Abstract
Vegetative forms of Bacillus anthracis replicate in tissues of an infected host and precipitate lethal anthrax disease. Upon host death, bacilli form dormant spores that contaminate the environment, thereby gaining entry into new hosts where spores germinate and once again replicate as vegetative forms. We show here that sortase C, an enzyme that is required for the formation of infectious spores, anchors BasI polypeptide to the envelope of predivisional sporulating bacilli. BasI anchoring to the cell wall requires the active site cysteine of sortase C and an LPNTA motif sorting signal at the C-terminal end of the BasI precursor. The LPNTA motif of BasI is cleaved between the threonine (T) and the alanine (A) residue; the C-terminal carboxyl group of threonine is subsequently amide linked to the side chain amino group of diaminopimelic acid within the wall peptides of B. anthracis peptidoglycan.
During infection, the cell wall envelope of gram-positive bacteria functions as a surface organelle for microbial interaction with host tissues (34). Many pathogenic strategies of bacteria utilize surface proteins to interact with extracellular matrices, specific host molecules, or target cells and thereby enable bacterial adherence to tissues, target cell invasion, or evasion of immune responses (14, 41). Many, but not all, surface proteins of gram-positive bacteria are anchored to the cell wall envelope by a mechanism requiring a C-terminal sorting signal with an LPXTG motif (10, 46, 47). These surface proteins are synthesized as precursors with N-terminal signal peptides in the bacterial cytoplasm (P1 precursor) (1, 5). After membrane translocation and signal peptide cleavage, the C-terminal sorting signal first retains the polypeptide in the cytoplasmic membrane (P2 precursor) (46). Membrane-anchored sortase A then cleaves the sorting signal between the threonine (T) and the glycine (G) residues (29, 33), generating an acylated enzyme with a thioester bond between the sortase active-site thiol group and the C-terminal carboxyl group of threonine (60, 61). Sortase acyl enzyme intermediates are resolved by nucleophilic attack of the amino group of cross-bridges within wall peptides, thereby anchoring the C terminus of surface proteins to the cell wall envelope (35, 58, 61, 62).
The cell wall of gram-positive bacteria is composed of peptidoglycan (43), a heteropolymeric macromolecule comprised of glycan strands and attached wall peptides (3, 16, 54, 55). In Bacillus species, including B. anthracis, glycan strands consist of the repeating disaccharide N-acetylmuramic acid-(β1-4)-N-acetylglucosamine (MurNAc-GlcNAc) with an average length of 10 to 20 disaccharide subunits (19, 64). Depending on the species, various degrees of hexosamine deacetylation have been observed (4, 17, 49). For example, the genome of B. anthracis encodes six GlcNAc deacetylases that together deacetylate 88% of GlcNAc residues in peptidoglycan to glucosamine (GlcNH2) (39, 66). In mature peptidoglycan a short peptide component, wall tetrapeptide (l-Ala-d-iGln-m-Dap-d-Ala) or tripeptide (l-Ala-d-iGln-m-Dap), is attached to the glycan strands via an amide bond between l-Ala and the lactyl moiety of MurNAc (49, 64). Some of the wall peptides within assembled peptidoglycan are cross-linked to neighboring wall peptides, i.e., the side chain amino group of meso-diaminopimelic acid (m-Dap) at position 3 of the wall peptide is amide linked to the carboxyl groups of d-Ala at position 4 of the neighboring wall peptide (49, 64). The degree of cross-linking can vary between Bacillus species and growth stages of bacilli. For vegetative forms of B. anthracis, 19% of m-Dap side chains in wall peptides are engaged in cross-linking (49).
B. anthracis encodes three sortases (26, 40). Sortase A cleaves the LPXTG motif sorting signals of seven surface proteins destined for the cell wall envelope of vegetative bacilli (15). Sortase B recognizes the NPKTG motif of BasK (IsdC), a heme-binding protein involved in uptake of iron during infection (24). Sortase C, whose genetic determinant (srtC) is located in the basI-srtC-sctR-sctS operon, is required for the formation of infectious spores in tissues of animals that have succumbed to anthrax disease (28). A hallmark of sporulation in Bacillus spp. is the formation of an asymmetrically positioned (polar) septum that divides the developing cell into dissimilarly sized progeny called the mother cell and the forespore, the smaller cell that develops into a mature spore (22, 36). basI specifies a protein substrate of sortase C with an LPNTA-motif sorting signal, whereas sctR-sctS encodes for a two-component regulatory system (28). A second gene specifying a polypeptide with a LPNTA-motif sorting signal, basH, is located elsewhere on the B. anthracis genome and is expressed by σF RNA polymerase only within the developing forespore (28). The srtC operon is regulated by the response regulator sctR, which activates srtC and basI expression 2 h prior to polar septation during the developmental program that leads to spore formation (28). Previous work indicated that srtC is expressed in the predivisional sporulating cell, and its protein product is at least in part inherited by both mother cell and spore envelopes (28). SrtC within forespores is responsible for the anchoring of BasH to the peptidoglycan of this compartment (28). This work left unresolved whether sortase C also anchors BasI to the envelope of forespores. Here we examined the subcellular location and anchor mechanism of BasI. In contrast to BasH, BasI is anchored to the predivisional cell envelope during sporulation and hence is not present in the forespore peptidoglycan. Anchoring of BasI to the cell wall absolutely requires its LPNTA motif sorting signal and the active-site cysteine of SrtC. Sortase C cleaves the LPNTA motif of BasI between the threonine (T) and the alanine (A) residues, and the C-terminal carboxyl group of threonine is amide linked to the side chain amino group of diaminopimelic acid within the wall peptides of B. anthracis peptidoglycan.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. anthracis strain Sterne (53) and mutant derivatives were used throughout the present study. Mutants were generated with the pLM4 shuttle vector as previously described (28). B. anthracis strain LAM5 contains an in-frame deletion of the srtC gene; its construction was also described previously (28). Strain LAM17 harbors mutations in sequences encoding the BasI LPNTA motif, changing it to TNLAP; pLM224 was used to introduce these modifications. Strain LAM18 harbors a mutation in srtC codon 181 that replaces the wild-type cysteine codon with an alanine codon (C181A); pLM225 was used to introduce the mutation. In strain LAM20, basI was modified by translational fusion with the cherry open reading frame inserted between basI codons 110 and 111, three codons upstream of the sequence that encodes BasI LPNTA motif; pLM232 was used to generate this strain.
B. anthracis chromosomal DNA was extracted with the Wizard Genomic DNA purification kit (Promega). Table 1 lists the primers used for plasmid construction. In order to introduce point mutations in B. anthracis, the basI-srtC-sctR region was amplified from B. anthracis Sterne chromosomal DNA with the P12 and P13 primers and cloned into pLM4 by using EcoRI and XmaI restriction sites. This generated pLM221, which was used as a template to introduce mutations by site-directed mutagenesis. PCR amplification with the primers P46 and P47 using pLM221 as a template introduced mutations in the BasI LPNTA motif, thereby generating pLM224. PCR amplification with the primers P81 and P82 introduced the C181A mutation in srtC, generating pLM225. To introduce a methionyl-hexahistidyl tag into BasI, DNA specifying basI promoter and an open reading frame up to three codons upstream of BasI LPNTA motif was amplified with P3/P4 primers, and the sequences encoding for the basI cell wall sorting signal, srtC and sctR, were amplified with the P5 and P13 primers, where P5 contained an appended sequence encoding for the MH6 tag (see Table 1). Both PCR products were cloned into pLM4 by using EcoRI, KpnI, and XmaI restriction sites, thereby generating pLM228. To generate a BasI-Cherry fusion, the cherry coding sequence was amplified with the primers P158 and P153 using pRSET-B-mCherry (50) as a template and cloned into pLM228 using KpnI; this generated pLM232. The construction of plasmids pLM218 and pLM226 is described elsewhere (28). PCRs were performed with Pfu DNA polymerase (Stratagene). Ligation products were transformed into E. coli K1077 Δ(dam dcm), and purified (nonmethylated) plasmid DNA was transformed into B. anthracis strains according to a previously developed protocol (20). Growth media were supplemented with kanamycin at 50 or 20 μg/ml to provide for plasmid selection in E. coli and B. anthracis, respectively.
TABLE 1.
Primers used in this study
| Primer | Restriction site | Nucleic acid sequence (5′-3′)a |
|---|---|---|
| P3 | KpnI | aaGGTACCcttatcgtcgtcatccttgtaatcttgtacagctggctttgctac |
| P4 | EcoRI | aaGAATTCcaatgaaatggagtaagtggag |
| P5 | KpnI | aaGGTACCatgcatcaccatcaccatcacggtgaaaaattaccaaacactg |
| P12 | EcoRI | aaGAATTCaggtttgaagaagagacccc |
| P13 | XmaI | aaCCCGGGgatctaatgtcgtaatccacg |
| P46 | None | gctgtacaaggtgaaaaaACaAACCTcGctCcatcaaataatgtagcaatg |
| P47 | None | cattgctacattatttgatgGagCgAGGTTtGTtttttcaccttgtacagc |
| P81 | None | cgatactaacactgacgactGCttacccatttgattatatagg |
| P82 | None | cctatataatcaaatgggtaaGCagtcgtcagtgttagtatcg |
| P153 | KpnI | aaaGGTACCcttgtacagctcgtccatgc |
| P158 | KpnI | aaaGGTACCagcaagggcgaggaggataac |
| P3784-N | NcoI | aaCCATGGatggaaatcagaaaaaaattagttg |
| P3784-C | BamHI | aaGGATCCttatttatcatcataccaccaatc |
Capital letters indicate the sequence of the restriction site inserted. For the sequences with no restriction site, small capital letters indicate nucleotide changes introduced. Underlined nucleotides encode for the MH6 tag.
pJB1 was used for expression of an untagged version of PlyL amidase (23). It was constructed by cloning the P3784-N/P3784-C PCR product into pET16b by using the NcoI and BamHI restriction sites. Ligation products were used to transform E. coli BL21(DE3) (Novagen) using 100 μg of ampicillin/ml for selection.
Microscopy.
B. anthracis LAM20(pLM218) overnight cultures were grown in brain heart infusion, diluted 1:10 into modG sporulation medium (21), and incubated at 30°C for 30 h. Culture aliquots were removed in hourly intervals and viewed by differential interference contrast or fluorescence microscopy using an Olympus Provis axial microscope (1,000-fold magnification) equipped with a Hamamatsu charge-coupled device camera.
Solubilization and detection of BasI.
B. anthracis strains harboring pLM218 or pLM228 were grown at 30°C up to T−1 in modG media (see Results). Strains harboring pLM226 were grown overnight at 30°C in LB broth either in the presence or in the absence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were collected by centrifugation (1 min, 16,000 × g), washed with 1 ml of sterile double-distilled water (ddH2O), suspended in 250 μl of UDS buffer (6 M urea, 1 mM dithiothreitol, 1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 9.5]) (48), and boiled for 10 min. B. anthracis murein sacculi were sedimented by centrifugation (1 min, 16,000 × g) and separated from the bacterial extract containing membrane and cytoplasmic proteins. Murein sacculi were washed with 1 ml of sterile ddH2O, suspended in 500 μl of 5% trichloroacetic acid, and boiled for 6 min. Precipitated material was centrifuged (1 min, 16,000 × g) and washed first with 500 μl of 0.5 M Tris-HCl (pH 6.3) and then twice with 50 mM Tris-HCl (pH 6.3)-1.5 mM MgCl2. Murein sacculi containing peptidoglycan and secondary wall polymers were subjected to mutanolysin digestion in 250 μl of buffer containing 50 mM Tris-HCl (pH 6.3), 1.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and 100 U of mutanolysin (Sigma). Digestion was carried out overnight at 37°C, and insoluble material was removed by centrifugation (1 min, 16,000 × g). The supernatant constituted the cell wall fraction and harbored mutanolysin-solubilized proteins. Aliquots (5 μl) of cell wall and bacterial extract samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and electrotransferred to polyvinylidene difluoride membrane. BasI was detected by immunoblotting using a polyclonal αBasI rabbit serum (28).
PlyL digestion of B. anthracis cell walls.
Four liters of B. anthracis Sterne (pLM228) cells were harvested at T−1 during sporulation and peptidoglycan was purified as previously described for SEB-MH6-CWSIsdC (27). Sedimented murein sacculi were suspended in 30 ml of an E. coli BL21(DE3)(pJB1) lysate. The lysate was obtained by growing cells in 1 liter of LB medium supplemented with 100 μg of ampicillin/ml to an optical density at 600 nm of 0.7, when the expression of amidase was induced by the addition of 1 mM IPTG. Cells were grown for 2 additional hours, collected by centrifugation (5 min, 6,000 × g), suspended in 30 ml of PlyL buffer, and lysed in a French pressure cell at 14,000 lb/in2. Digestion was carried out overnight at 37°C, and insoluble material was removed by centrifugation (10 min, 33,000 × g).
Mutanolysin digestion of B. anthracis cell walls.
For mutanolysin digestion, cell wall preparations were obtained according to a previously described protocol for purification of B. subtilis spore peptidoglycan, modified to retain cell wall-anchored proteins (38). A colony of B. anthracis Sterne harboring pLM228 was inoculated into LB broth containing kanamycin at 20 μg/ml and grown overnight at 30°C. Cells were diluted 1:50 into 4 liters of modG sporulation medium, grown until T−1 (optical density at 600 nm of 3.0), and harvested by centrifugation at 10,000 × g for 5 min. Sedimented cells were washed once with 100 ml of sterile ddH2O, suspended in 50 ml of UDS buffer, and boiled for 15 min. B. anthracis murein sacculi were sedimented by centrifugation (10 min, 10,000 × g), washed with 40 ml of sterile ddH2O, suspended in 10 ml of 5% trichloroacetic acid, and boiled for 6 min. Precipitated murein sacculi were again sedimented by centrifugation (10 min, 10,000 × g) and washed first with 40 ml of 0.5 M Tris-HCl (pH 6.3) and then twice with 50 mM Tris-HCl (pH 6.3)-1.5 mM MgCl2. After these washes, the murein sacculi were subjected to mutanolysin digestion overnight at 37°C in 30 ml of buffer containing 50 mM Tris-HCl (pH 6.3), 1.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and 10,000 U of mutanolysin. Insoluble material was removed by centrifugation (10 min, 33,000 × g), and the soluble supernatant was subjected to Ni-nitrilotriacetic acid (NTA) affinity chromatography.
Purification of BasIMH6.
Cytosolic extracts of bacilli (obtained during bead-beating, see above) and cell wall lysates (obtained by mutanolysin or PlyL digestion) were adjusted to pH 7.5 with 1 M Na2HPO4 prior to purification of BasIMH6 on Ni-NTA affinity chromatography as previously described for SEB-MH6-CWSIsdC (27).
Preparation of C-terminal anchor peptides.
Purified BasIMH6 was methanol-chloroform precipitated and cleaved with cyanogen bromide, and anchor peptides were purified by a second round of Ni-NTA affinity chromatography and separated by high-pressure liquid chromatography (HPLC) as previously described for SEB-MH6-CWSIsdC (27).
MALDI-MS.
Dried HPLC fractions containing peptides of interest were suspended in 15 μl of CH3CN-water-trifluoroacetic acid (TFA; 30:70:0.1). Matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) spectra were obtained on a reflectron time-of-flight (TOF) instrument (ABI Biosystems) in the reflection mode. Samples (0.5 μl) were cospotted with 0.5 μl of matrix (α-cyano-4-hydroxycinnamic acid; Sigma) at 10 mg/ml in CH3CN-water-trifluoroacetic acid (70:30:0.1). All samples were externally calibrated to a standard of bovine insulin.
RESULTS
BasI is targeted to the envelope of sporulating B. anthracis cells.
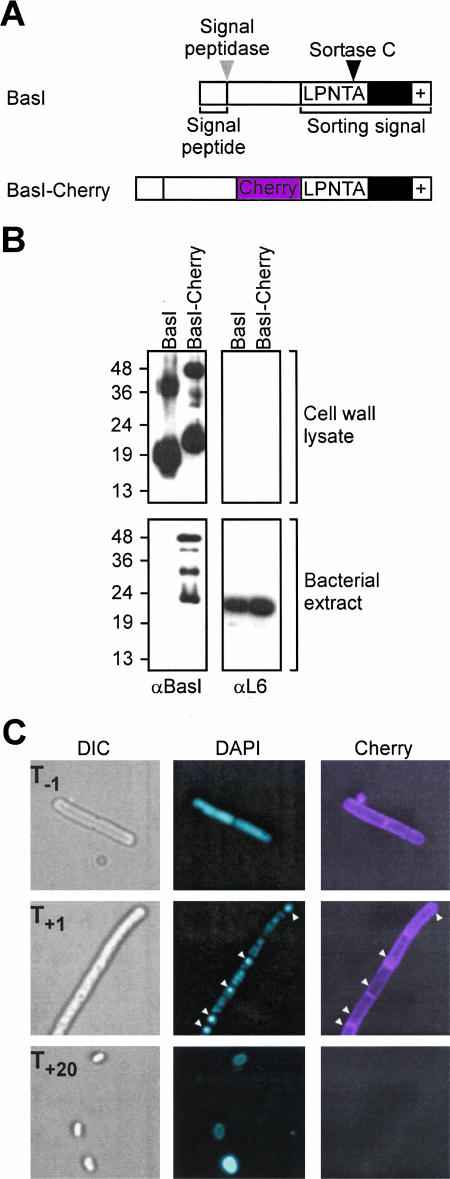
In a previous study, we observed that B. anthracis expresses the basI-srtC-sctR-sctS operon during sporulation at T−1, 2 h prior to the formation of the polar septum that divides sporulating bacilli into the mother cell (bigger compartment) and forespore (smaller compartment) (T+1) (28). After septation, sortase C (the product of srtC) is found in both compartments (28). basH, also encoding a sortase C substrate with LPNTA motif, is, however, expressed in the developing forespore by σF RNA polymerase (28). The presence of SrtC in the forespore envelope, observed in fluorescence microscopy experiments using a SrtC-Cherry reporter (28), is likely required for the anchoring of BasH to the spore cell wall. However, the subcellular location of BasI during sporulation remained unknown. To determine whether this polypeptide is targeted to the cell wall envelope of predivisional cells, mother cells, or forespores, we constructed a BasI-Cherry fluorescent reporter protein by the insertion of the cherry coding sequence between basI codons 110 and 111. The mutant strain LAM20 expresses a BasI-Cherry fusion that harbors an N-terminal signal peptide (residues 1 to 24), the Cherry fluorescent protein domain (residues 113 to 346), and a C-terminal LPNTA sorting signal (residues 359 to 393) (Fig. 1A). We then tested whether the fusion protein was targeted to the cell wall compartment of sporulating cells, as is observed for wild-type BasI (28). The murein sacculi of sporulating B. anthracis, strains Sterne (wild type) or LAM20, were treated with mutanolysin, a muramidase that cleaves the repeating disaccharide MurNAc-GlcNAc within peptidoglycan (7, 65). Proteins released by muramidase from cell wall envelopes were separated on SDS-15% PAGE and immunoreactive signals were detected with a BasI-specific antibody (Fig. 1B). Mutanolysin-released BasI species migrated with apparent molecular mass of 17 and 35 kDa, more slowly than the molecular mass of BasI that can be deduced from the mature polypeptide. After signal peptide (37) and sorting signal cleavage (28), the calculated mass of BasI (Asp25-Thr117) is 10,371 Da. Thus, the molecular mass of the more slowly migrating BasI species must be accounted for by the molecular masses of BasI polypeptide and the cell wall fragments to which this polypeptide is linked (see below). Immunoblotting of proteins released from the envelope of B. anthracis LAM20 revealed two predominant BasI-Cherry species that migrated at 20 to 22 kDa and 42 kDa, respectively (Fig. 1B). After signal peptide and sorting signal cleavage, the calculated mass of BasI-Cherry (Asp25-Thr362) is 39,191 Da. We presume the 42-kDa species represents mature, anchored BasI-Cherry, whereas the faster-migrating species (20 to 22 kDa) are likely caused by the degradation of anchored BasI-Cherry products in the cell wall envelope. Moreover, BasI-Cherry cytoplasmic precursors are also subjected to a similar degradation, as judged from immunoblotting of LAM20 bacterial extracts. To rule out the presence of cytoplasmic contaminating material, cell wall extracts were subjected to immunoblotting with L6-specific antiserum. This experiment demonstrated that BasI-Cherry is targeted to the cell wall compartment of sporulating cells, and therefore the fusion protein is a valid surrogate for the study of BasI localization.
FIG. 1.
BasI is anchored to the envelope of B. anthracis cells during sporulation. (A) Diagram illustrating the primary structure of BasI with a cleavable N-terminal signal peptide and C-terminal sorting signal, encompassing the LPNTA motif, hydrophobic domain (black box), and positively charged tail (+). The sorting signal of BasI is cleaved by sortase C between the threonine (T) and the alanine (A) residues of the LPNTA motif. BasI-Cherry harbors a translational fusion of the Cherry fluorescence protein inserted three residues upstream of the LPNTA sorting signal. The coding sequence for the translational hybrid basI-cherry was recombined into the basI locus of B. anthracis Sterne (wild type), thereby generating strain LAM20. (B) Immunoblotting of proteins in mutanolysin cell wall lysates and bacterial extracts of B. anthracis wild type (BasI) and LAM20 (BasI-Cherry) during sporulation (T−1) with BasI-specific antiserum identified anchored products for both BasI and BasI-Cherry in the cell wall envelope, as well as BasI-Cherry precursors. L6 antiserum was used to control cell wall fractions for the presence of contaminating cytoplasmic material. Numbers indicate the mobility of protein molecular mass markers in kilodaltons after SDS-15% PAGE. (C) LAM20(pLM218) cells overexpressing the SctR response regulator were analyzed by fluorescence microscopy during sporulation in modG media. The expression of BasI-Cherry commenced at T−1, 2 h prior to formation of the polar septum. BasI-Cherry fluorescence was detected in the envelope of sporulating bacilli. At T+1, upon formation of the polar septum, BasI-Cherry was found in the envelope of predivisional cells but not in the forespore. DAPI staining of the chromosomal DNA of mother cells and the condensed chromosomal DNA of forespores (arrowheads) and spores was used to reveal the location of both cellular compartments. Upon disintegration of mother cells and release of endospores at T+20, BasI-Cherry fluorescence was no longer detected.
To analyze BasI-Cherry localization, cultures of B. anthracis LAM20 (basI-cherry) were subjected to sporulation in modG media. Both differential interference contrast and fluorescence microscopy images were captured to reveal BasI-Cherry or DAPI (4′,6′-diamidino-2-phenylindole)-stained chromosomal DNA of mother cells and the condensed chromosome of forespores and spores (Fig. 1C). BasI-Cherry fluorescent signals were first detected at T−1, 2 h prior to the formation of polar septa and the appearance of highly condensed forespore DNA. The fluorescence signal of BasI-Cherry was found in the envelope of sporulating B. anthracis LAM20 cells but not in either the forespore envelope (T+1) or the mature spores (T+20) (Fig. 1C). These observations are consistent with the hypothesis that the reporter protein BasI-Cherry, and likely wild-type BasI, are anchored to the cell wall envelope of predivisional sporulating cells. At the time of polar septation (T+1) BasI molecules are presumably anchored and no longer synthesized, thereby preventing BasI anchoring to forespore cell wall.
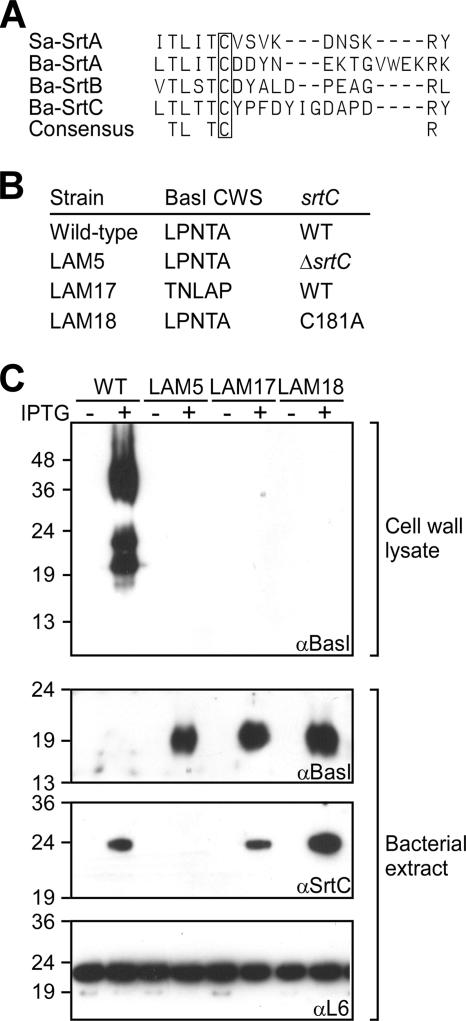
Sortase C with an active-site cysteine and the LPNTA sorting signal of BasI are required for anchoring of the polypeptide to the cell wall envelope.
Purified SrtCΔN, with a six-histidyl replacing the N-terminal signal peptide of sortase C, cleaves LPNTA motif peptide between threonine (T) and alanine (A) (28). Cleavage is blocked by methyl-methane thiosulfonate, which reacts with the thiol group to form a disulfide bond (51), suggesting the cysteine residue of sortase C (Cys181) functions as an active site in vitro, similar to the cysteine (Cys184) of S. aureus sortase A (28, 60, 63) (Fig. 2A). To examine the requirement of SrtC Cys181 for in vivo anchoring of BasI, B. anthracis strain LAM18, carrying an alanine substitution at codon 181 of srtC, was generated (C181A, Fig. 2B). basI-srtC expression was achieved by IPTG induction of sctR, encoding the response regulator and transcriptional activator of basI-srtC, via the Pspac promoter on plasmid pLM226. BasI immunoreactive species were not detected in cell wall lysates of B. anthracis LAM18 (SrtCC181A); however, immunoblotting revealed accumulation of the BasI precursor in bacterial extracts of this strain (Fig. 2C). In contrast, BasI was found in cell wall envelope lysates, but not in bacterial extracts, of wild-type bacilli (Fig. 2C). Both wild-type bacilli and LAM18 carrying the srtCC181A substitution expressed sortase C, which could be detected with SrtC-specific antibody in bacterial extracts (Fig. 2C). B. anthracis LAM5, with a deletion of the srtC gene, also failed to anchor BasI to the cell wall envelope and accumulated BasI precursor in bacteria. As expected, SrtC immunoreactive species were not detected in bacterial extracts of strain LAM5. Immunoblotting with antibodies to ribosomal protein L6 was used as a control for proper sample loading. To determine whether sortase C-mediated anchoring of BasI to the cell wall envelope also requires the C-terminal sorting signal of the BasI precursor, we generated strain LAM17, in which the basI coding sequence for the LPNTA motif was scrambled to TNLAP. Immunoblotting experiments revealed precursor species in bacterial extracts; however, BasITNLAP was not found in the cell wall envelope of sporulating bacilli (Fig. 2C). In sum, BasI anchoring to the cell envelope of sporulating bacilli requires both the active-site cysteine of sortase C and the C-terminal sorting signal of BasI precursor.
FIG. 2.
srtC and basI requirements for the anchoring of BasI. (A) Alignment to reveal the conservation of cysteine in the active site of Staphylococcus aureus sortase A (Sa-SrtA), B. anthracis sortase A (Ba-SrtA), B. anthracis sortase B (Ba-SrtB), and B. anthracis sortase C (Ba-SrtC). In Ba-SrtC, the conserved cysteine occupies residue 181 (Cys181). (B) B. anthracis wild-type (WT) and three mutant strains were analyzed for the anchoring of BasI to the cell wall envelope of sporulating cells. Strain LAM5 harbors an in-frame deletion of the srtC gene (ΔsrtC), strain LAM17 encodes a mutant basI in which the coding sequence for its LPNTA motif has been scrambled to TNLAP, and strain LAM18 harbors a srtCC181A allele in which the active-site Cys181 codon (UGU) has been replaced with an alanine codon (GCU). All B. anthracis strains were transformed with plasmid pLM226, which provides for IPTG-inducible expression of the SctR response regulator and then expression of basI-srtC. (C) Where indicated, IPTG was added to sporulating bacilli in order to induce the expression of basI-srtC. Immunoblotting with antisera specific for BasI, SrtC, or ribosomal protein L6 was used to detect polypeptides in mutanolysin-degraded murein sacculi (cell wall lysate) or in bacterial extracts after separation of proteins on SDS-PAGE and electroblotting of samples to polyvinylidene difluoride membranes. The numbers indicate the mobility of protein molecular mass markers in kilodaltons after SDS-15% PAGE.
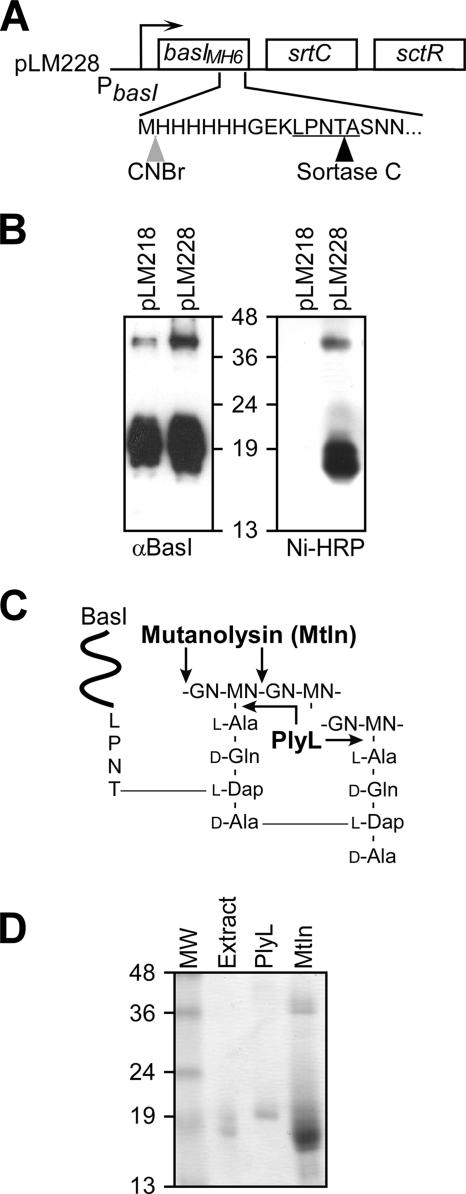
Purification of BasIMH6 from the cell wall of sporulating bacilli.
To analyze the cell wall anchor structure of BasI, we engineered BasIMH6 with an insertion of methionyl-six histidyl (MH6) upstream of the LPNTA motif sorting signal of BasI. To achieve high anchoring efficiency, we overexpressed BasIMH6, along with sortase C and the SctR response regulator, using plasmid pLM228 (Fig. 3A). To test whether B. anthracis anchors BasIMH6 similarly to wild-type BasI, the cell walls of bacilli harboring pLM218 or pLM228 were analyzed by immunoblotting to compare the anchoring pattern of both proteins. By overexpressing the SctR response regulator, pLM218 provides for expression of chromosomal wild-type basI, whereas pLM228 expresses chromosomal wild-type basI and plasmid-encoded basIMH6. Cells were grown in sporulating media until T−1, sedimented by centrifugation, and digested with mutanolysin (Fig. 3B). Cell wall lysates were subjected to immunoblotting with BasI-specific antibodies, which revealed similar SDS-PAGE mobility patterns for immunoreactive BasI and BasIMH6 species (Fig. 3B). Detection of the engineered protein with nickel-horseradish peroxidase demonstrated that BasIMH6 is anchored to the envelope of sporulating cells in a fashion similar to that of BasI and can therefore be used as a reporter to study the cell wall anchor structure of BasI.
FIG. 3.
Purification of BasIMH6 from the cell wall envelope of sporulating B. anthracis. (A) In order to purify anchored BasI, a methionyl-six-histidyl tag was inserted three residues upstream of the BasI LPNTA motif and cell wall sorting signal. The BasIMH6 variant is encoded by plasmid pLM228, which also includes srtC and sctR, the latter providing for the expression of both basIMH6 and srtC during sporulation. (B) To validate the use of BasIMH6 as a surrogate for the study of BasI anchoring, cell wall fractions of wild-type bacilli carrying pLM218 and pLM228 were compared for the presence of BasI and BasIMH6 by Western blotting with anti-BasI, which detects both versions of the surface protein, and nickel-horseradish peroxidase, which only detects BasIMH6. The pattern of mutanolysin-released anchored species was similar for both BasI variants. (C) Cell wall anchor structure of B. anthracis BasI and cleavage sites of two murein hydrolases used in the present study: mutanolysin and PlyL amidase. (D) Coomassie blue-stained SDS-PAGE of Ni-NTA-purified BasIMH6 released from murein sacculi by digestion with mutanolysin (Mtln) or PlyL amidase. As a control, cytoplasmic precursors of BasIMH6 were purified from bacterial extracts. The numbers indicate the mobility of protein molecular mass markers in kilodaltons after SDS-15% PAGE.
Cell wall-anchored BasIMH6 was solubilized from purified peptidoglycan of B. anthracis Sterne (pLM228) cells with two different murein hydrolases and the released polypeptides were subjected to Ni-NTA affinity chromatography. Mutanolysin cleavage of the glycan strands (Fig. 3C) released a spectrum of BasIMH6 species that migrated as two clusters (17 to 25 kDa and 36 to 45 kDa) on Coomassie-stained SDS-PAGE, a finding in agreement with the migration patterns of BasI reported in Fig. 1 (Mtln, Fig. 3D). Since both of the spectra of BasIMH6 species copurified with peptidoglycan and require a murein hydrolase for solubility, we conclude that they represent anchored products that may or may not harbor peptidoglycan with higher degrees of cross-linking attached to its C-terminal end. Previous work determined that 19% of wall peptides within B. anthracis peptidoglycan are cross-linked to neighboring wall peptides (49). PlyL, a presumed amidase that is thought to cleave the amide bond between MurNAc-l-Ala (23), was expressed in E. coli and bacterial lysates harboring the enzyme were used to cleave the cell wall envelope of sporulating bacilli. Purified BasIMH6 species that had been liberated by PlyL digestion from the cell wall envelope of bacilli migrated mostly as a single discrete species with mobility of 19 kDa on SDS-PAGE (PlyL, Fig. 3D). To determine the effect of attached peptidoglycan on the electrophoretic mobility of mutanolysin- and PlyL-released BasIMH6, we purified BasIMH6 precursors from bacterial extracts and analyzed their mobility by gel electrophoresis. Coomassie blue-stained SDS-PAGE gels revealed two BasIMH6 species with apparent molecular masses of 19 and 17 kDa (extract, Fig. 3D), whose mobility on SDS-PAGE corresponded to the P1 precursor (BasIMH6 with N-terminal signal peptide and C-terminal sorting signal [18,155 Da]) and the P2 precursor (BasIMH6 with only the C-terminal sorting signal [15,695 Da]), respectively (26). Cleavage of the glycan strands or wall peptides within cross-linked peptidoglycan is known to release distinct spectra of anchored proteins with different mobility on SDS-PAGE (45). In agreement with earlier observations, the P1 and P2 precursors, PlyL- and mutanolysin-solubilized BasIMH6, each display different migration patterns (masses) on SDS-PAGE (29). We conclude that the different migration patterns must be due to different amounts of peptidoglycan attached to the C terminus of BasIMH6 after the polypeptide has been solubilized by PlyL or mutanolysin (see below).
Cell wall anchor structure of PlyL-solubilized BasIMH6.
BasIMH6 was solubilized by PlyL digestion from the cell wall envelope of sporulating bacilli and purified by affinity chromatography on Ni-NTA. After cleavage of purified BasIMH6 at methionine residues with CNBr (52), C-terminal peptides harboring the cell wall anchor structures were purified by affinity chromatography on Ni-NTA and subjected to reversed-phase (RP)-HPLC on a C18 column. PlyL-released anchor peptides eluted at 28% CH3CN-0.01% TFA and were subjected to MALDI-TOF MS (Fig. 4A). A complex set of ion signals was observed. Four predominant ion signals—m/z 1,751.98, 1,779.98, 1,794.98, and 1,807.98—were explained as C-terminal anchor peptides linked to diaminopimelic acid (H6GEKLPNT-m-Dap). meso-Diaminopimelic acid (m-Dap) is a diamino acid at position 3 of the Bacillus cell wall pentapeptide precursor (l-Ala-d-iGln-m-Dap-d-Ala-d-Ala), and its side chain amino group is also involved in peptidoglycan cross-linking by forming an amide bond with d-Ala at position four of neighboring wall peptides [l-Ala-d-iGln-(l-Ala-d-iGln-m-Dap-d-Ala-)m-Dap-d-Ala-d-Ala] (44). The predicted m/z of the anchor peptide H6GEKLPNT-m-Dap is 1,752.84, and its formylated, diformylated, and carbamylated forms are calculated to be 1,780.85, 1,808.86, and 1,795.87 (Table 2). Formylation and carbamylation of α-NH2 groups in cell wall anchor peptides are known to occur during CNBr cleavage (70% formic acid) and affinity chromatography (urea buffer) (27). To examine the validity of our prediction, the parent ion at m/z 1,779.98 was subjected to collisionally induced dissociation (CID) in a MALDI-TOF tandem MS experiment, and daughter ion spectra were collected (Fig. 4C). Table 3 summarizes the observed ions, their structural interpretation, and fragmentation patterns, which are in agreement with the peptide structure H6GEKLPNT-m-Dap. The presence of a single Dap linked to the C terminus of the BasIMH6 anchor peptide is, however, not in agreement with the predicted amidase function of PlyL. We would have expected to find anchor peptides encompassing tri- or tetrapeptides [l-Ala-d-iGln-(H6GEKLPNT-)m-Dap and l-Ala-d-iGln-(H6GEKLPNT-)m-Dap-d-Ala)]. It should be noted that our protocol used recombinant PlyL from crude E. coli lysates as an enzyme preparation. Thus, cleavage of wall peptides between d-iGln-m-Dap may be a feature of contaminating E. coli enzymes that can act on the primary cleavage products of PlyL. In agreement with this view, several less-abundant ion signals of PlyL-released BasIMH6 anchor peptides were identified and interpreted to represent cell wall tetrapeptides linked to the C-terminal end of the polypeptide chain (Table 2).
FIG. 4.
MS of PlyL-released BasIMH6 anchor peptides. (A) MALDI-TOF spectrum of BasIMH6 anchor peptides released from B. anthracis murein sacculi. Isolated cell wall preparations were treated with extracts of recombinant E. coli harboring PlyL amidase, and cell wall lysates were subjected to Ni-NTA affinity chromatography. Purified BasIMH6 was cleaved with CNBr at methionine residues to generate C-terminal anchor peptides, which were isolated by a second chromatography step on Ni-NTA and further purified by RP-HPLC on a C18 column. Anchor peptides eluted at 30% CH3CN and were subjected to MALDI-TOF. Major ion signals are labeled; please see Table 2 for a listing of all ions. (B) Deduced anchor structures and calculated m/z values for species released by PlyL digestion (see Table 2 for mass measurements). Most ions detected by MALDI-TOF were modified by formyl or carbamyl at their NH2 groups (arrows), and the calculated mass of formylated (the most common modification) ion is indicated. Cell wall tri- or tetrapeptides lacking or containing a terminal d-Ala (between parentheses), respectively, were identified in BasI anchor structures. (C) CID and MALDI-TOF/TOF analysis of the m/z 1,779.98 identified in panel A. Please see Table 3 for a listing of daughter ions and their structural interpretation.
TABLE 2.
Summary of ions produced during MS of PlyL-released BasIMH6 anchor peptides
|
m/z
|
Δcalc-obsb | Proposed structure | Modificationc | |
|---|---|---|---|---|
| Calculateda | Observed | |||
| 1,752.84 | 1,751.98 | 0.86 | HHHHHHGEKLPNT-m-Dap | None |
| 1,780.85 | 1,779.98 | 0.87 | HHHHHHGEKLPNT-m-Dap | HC(O)- |
| 1,795.87 | 1,794.98 | 0.89 | HHHHHHGEKLPNT-m-Dap | NH2-C(O)- |
| 1,808.86 | 1,807.98 | 0.88 | HHHHHHGEKLPNT-m-Dap | 2 × HC(O)- |
| 1,823.88 | 1,823.00 | 0.88 | HHHHHHGEKLPNT-m-Dap-d-Ala | None |
| 1,851.89 | 1,851.03 | 0.86 | HHHHHHGEKLPNT-m-Dap-d-Ala | HC(O)- |
| 1,866.91 | 1,866.04 | 0.87 | HHHHHHGEKLPNT-m-Dap-d-Ala | NH2-C(O)- |
| 1,979.95 | 1,980.09 | −0.14 | d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala | HC(O)- |
| 1,994.97 | 1,995.09 | −0.12 | d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala | NH2-C(O)- |
| 2,022.98 | 2,022.14 | 0.84 | l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala | None |
| 2,050.99 | 2,050.14 | 0.85 | l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala | HC(O)- |
| 2,066.01 | 2,065.16 | 0.85 | l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala | NH2-C(O)- |
| 2,095.01 | 2,094.25 | 0.76 | [HHHHHHGEKLPNT-m-Dap-d-Ala]m-Dap-d-Ala | HC(O)- |
| 2,123.02 | 2,122.19 | 0.83 | [HHHHHHGEKLPNT-m-Dap-d-Ala]m-Dap-d-Ala | 2 × HC(O)- |
| 2,195.06 | 2,195.23 | −0.17 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | None |
| 2,223.07 | 2,222.24 | 0.83 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | HC(O)- |
| 2,238.09 | 2,237.23 | 0.86 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | NH2-C(O)- |
| 2,251.08 | 2,250.24 | 0.84 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | 2 × HC(O)- |
| 2,266.10 | 2,265.25 | 0.85 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap-d-Ala | None |
| 2,294.11 | 2,294.25 | −0.14 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap-d-Ala | HC(O)- |
| 2,309.13 | 2,309.28 | −0.15 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap-d-Ala | NH2-C(O)- |
| 2,322.12 | 2,322.29 | −0.17 | [l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap-d-Ala | 2 × HC(O)- |
| 2,422.17 | 2,422.34 | −0.17 | l-Ala-d-iGln[l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | HC(O)- |
| 2,450.18 | 2,450.32 | −0.14 | l-Ala-d-iGln[l-Ala-d-iGln(HHHHHHGEKLPNT-)m-Dap-d-Ala]m-Dap | 2 × HC(O)- |
Calculations are based on masses obtained with the MS-Product in the ProteinProspector 4.0.5 internet tool (http://prospector.ucsf.edu/ucsfhtml4.0/msprod.htm) (9).
That is, the difference between the calculated and observed ion masses.
Modification of αNH2 groups. Formylation, HC(O)-; carbamylation, NH2-C(O)-.
TABLE 3.
Summary of daughter ions produced during MS/MS of the PlyL-released m/z 1,779.98 parent ion
|
m/z
|
Δcalc-obsb | Proposed structurec | Ion typed | |
|---|---|---|---|---|
| Calculateda | Observed | |||
| 110.07 | 110.06 | 0.01 | H | ie |
| 275.13 | 275.19 | −0.06 | HH | a2 |
| 303.12 | 303.21 | −0.09 | HH | b2 |
| 412.18 | 412.27 | −0.09 | HHH | a3 |
| 440.18 | 440.29 | −0.11 | HHH | b3 |
| 452.23 | 452.36 | −0.13 | HGEK | i |
| 503.25 | 502.37 | 0.88 | PNT-m-Dap | y4 |
| 549.24 | 549.36 | −0.12 | HHHH | a4 |
| 565.31 | 565.42 | −0.11 | HGEKL | i |
| 577.24 | 577.34 | −0.10 | HHHH | b4 |
| 589.28 | 589.41 | −0.13 | HHGEK | i |
| 686.30 | 686.39 | −0.09 | HHHHH | a5 |
| 702.37 | 702.46 | −0.09 | HHGEKL | i |
| 714.30 | 714.37 | −0.07 | HHHHH | b5 |
| 726.34 | 726.43 | −0.09 | HHHGEK | i |
| 823.36 | 823.40 | −0.04 | HHHHHH | a6 |
| 851.36 | 851.37 | −0.01 | HHHHHH | b6 |
| 908.34 | 908.38 | −0.04 | HHHHHHG | b7 |
| 930.49 | 929.49 | 1.00 | GEKLPNT-m-Dap | y8 |
| 1,009.42 | 1,009.31 | 0.11 | HHHHHHGE | a8 |
| 1,037.42 | 1,037.30 | 0.12 | HHHHHHGE | b8 |
| 1,054.45 | 1,054.32 | 0.13 | HHHHHHGE | c8 |
| 1,067.55 | 1,067.42 | 0.13 | HGEKLPNT-m-Dap | y9 |
| 1,137.52 | 1,137.29 | 0.23 | HHHHHHGEK | a9 |
| 1,165.51 | 1,165.29 | 0.22 | HHHHHHGEK | b9 |
| 1,204.61 | 1,204.35 | 0.26 | HHGEKLPNT-m-Dap | y10 |
| 1,250.60 | 1,250.30 | 0.30 | HHHHHHGEKL | a10 |
| 1,278.60 | 1,278.24 | 0.36 | HHHHHHGEKL | b10 |
| 1,341.67 | 1,341.25 | 0.42 | HHHGEKLPNT-m-Dap | y11 |
| 1,392.68 | 1,392.21 | 0.47 | HHHHHHGEKLP | c11 |
| 1,478.72 | 1,478.14 | 0.58 | HHHHGEKLPNT-m-Dap | y12 |
| 1,489.69 | 1,489.07 | 0.62 | HHHHHHGEKLPN | b12 |
| 1,607.77 | 1,606.90 | 0.87 | HHHHHHGEKLPNT | c13 |
| 1,615.78 | 1,614.87 | 0.91 | HHHHHGEKLPNT-m-Dap | y13 |
Calculations are based on average masses obtained with the MS-Product in the ProteinProspector 4.0.5 internet tool (http://prospector.ucsf.edu/ucsfhtml4.0/msprod.htm) (9).
Thate is, the difference between the calculated and observed masses of daughter ions.
N-terminal daughter ions contain a formylated N-terminal histidyl residue.
The nomenclature used refers to the NH2-and CO2H-terminal cleavage fragments according to Biemann (6).
Internal ion; the calculated mass corresponds to the sum of the residue masses (M+H+).
Ion signals indicating cross-linking between anchor peptides and neighboring cell wall peptides were also detected (Table 2). Anchor peptides with ion signals at m/z 2,195.23, 2,222.24, 2,237.23, and 2,250.24 were explained as anchor peptides linked to cell wall tetrapeptides that were in turn tethered to diaminopimelic acid within another wall peptide ([l-Ala-d-iGln(H6GEKLPNT-)m-Dap-d-Ala]m-Dap, calculated m/z = 2,195.06) and its formylated and carbamylated forms (Table 2). The predicted structure of this compound was also validated in a tandem MS experiment (data not shown). Degradation products of cross-linked wall peptides tethered to the C-terminal end of BasIMH6 were also detected (Table 2). Taken together, these results demonstrate that during anchoring of BasIMH6, the polypeptide chain is cleaved by sortase C between threonine and alanine of the LPNTA motif and that the carboxyl group of threonine is amide linked to the side chain amino group of diaminopimelic acid within wall peptides that may or may not be cross-linked to neighboring wall peptides in the murein sacculus of the B. anthracis envelope.
Cell wall anchor structure of muramidase-solubilized BasIMH6.
The glycan strands of isolated peptidoglycan were cut with mutanolysin at the β(1-4) glycosidic bond between the repeating disaccharide MurNAc-GlcNAc (Fig. 5B) and anchor peptides were purified. The amino sugars of C-terminal anchor peptides were reduced by sodium borohydride treatment, and glycopeptides were separated by RP-HPLC on a C18 column. Mutanolysin-released anchor peptides eluted at 28% CH3CN-0.01% TFA and were subjected to MALDI-TOF MS. Two sets of ions were detected (Fig. 5A). The compound at m/z 2,418.38 represented the most abundant ion of the first cluster of compounds, and its structure was explained as a formylated C-terminal anchor peptide linked to murein disaccharide tripeptide:GlcNH2-(β1-4)-MurNAc-[L-Ala-d-iGln-(H6GEKLPNT-)m-Dap], calculated m/z = 2,418.54) (Fig. 5B and Table 4). To test this prediction, the compound was subjected to CID in a MALDI-TOF/TOF experiment. Consistent with previous reports using MS to analyze the structure of peptidoglycan fragments (2, 27), the most abundant CID daughter ions resulted from the breakage of the β(1-4) glycosidic bond between GlcNH2 and MurNAc (Table 5, observed m/z = 2,255.91, calculated m/z = 2,255.93). However, many other, less-abundant ions were also detected and could be structurally assigned to N- or C-terminal CID fragments of the predicted structure (Table 5). The lack of amino acetylation of glucosamine (GlcNH2) residues and the abundance of wall tripeptides corroborate previous work on the structure of peptidoglycan for members of the B. cereus family (49, 66). Nevertheless, anchor peptides containing C-terminal tetrapeptides were found as well (Fig. 5A and Table 4).
FIG. 5.
MS of mutanolysin-released BasIMH6 anchor peptides. (A) MALDI-TOF spectrum of BasIMH6 anchor peptides released from B. anthracis murein sacculi. Cell wall preparations were treated with mutanolysin, and lysates were subjected to Ni-NTA affinity chromatography. Purified BasIMH6 was cleaved with CNBr at methionyl to generate C-terminal anchor peptides, which were isolated by a second chromatography step on Ni-NTA and further purified by RP-HPLC. Anchor peptides eluted at 28% CH3CN and were subjected to MALDI-TOF. (B) Proposed structures and their calculated m/z values for species released by mutanolysin digestion. Major ion signals are labeled; please see Table 4 for a listing of all detected ions. Ion signals detected by MALDI-TOF in panel A correspond to formylated or carbamylated species with modified glucosamine- and/or alpha-amino groups (arrows), and the calculated mass of formylated (most common modification) ion is indicated. Glucosamine residues in BasIMH6 anchor peptides were deacetylated (GlcNH2). Both cell wall tri- or tetrapeptides, with or without the terminal d-Ala (between parentheses), were found to be linked to the C-terminal end of BasIMH6. (C) CID and MALDI-TOF/TOF of the m/z 2,418.38 identified in panel A. Please see Table 5 for a listing of daughter ions and their structural interpretation.
TABLE 4.
Summary of ions produced during MS of mutanolysin-released BasIMH6 anchor peptides
|
m/z
|
Δcalc-obsb | Proposed structure | Modificationc | |
|---|---|---|---|---|
| Calculateda | Observed | |||
| 2,390.53 | 2,390.37 | 0.16 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap] | None |
| 2,418.54 | 2,418.38 | 0.16 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap] | HC(O)- |
| 2,433.56 | 2,433.41 | 0.15 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap] | NH2-C(O)- |
| 2,446.55 | 2,446.38 | 0.17 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap] | 2 × HC(O)- |
| 2,461.61 | 2,461.40 | 0.21 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-d-Ala] | None |
| 2,489.62 | 2,489.41 | 0.22 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-d-Ala] | HC(O)- |
| 2,517.63 | 2,518.43 | −0.80 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-d-Ala] | 2 × HC(O)- |
| 2,532.61 | 2,532.41 | −0.20 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-d-Ala] | 2 × HC(O)-; NH2-C(O)- |
| 3,270.43 | 3,270.90 | −0.47 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | None |
| 3,298.44 | 3,298.90 | −0.46 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | HC(O)- |
| 3,326.45 | 3,326.83 | −0.38 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | 2 × HC(O)- |
| 3,341.51 | 3,341.97 | −0.46 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-d-Ala}d-Ala] | None |
| 3,354.46 | 3,354.92 | −0.46 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | 3 × HC(O)- |
| 3,369.52 | 3,368.85 | 0.67 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | HC(O)- |
| 3,384.54 | 3,383.91 | 0.63 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | NH2-C(O)- |
| 3,397.53 | 3,397.89 | −0.36 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-}d-Ala] | 2 × HC(O)- |
Calculations are based on masses obtained with the MS-Product in the ProteinProspector 4.0.5 internet tool (http://prospector.ucsf.edu/ucsfhtml4.0/msprod.htm) (9).
That is, the difference between the calculated and observed ion masses.
Modification of NH2 groups, see Fig. 5B: formylation, HC(O)-; and carbamylation, NH2-C(O)-.
TABLE 5.
Summary of daughter ions produced during MS/MS of the mutanolysin-released m/z 2,418.38 parent ion
|
m/z
|
Δcalc-obsb | Proposed structurec | Ion typed | |
|---|---|---|---|---|
| Calculateda | Observed | |||
| 275.13 | 275.23 | −0.10 | HH | a2 |
| 412.18 | 412.24 | −0.06 | HHH | a3 |
| 440.18 | 440.38 | −0.20 | HHH | b3 |
| 549.24 | 549.28 | −0.04 | HHHH | a4 |
| 577.24 | 577.52 | −0.28 | HHHH | b4 |
| 714.30 | 714.56 | −0.26 | HHHHH | b5 |
| 726.34 | 726.69 | −0.35 | HHHGEK | ie |
| 823.36 | 823.63 | −0.27 | HHHHHH | a6 |
| 851.36 | 851.61 | −0.25 | HHHHHH | b6 |
| 908.38 | 908.61 | −0.23 | HHHHHHG | b7 |
| 1,009.42 | 1,009.67 | −0.25 | HHHHHHGE | a8 |
| 1,037.42 | 1,037.65 | −0.23 | HHHHHHGE | b8 |
| 1,137.52 | 1,137.73 | −0.21 | HHHHHHGEK | a9 |
| 1,165.51 | 1,165.69 | −0.18 | HHHHHHGEK | b9 |
| 1,233.58 | 1,233.39 | 0.19 | HHHHHHGEKL(-NH3) | i |
| 1,252.60 | 1,251.73 | 0.87 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(LPNT-)m-Dap] | y8 |
| 1,261.57 | 1,261.57 | 0.00 | HHHHHHGEKL(-NH3) | b10-NH3 |
| 1,278.60 | 1,278.73 | −0.13 | HHHHHHGEKL | b10 |
| 1,489.69 | 1,490.69 | −1.00 | HHHHHHGEKLPN | b12 |
| 1,544.74 | 1,544.66 | 0.08 | HHHHHHGEKLPNT(-H2O) | i |
| 1,703.82 | 1,704.63 | −0.81 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HGEKLPNT-)m-Dap] | y12 |
| 1,716.82 | 1,716.50 | 0.32 | HHHHHHGEKLPNT-m-Dap-(-H2O) | i |
| 1,779.84 | 1,779.53 | 0.31 | HHHHHHGEKLPNT-m-Dap | c14 |
| 1,840.88 | 1,841.50 | −0.62 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHGEKLPNT-)m-Dap] | y13 |
| 1,890.89 | 1,890.41 | 0.48 | d-iGln-(HHHHHHGEKLPNT)m-Dap | b15 |
| 1,908.90 | 1,908.41 | 0.59 | l-Ala-d-iGln-(HHHHHHGEKLPNT)m-Dap | c15 |
| 1,977.94 | 1,978.42 | −0.48 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHGEKLPNT-)m-Dap] | y14 |
| 2,052.00 | 2,051.34 | 0.66 | lactoyl-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap] | -f |
| 2,115.00 | 2,115.23 | −0.23 | GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(HHHHGEKLPNT-)m-Dap] | y15 |
| 2,255.91 | 2,255.93 | −0.02 | MurNAc-[l-Ala-d-iGln-(HHHHHHGEKLPNT-)m-Dap]h | -g |
Calculations are based on masses obtained with the MS-Product in the ProteinProspector 4.0.5 internet tool (http://prospector.ucsf.edu/ucsfhtml4.0/msprod.htm) (9).
That is, the difference between the calculated and observed masses of daughter ions.
N-terminal histidyl residues are formylated.
The nomenclature used refers to the NH2-and CO2H-terminal cleavage fragments according to Biemann (6); exceptions are indicated below. Ion number is calculated considering the histidine tail as the main NH2 terminus.
Internal ion.
The y-type ion originated from the cleavage of the ether bond between N-acetylglucosamine and lactoyl groups of N-acetylmuramitol (56).
The y-type ion originated from the cleavage of the β1-4 O-glycosidic bond between N-acetylglucosamine and N-acetylmuramitol (56).
Main peak.
The second cluster of ions comprised BasIMH6 anchor species that were linked to cross-linked peptidoglycan, i.e., murein tetrapeptide-tripeptide and tetrapeptide-tetrapeptide (Fig. 5A and Table 4). The most abundant compound of this cluster, m/z 3,341.97, could be assigned the structure GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-(H6GEKLPNT-)m-Dap-{GlcNH2-(β1-4)-MurNAc-[l-Ala-d-iGln-m-Dap-d-Ala-d-Ala]} (calculated m/z = 3,341.43). MALDI-TOF/TOF analysis of this ion corroborated our hypothesis (data not shown). Anchor peptides linked to murein subunits with higher degrees of cross-linking could not be detected. In all cases examined, anchor species modified by formylation and/or carbamylation on either αNH2 or glucosamine NH2 groups (Fig. 5B) were observed. In summary, the cell wall anchor structure of BasIMH6 revealed that the polypeptide is linked to B. anthracis peptidoglycan subunits through the formation of an amide bond between its C-terminal threonine and the side chain amino group of diaminopimelic acid. The low degree of cross-linking observed for BasIMH6 anchor peptides is in agreement with the overall low degree of cross-linking in the peptidoglycan of B. anthracis.
DISCUSSION
The function of many sortases is the anchoring of surface proteins to the peptidoglycan cell wall envelope of gram-positive bacteria (26). Three classes of sortases catalyze such reactions. Sortase A recognizes its substrates by the presence of an LPXTG motif sorting signal, cleaving polypeptides between the conserved threonine (T) and glycine (G) residues (60). Sortase A performs a transpeptidation reaction whereby the C-terminal threonine of the LPXTG bearing polypeptide is amide linked to cell wall cross-bridges (61). Cross-bridges are heterogeneous in structure, and surface protein anchoring has been described for the pentaglycine cross-bridges of staphylococci (27, 35, 45, 58, 59), as well as for the listerial diaminopimelic acid cross-bridge (11). In spite of such structural heterogeneity, the availability of the cross-bridge amino group is a conserved prerequisite feature of the sorting pathway (57); the amino group functions as a nucleophile and resolves the thioester linked acyl enzyme intermediate between the cysteine thiol of sortase and the carboxyl group of threonine at the C-terminal end of the polypeptide chain (31). Sortase B anchors proteins to cell wall cross-bridges under iron starvation conditions and cleaves the NPQTN or NPKTG motif of S. aureus or B. anthracis IsdC, respectively (24, 27, 30, 32). Sortase B anchored products appear to assume a unique location in murein sacculi, enabling passage of heme-iron from the surface across the cell wall envelope layer for delivery to membrane transporters, a distance of up to 100 nm (25). Sortase C has been shown to specifically cleave the LPNTA motif in B. anthracis BasI and BasH, presumably anchoring these polypeptides during sporulation (28). Here we reveal the BasI anchor structure and demonstrate for the first time that sortase C links the C-terminal threonine (T) of BasI to the amino group of diaminopimelic acid cross-bridges in the cell wall envelope of B. anthracis sporulating cells. This report presents the first surface protein anchor structure for Bacillus species. Our findings further corroborate the universality of the sortase transpeptidation mechanism. However, the data also identify two unique features of sortase C. This enzyme not only cleaves polypeptides between threonine and alanine (instead of threonine and glycine) but sortase C also acts on two different substrates in two different subcellular compartments: the envelope of predivisional sporulating cells (BasI) and the peptidoglycan of developing forespores (BasH).
A second gene encoding a sortase C substrate with an LPNTA motif sorting signal, basH, is expressed in the B. anthracis forespore after formation of the polar septum (28). Its product, BasH, appears to be targeted exclusively to the forespore envelope, since fluorescent reporter BasH-Cherry can be found only in the spore envelope and not in mother cell envelopes (L. A. Marraffini and O. Schneewind, unpublished data). In contrast, BasI-Cherry fluorescence is only observed in the predivisional cell envelope and not in the envelope of forespores. Fluorescence microscopy experiments with SrtC-Cherry detected sortase C first in the envelope of predivisional sporulating cells at T-1 (28). Upon the formation of polar septa at T+1, SrtC-Cherry fluorescence was also observed in the forespore envelope and even in mature spores that have been released after mother cell disintegration at T+20. We hypothesize that BasI is expressed and anchored at T−1 and that anchoring reaches completion sometime after T−1. During a later stage of this developmental program, at T+1, BasI molecules that have been previously anchored remain in the envelope of sporulating cells. However, since BasI may no longer be synthesized and forespore peptidoglycan turned over, the polypeptide is not found in the forespore envelope. These results support the notion that different surface proteins decorate different compartments of the sporulating cell, with BasI present in predivisional cells and BasH present in forespores. On the other hand, sortase C, which is responsible for the targeting of BasI and BasH, is present in both predivisional cells and forespores.
Sortase C is a member of a distinct class of sortases (class D) that is found in spore-forming bacteria (bacilli, clostridia, and actinomycetales) (12). Previous work examined the role of class D sortases in Streptomyces coelicolor, a gram-positive, filamentous soil bacterium. S. coelicolor forms mycelia that submerge in a liquid or even in a semiliquid environment. The mycelia eventually differentiate to generate aerial hyphae that septate and then develop chains of spores for bacterial dissemination in air. Hyphal surfaces are highly hydrophobic, a property that promotes outgrowth into the air and dispersion of spores, which eventually give rise to new mycelia (18). Six surface proteins named chaplins are involved in aerial hyphae formation, and three of these are synthesized as precursors carrying N-terminal signal peptides and C-terminal sorting signals that require a sortase C homolog for proper anchoring to the bacterial envelope (8, 13). Presumably, these polypeptides function to lower aqueous surface tension for the emergence of aerial hyphae into air (8, 13). In this regard, it seems noteworthy that B. anthracis sortase C is also required for spore formation in infected guinea pig tissues or sheep blood under conditions of low oxygen concentration (28). Perhaps one or both of the sortase C anchored products of B. anthracis, BasI in the envelope of predivisional cells or BasH in the forespore envelope, fulfill functions analogous to those of chaplins. If acting as surfactants, this may allow sporulating bacilli access to air-tissue interfaces in anthrax carcasses or access to oxygen, which is essential for the completion of spore development (42). Our ongoing experiments are designed to examine basI and basH contributions to spore formation in carcasses.
Acknowledgments
We thank Jonathan Budzik for the construction of pJB1 and members of our laboratory for discussion and critical review of the manuscript. The University of Chicago Proteomics Core Laboratory is gratefully acknowledged for assistance with the mass spectrometry experiments.
This study was supported by U.S. Public Health Service grants AI38897 and AI69227 to O.S. L.A.M. is supported by the William Rainey Harper Fellowship of The University of Chicago. O.S. acknowledges membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE, National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Abrahmsen, L., T. Moks, B. Nilsson, U. Hellman, and M. Uhlen. 1985. Analysis of signals for secretion in the staphylococcal protein A gene. EMBO J. 4:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeloa, A., J. E. Hugonnet, A. C. Sentilhes, N. Josseaume, L. Dubost, C. Monsempes, D. Blanot, J. P. Brouard, and M. Arthur. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 279:41546-41556. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, A. R., J. J. Armstrong, J. Baddiley, and J. B. Hay. 1961. Teichoic acids and the structure of bacterial cell walls. Nature 191:570-572. [DOI] [PubMed] [Google Scholar]
- 4.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae, T., and O. Schneewind. 2003. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185:2910-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biemann, K. 1992. Mass spectrometry of peptides and proteins. Annu. Rev. Biochem. 61:977-1010. [DOI] [PubMed] [Google Scholar]
- 7.Calandra, G. B., and R. Cole. 1980. Lysis and protoplast formation of group B streptococci by mutanolysin. Infect. Immun. 28:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claessen, D., R. Rink, W. de Jong, J. Siebring, P. de Vreugd, F. G. Boersma, L. Dijkhuizen, and H. A. Wosten. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 10.Cossart, P., and R. Jonquieres. 2000. Sortase, a universal target for therapeutic agents against gram-positive bacteria? Proc. Natl. Acad. Sci. USA 97:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 12.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 13.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17:1727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 15.Gaspar, A. H., L. A. Marraffini, E. M. Glass, K. L. DeBord, H. Ton-That, and O. Schneewind. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghuysen, J.-M. 1968. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 32:425-464. [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, H., Y. Araki, and E. Ito. 1973. Occurrence of glucosamine residues with free amino groups in cell wall peptidoglycan from bacilli as a factor responsible for resistance to lysozyme. J. Bacteriol. 113:592-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, R. C. 1970. Autolysis of isolated cell walls of Bacillus licheniformis N.C.T.C. 6346 and Bacillus subtilis Marburg Strain 168. Separation of the products and characterization of the mucopeptide fragments. Biochem. J. 119:849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, H. S., D. Sherman, F. Johnson, and A. I. Aronson. 2004. Characterization of a major Bacillus anthracis spore coat protein and its role in spore inactivation. J. Bacteriol. 186:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, H. U., and J. M. Goepfert. 1974. A sporulation medium for Bacillus anthracis. J. Appl. Bacteriol. 37:265-267. [DOI] [PubMed] [Google Scholar]
- 22.Losick, R., and P. Stragier. 1992. Criss-cross regulation of cell-type specific gene expression during development in Bacillus subtilis. Nature 355:601-604. [DOI] [PubMed] [Google Scholar]
- 23.Low, L. Y., C. Yang, M. Perego, A. Osterman, and R. C. Liddington. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433-35439. [DOI] [PubMed] [Google Scholar]
- 24.Maresso, A. W., T. J. Chapa, and O. Schneewind. 2006. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J. Bacteriol. 188:8145-8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maresso, A. W., and O. Schneewind. 2006. Iron acquisition and transport in Staphylococcus aureus. Biometals 19:193-203. [DOI] [PubMed] [Google Scholar]
- 26.Marraffini, L. A., A. C. DeDent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marraffini, L. A., and O. Schneewind. 2005. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J. Biol. Chem. 280:16263-16271. [DOI] [PubMed] [Google Scholar]
- 28.Marraffini, L. A., and O. Schneewind. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol. Microbiol. 62:1402-1417. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 30.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 31.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 32.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 34.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J. Biol. Chem. 273:29135-29142. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot. Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 38.Popham, D. L., and P. Setlow. 1993. The cortical peptidoglycan from spores of Bacillus megaterium and Bacillus subtilis is not highly cross-linked. J. Bacteriol. 175:2767-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Psylinakis, E., I. G. Boneca, K. Mavromatis, A. Deli, E. Hayhurst, S. J. Foster, K. M. Varum, and V. Bouriotis. 2005. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 280:30856-30863. [DOI] [PubMed] [Google Scholar]
- 40.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baille, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. T. Cline, C. Redmond, J. E. Thwaite, O. WHite, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 41.Rivas, J. M., P. Speziale, J. M. Patti, and M. Hook. 2004. MSCRAMM-targeted vaccines and immunotherapy for staphylococcal infection. Curr. Opin. Drug Discov. Dev. 7:223-227. [PubMed] [Google Scholar]
- 42.Roth, N. G., D. H. Livery, and H. M. Hodge. 1955. Influence of oxygen uptake and age of culture on sporulation of Bacillus anthracis and Bacillus globigii. J. Bacteriol. 69:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salton, M. R. J. 1952. Cell wall of Micrococcus lysodeikticus as the substrate of lysozyme. Nature 170:746-747. [DOI] [PubMed] [Google Scholar]
- 44.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 46.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface protein of gram-positive bacteria. EMBO 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 48.Sedlak, M., T. Walter, and A. Aronson. 2000. Regulation by overlapping promoters of the rate of synthesis and deposition into crystalline inclusions of Bacillus thuringiensis delta-endotoxins. J. Bacteriol. 182:734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Severin, A., K. Tabei, and A. Tomasz. 2004. The structure of the cell wall peptidoglycan of Bacillus cereus RSVF1, a strain closely related to Bacillus anthracis. Microb. Drug Resist. 10:77-82. [DOI] [PubMed] [Google Scholar]
- 50.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 51.Smith, D. J., E. T. Maggio, and G. L. Kenyon. 1975. Simple alkanethiol groups for temporary blocking of sulfhydryl groups of enzymes. Biochemistry 14:764-771. [DOI] [PubMed] [Google Scholar]
- 52.Steers, E., Jr., G. R. Craven, C. B. Anfinsen, and J. L. Bethune. 1965. Evidence for nonidentical chains in the beta-galactosidase of Escherichia coli K-12. J. Biol. Chem. 240:2478-2484. [PubMed] [Google Scholar]
- 53.Sterne, M. 1937. Avirulent anthrax vaccine. Onderstepoort J. Vet. Sci. Animal Ind. 21:41-43. [PubMed] [Google Scholar]
- 54.Strominger, J. L. 1968. Penicillin-sensitive enzymatic reactions in bacterial cell wall synthesis. Harvey Lectures 64:179-213. [PubMed] [Google Scholar]
- 55.Strominger, J. L., and J.-M. Ghuysen. 1967. Mechanisms of enzymatic bacteriolysis. Science 156:213-221. [DOI] [PubMed] [Google Scholar]
- 56.Sydner, A. P. 2000. Interpreting protein mass spectra: a comprehensive resource. Oxford University Press, Washington, DC.
- 57.Tipper, D. J., and J. L. Strominger. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-alanine. Proc. Natl. Acad. Sci. USA 54:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 59.Ton-That, H., H. Labischinski, B. Berger-Bächi, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. III. The role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J. Biol. Chem. 273:29143-29149. [DOI] [PubMed] [Google Scholar]
- 60.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ton-That, H., H. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. I. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 62.Ton-That, H., S. K. Mazmanian, L. Alksne, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. II. Cysteine 184 and histidine 120 of sortase A form a thiolate imidazolium ion pair for catalysis. J. Biol. Chem. 277:7447-7452. [DOI] [PubMed] [Google Scholar]
- 63.Ton-That, H., and O. Schneewind. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 274:24316-24320. [DOI] [PubMed] [Google Scholar]
- 64.Warth, A. D., and J. L. Strominger. 1971. Structure of the peptidoglycan from vegetative cell walls of Bacillus subtilis. Biochemistry 10:4349-4358. [DOI] [PubMed] [Google Scholar]
- 65.Yokogawa, K., S. Kawata, S. Nishimura, Y. Ikeda, and Y. Yoshimura. 1974. Mutanolysin, bacteriolytic agent for cariogenic streptococci: partial purification and properties. Antimicrob. Agents Chemother. 6:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zipperle, G. F., Jr., J. W. Ezzell, Jr., and R. J. Doyle. 1984. Glucosamine substitution and muramidase susceptibility in Bacillus anthracis. Can. J. Microbiol. 30:553-559. [DOI] [PubMed] [Google Scholar]