membrane protein, in addition to activating the expression of the CFA/I pilin genes (3). This suggests that the CfaD regulon may be larger than previously realized. Since CfaD is a DNA binding protein, we sought to identify additional CfaD-regulated genes by searching the genome of ETEC strain H10407 for sites similar to known CfaD/Rns binding sites. We first aligned the two binding sites located upstream of cfaAp to previously reported Rns binding sites at the promoters for the CS1 pilus (24), nlpA (3), and rns (25) (Fig. 2). Because CfaD/Rns binding sites are not palindromic, the sequences shown are found on the coding or noncoding strands of DNA as stated. Only the central core of 12 nucleotides within each binding site were used in the alignment because these nucleotides have the greatest conservation. The alignment was then used to write a text search string, “[CTG][AG][TA][TA][TA][TAG][ATC][ATG]TAT[CT],” in the form of Regular Expression (26), where each position is represented by a nucleotide set, enclosed by brackets, or a single character. At each position, potential binding sites must match one character within each enclosed set as well as exact matches to “TAT” at positions 9, 10, and 11. With this approach, two potential binding sites were identified upstream of a previously uncharacterized gene, cexE (Fig. 2). The site most proximal to cexE, cexEo1, is identical to the CfaD/Rns binding site cfaAo1 upstream of the CFA/I pilus promoter (Fig. 1B and 2B).
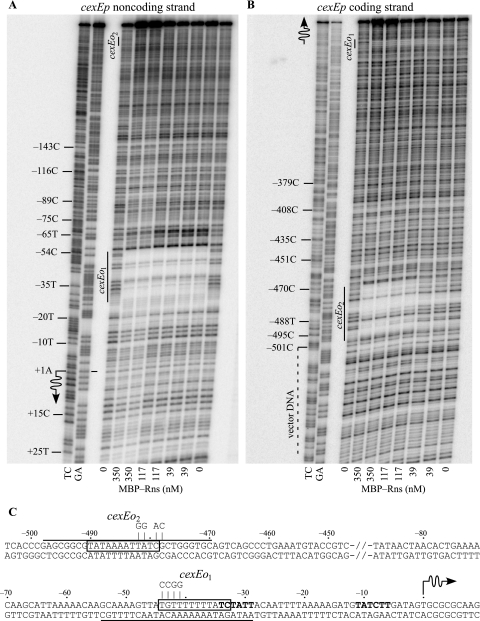
We next determined that MBP-Rns binds to both of the predicted CfaD/Rns binding sites by in vitro DNase I footprinting (Fig. 3). The predicted site centered at position −38, cexEo1, was fully encompassed by the MBP-Rns DNase I footprint, which extends from positions −29 to −54 (Fig. 3A) (numbering relative to the transcription start site of cexE) (see below). A second DNase I footprint was observed between positions −470 and −498, which corresponds to site cexEo2 (Fig. 3B). Additional DNase I footprinting experiments revealed that cexEo1 and cexEo2 are the only MBP-Rns binding sites within the region examined, from position −501 through +7.
FIG. 3.
Locations of CfaD/Rns binding sites upstream of cexEp. Numbering is relative to the transcription start site of cexEp, whose location is indicated by a wavy arrow. DNase I protection of MBP-Rns bound to the (A) noncoding and (B) coding strands of cexEp. CfaD/Rns binding sites are indicated as cexEo1 and cexEo2. Lanes labeled GA and TC contain Maxam-Gilbert sequencing ladders. (C) Sequence of cexEp and upstream region. The underlined sequence indicates the extent of MBP-Rns DNase I footprints on the noncoding or coding strand. Boxed sequences indicate nucleotides that are conserved among various CfaD/Rns binding sites. Probable −35 and −10 hexamers are shown in boldface type. Point mutations, shown above each binding site, were introduced into cexEo1 and cexEo2 so that the function of each site could be evaluated in vivo.
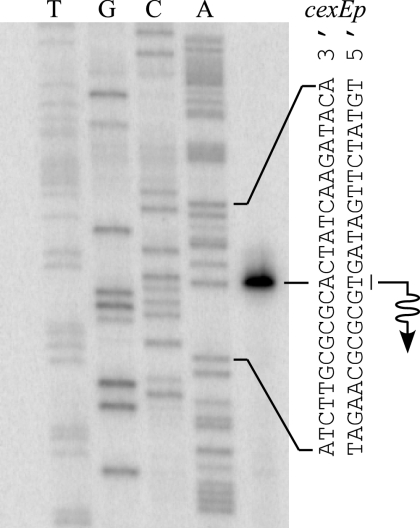
Transcription start site of cexE.
We next mapped the transcription start site of the cexE promoter, cexEp, by primer extension because the position of a regulator's binding sites relative to a promoter often provides an indication of the regulator's function (8). In the presence of Rns, we found that cexE mRNA begins at a thymine nucleotide that is 48 bp upstream of the first ATG codon of cexE (Fig. 4). Thus, the DNase I footprint of MBP-Rns bound to site cexEo1 encompasses the −35 hexamer of cexEp (Fig. 3C). This is similar to the location of site cfaAo1 at the CfaD-activated CFA/I pilus promoter (Fig. 1B). We did not map the transcription start site of cexEp in the absence of Rns because the cexE promoter has low activity in the absence of an activator (Table 1).
FIG. 4.
Transcription start site of cexEp. Primer extension was used to map the Rns-independent transcription start site of cexEp with reverse transcriptase and RNA harvested from strain GPM1070/ pGPMRns (Rns+) growing logarithmically in LB medium. A dideoxy chain-terminated sequencing ladder was generated with the same primer used for reverse transcription. The first nucleotide of the cexE mRNA is underlined, and the wavy arrow indicates the direction of transcription.
TABLE 1.
CfaD and Rns activate expression of cexE
| lacZ reporter constructb | Μean β-galactosidase activity (Miller units) ± SDa
|
|||||
|---|---|---|---|---|---|---|
| Vector | CfaD | rns::kan | Rns | Vectorc | Rnsc | |
| cexEp(−501 to +312) | BD | 6,599 ± 314 | BD | 6,613 ± 391 | 7 ± 3 | 10,587 ± 605 |
| cexEp(−501 to +312) cexEo1-1 | BD | 136 ± 7 | BD | 384 ± 36 | 18 ± 3 | 415 ± 27 |
| cexEp(−501 to +312) cexEo2-1 | BD | 4,110 ± 193 | BD | 4,527 ± 351 | 16 ± 4 | 5,736 ± 261 |
| cexEp(−421 to +312) ΔcexEo2 | 15 ± 5 | 5,582 ± 472 | BD | 2,084 ± 171 | 11 ± 6 | 7,539 ± 200 |
| cexEp(−174 to +312) ΔcexEo2 | 45 ± 5 | 5,764 ± 212 | BD | 3,809 ± 320 | 101 ± 8 | 7,483 ± 293 |
Values are reported in Miller units (22) (means and standard deviations) (n ≥ 3). BD, below the limit of detection.
Numbering relative to the transcription start site of cexEp.
Low-copy-number plasmid replicon.
Expression of cexE is CfaD dependent.
To determine if the expression of cexE is activated by CfaD or Rns, cexEp::lacZ reporters were constructed and integrated into the chromosome of K-12 strain MC4100. Each reporter construct carries a transcriptional terminator upstream of the promoter fragment to prevent readthrough from promoters in the chromosome. Reporter strains were then transformed with plasmids expressing CfaD, Rns, or negative control plasmids and assayed for the expression of β-galactosidase (Table 1). All five reporter constructs had very low levels of β-galactosidase expression in the absence of CfaD and Rns. In some cases, the expression of β-galactosidase was below the limits of detection when cells were harvested during the log phase of growth. However, we observed significant β-galactosidase expression in the presence of CfaD or Rns, demonstrating that cexEp is activated by these virulence regulators (Table 1).
Point mutations, shown in Fig. 3C, were introduced into the conserved core sequence of each of the two CfaD/Rns binding sites to determine if both binding sites are required for the activation of cexEp. Reporter cexEp(−501 to +312)::lacZ cexEo1-1 carries four point mutations in the promoter-proximal binding site. These mutations reduced CfaD-dependent expression by 98% and Rns-dependent expression by 94% but had no detectable effect upon CfaD- and Rns-independent expression (Table 1). Mutations within the distal upstream site, cexEo2-1, also reduced CfaD- and Rns-dependent expression from cexEp, but in each case, the reduction was less than 40%. These effects were also observed when Rns was expressed from a plasmid with a low copy number (Table 1). This suggests that cexEo2 is functional in vivo despite its distance from the promoter and the fact that it appears to be a low-affinity site in vitro. We did observe higher levels of expression from cexEp(−501 to +312)::lacZ when Rns was expressed from a low-copy-number replicon versus a high-copy-number replicon even though it was expressed from the same promoter, lacp, in both cases. However, strains with low-copy-number replicons could not be cultured in the same medium as those with high-copy-number replicons, and therefore, the absolute levels of β-galactosidase expression are not directly comparable.
The distal upstream binding site, cexEo2, was deleted from reporters cexEp(−421 to +312)::lacZ and cexEp(−174 to +312)::lacZ. These deletions reduced both CfaD- and Rns-dependent expression of β-galactosidase, although we observed that the various strains had considerable variations in expression (Table 1). The reasons for this variation are unclear, but the reduction in CfaD- and Rns-dependent expression from cexEp upon the deletion of cexEo2 is consistent with the effect of point mutations in that site. Taken together, these results demonstrate that CfaD and Rns activate the expression of cexE and that both DNA binding sites are required for full activation.
CexE is exported from the cytoplasm.
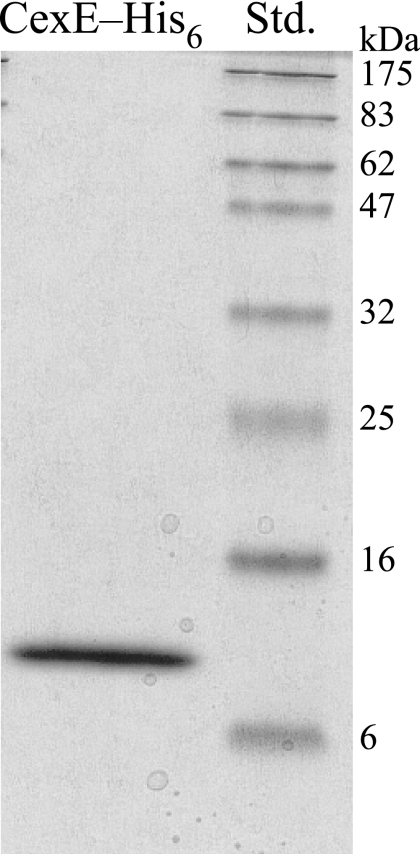
CexE (GenBank accession no. ABM92275) is a 120-amino-acid, 12.6-kDa protein. The first 19 amino acids of CexE have many of the features of a signal peptide: lysines at residues 2 and 3, which would impart a net positive charge; an α-helical hydrophobic region at residues 4 through 12; and residues 13 through 19, comprising the recognition site for a signal peptidase (2, 34). These features suggest that CexE is transported across the cytoplasmic membrane via the general secretory pathway (27). To determine if CexE is an exported protein, a hexahistidine tag was added to its carboxy terminus, and the epitope-tagged protein was expressed in strain BL21(DE3)/pGPM1034. After 4 h of expression, the tagged protein was purified from the growth medium by immobilized metal ion affinity chromatography (Fig. 5). The first 10 amino acids of the purified protein were determined to be “GGGNSERPPS” by Edman degradation, which is identical to residues 20 through 29 of CexE. Thus, CexE is transported from the cytoplasm by the general secretory pathway, and its signal peptide is removed by a signal peptidase. Although we purified CexE-His6 from the culture medium, analysis of the cell pellet revealed that the majority of the protein remained associated with the cell, and approximately 50% of the cell-associated protein can be released from the periplasm by chloroform shock (1) (data not shown).
FIG. 5.
Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel of purified CexE-His6. CexE-His6 was purified from the culture medium of an E. coli strain expressing the fusion protein by immobilized metal ion affinity chromatography. The amino terminus of the purified protein was determined to be “GGGNSERPPS” by Edman degradation, demonstrating that the amino terminus of CexE contains a signal peptide and that its first 19 amino acids are removed during translocation from the cytoplasm. The expected molecular mass of CexE-His6 after removal of its signal peptide is 11.7 kDa. Std., protein standards.
DISCUSSION
Pili function as the primary adherence factors of ETEC (15), and some of the most prevalent pilus serotypes require a transcriptional activator for the expression of pilin genes. These include the CFA/I pilus, which is regulated by CfaD (6, 32), and the Rns-regulated CS1, CS2, CS3, and CS4 pili (4, 5, 11). CfaD and Rns are fully interchangeable and recognize the same DNA binding sites (23). This allowed us to use an MBP-Rns fusion protein in lieu of CfaD to identify the activator's binding sites at the CFA/I pilus promoter. DNase I footprinting revealed two DNA binding sites, site cfaAo1, from positions −23 to −56, and site cfaAo2, from positions −73 to −103 (numbering relative to the transcription start site) (18). The arrangement of CfaD's binding sites at cfaAp is very similar to those of Rns at the CS1 pilus promoter, cooBp (Fig. 2A) (24). At both promoters, the activators bind to a site that extends into the −35 hexamer. However, Rns binds to a second site, site II, between positions −93 and −129, that is further upstream from the CS1 promoter than site cfaAo2 at the CFA/I promoter. Site cfaAo2 is not essential for the activation of the CFA/I pilus promoter because it was fortuitously deleted from cfaAp::lacZ reporters in two previous studies without abolishing CfaD-dependent expression of β-galactosidase (18, 19). However, it is likely that CfaD binding to site cfaAo2 contributes to the activation of cfaAp, as has been shown for Rns binding to the promoter-distal site at the CS1 promoter (24).
The identification of cfaAo1 and cfaAo2 has increased the number of known CfaD/Rns binding sites from seven to nine (Fig. 2). The recent availability of the genomic sequence from ETEC strain H10407 has made it possible to exploit this knowledge for the identification of additional genes within the CfaD regulon. By using custom-designed software that accepted a search parameter that represents the full range of CfaD/Rns binding sites, we were able to identify several dozen potential CfaD binding sites in the genome of H10407. Although some of these predicted sites have proven to be nonfunctional, others have not (G. P. Munson, unpublished data). In this study, we have shown that two of these predicted sites are bound by MBP-Rns in vitro and that they are involved in the activation of a previously uncharacterized gene, which we have given the mnemonic cexE, for CfaD-dependent expression, extracytoplasmic protein. Like the CFA/I and CS1 pilus promoters, CfaD/Rns has a binding site that extends into the −35 hexamer of cexEp. Curiously, the second binding site (cexEo2, between positions −470 and −498) is much further upstream of cexEp than site cfaAo2 at cfaAp or site II at the CS1 pilus promoter (Fig. 2A). Despite the distance of cexEo2 from cexEp, point mutations within cexEo2 reduced CfaD- and Rns-dependent expression of β-galactosidase from cexEp::lacZ reporters (Table 1). However, deletion of cexEo2 did not abolish CfaD and Rns activation of cexEp. In contrast, expression from cexEp was nearly abolished by point mutations within the promoter-proximal site cexEo1. Thus, both sites are functional in vivo, and, as expected, the site closest to RNA polymerase has the greater contribution towards transcriptional activation than the distal site.
In this study, we have shown that CexE is an extracytoplasmic protein whose secretory signal peptide is cleaved from a CexE-His6 fusion protein between residues Ala19 and Gly20. We initially purified CexE-His6 from the culture medium of a strain expressing the fusion protein, but we have subsequently found that the majority of the protein remains cell associated, with at least 50% within the periplasm. Thus, the relatively small fraction of extracellular CexE-His6 that we recovered was most likely the result of leakage from the periplasm or cell lysis rather than specific transport across the outer membrane. However, we cannot yet exclude the possibility that CexE will be further processed and/or exported from the periplasm by an ETEC system that laboratory strains of E. coli lack or under growth conditions that we have not replicated.
In addition to its presence in H10407, we have found the cexE locus in CFA/I+ strains unrelated to H10407 (data not shown). The locus was cloned and sequenced (GenBank accession no. EF205439) from two of those strains, 27D (O126:nonmotile CfaD+ CFA/I+ STIb+) and G427 (O28:H12 CfaD+ CFA/I+ STIa+ LT+), and found to be identical to that of H10407 (O78:H11 CfaD+ CFA/I+ STIb+ LT+). However, we have also found CFA/I+ strains that lack cexE (M. C. Pilonieta and G. P. Munson, unpublished data). This indicates that the function of CexE is probably unrelated to the assembly of the CFA/I pilus and is further supported by experiments that have shown that the expression of the pilin genes in K-12 strains results in CFA/I+ bacteria (6, 29).
CexE has no homology to any characterized protein or domain that would provide clues as to its function other than the presence of a secretory signal peptide at its amino terminus. In fact, there is currently only one protein, pCoo087 (GenBank accession no. CAI79570), with significant homology to CexE. The gene, orf087, encoding this protein is carried on the 98-kb pCoo virulence plasmid (accession no. CR942285) in a Rns+ CS1+ ETEC strain (14). Like CexE, pCoo087 has a potential secretory signal peptide at its amino terminus with a predicted cleavage site between residues Ala19 and Gly20. Cleavage would produce an 11-kDa protein, similar in size to CexE after cleavage of its signal peptide. Unlike cexE, the expression of orf087 is not activated by Rns or CfaD (M. D. Bodero and G. P. Munson, unpublished data). Although the function of CexE is unknown, the fact that its expression is dependent upon the CfaD virulence regulator and the fact that it is found only in ETEC strains suggest that it may be an ETEC virulence factor. Therefore, future studies will seek to identify the function of this novel ETEC protein.
Acknowledgments
We thank L. Peruski and S. Savarino for providing ETEC strains. We also thank I. Henderson, M. Pallen, and the Sanger Institute for providing internet access to the genome of H10407 prior to publication. Protein sequencing was performed by the staff of the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
This research was supported by NIH NIAID Public Health Service award AI 057648 and the University of Miami Miller School of Medicine.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Ames, G. F., C. Prody, and S. Kustu. 1984. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J. Bacteriol. 160:1181-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bodero, M. D., M. C. Pilonieta, and G. P. Munson. 2007. Repression of the inner membrane lipoprotein NlpA by Rns in enterotoxigenic Escherichia coli. J. Bacteriol. 189:1627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron, J., D. R. Maneval, J. B. Kaper, and J. R. Scott. 1990. Association of rns homologs with colonization factor antigens in clinical Escherichia coli isolates. Infect. Immun. 58:3442-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caron, J., and J. R. Scott. 1990. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 58:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 8.Collado-Vides, J., B. Magasanik, and J. D. Gralla. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55:371-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre, D., S. Ludi, M. Stoffel, J. Frey, M. P. Horn, G. Dietrich, S. Spreng, and J. F. Viret. 2006. Expression of enterotoxigenic Escherichia coli colonization factors in Vibrio cholerae. Vaccine 24:4354-4368. [DOI] [PubMed] [Google Scholar]
- 12.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froehlich, B., L. Husmann, J. Caron, and J. R. Scott. 1994. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 176:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Froehlich, B., J. Parkhill, M. Sanders, M. A. Quail, and J. R. Scott. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 16.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordi, B. J., B. Dagberg, L. A. de Haan, A. M. Hamers, B. A. van der Zeijst, W. Gaastra, and B. E. Uhlin. 1992. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 11:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordi, B. J., B. A. van der Zeijst, and W. Gaastra. 1994. Regions of the CFA/I promoter involved in the activation by the transcriptional activator CfaD and repression by the histone-like protein H-NS. Biochimie 76:1052-1054. [DOI] [PubMed] [Google Scholar]
- 20.McBroom, A. J., A. P. Johnson, S. Vemulapalli, and M. J. Kuehn. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVeigh, A., A. Fasano, D. A. Scott, S. Jelacic, S. L. Moseley, D. C. Robertson, and S. J. Savarino. 2000. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 68:5710-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 26.Neuburg, M. 2001. REALbasic: the definitive guide, 2nd ed. O'Reilly, Sebastopol, CA.
- 27.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakellaris, H., G. P. Munson, and J. R. Scott. 1999. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc. Natl. Acad. Sci. USA 96:12828-12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Savelkoul, P. H., G. A. Willshaw, M. M. McConnell, H. R. Smith, A. M. Hamers, B. A. van der Zeijst, and W. Gaastra. 1990. Expression of CFA/I fimbriae is positively regulated. Microb. Pathog. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 32.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 34.von Heijne, G. 1990. The signal peptide. J. Membr. Biol. 115:195-201. [DOI] [PubMed] [Google Scholar]
- 35.Wadood, A., M. Dohmoto, S. Sugiura, and K. Yamaguchi. 1997. Characterization of copy number mutants of plasmid pSC101. J. Gen. Appl. Microbiol. 43:309-316. [DOI] [PubMed] [Google Scholar]
- 36.Willshaw, G. A., H. R. Smith, and B. Rowe. 1983. Cloning of regions encoding colonisation factor antigen 1 and heat-stable enterotoxin in Escherichia coli. FEMS Microbiol. Lett. 16:101-106. [Google Scholar]
- 37.World Health Organization. 2006. Future directions for research on enterotoxigenic Escherichia coli vaccines in developing countries. Wkly. Epidemiol. Rec. 81:97-104. [PubMed] [Google Scholar]