Abstract
In an effort to identify key domains of the Pseudomonas aeruginosa MexAB-OprM drug efflux system involved in component interactions, extragenic suppressors of various inactivating mutations in individual pump constituents were isolated and studied. The multidrug hypersusceptibility of P. aeruginosa expressing MexB with a mutation in a region of the protein implicated in oligomerization (G220S) was suppressed by mutations in the α/β domain of MexA. MexB(G220S) showed a reduced ability to bind MexA in vivo while representative MexA suppressors (V66M and V259F) restored the MexA-MexB interaction. Interestingly, these suppressors also restored resistance in P. aeruginosa expressing OprM proteins with mutations at the proximal (periplasmic) tip of OprM that is predicted to interact with MexB, suggesting that these suppressors generally overcame defects in MexA-MexB and MexB-OprM interaction. The multidrug hypersusceptibility arising from a mutation in the helical hairpin of MexA implicated in OprM interaction (V129M) was suppressed by mutations (T198I and F439I) in the periplasmic α-helical barrel of OprM. Again, the MexA mutation compromised an in vivo interaction with OprM that was restored by the T198I and F439I substitutions in OprM, consistent with the hairpin domain mediating MexA binding to this region of OprM. Interestingly, these OprM suppressor mutations restored multidrug resistance in P. aeruginosa expressing MexB(G220S). Finally, the oprM(T198I) suppressor mutation enhanced the yields of all three constituents of a MexA-MexB-OprM(T198I) pump as detected in whole-cell extracts. These data highlight the importance of MexA and interactions with this adapter in promoting MexAB-OprM pump assembly and in stabilizing the pump complex.
Pseudomonas aeruginosa is an opportunistic human pathogen characterized by an innate resistance to multiple antimicrobials (10), resistance increasingly attributable to the operation of broadly specific, multidrug efflux systems of the resistance-nodulation-division (RND) family (33, 34). Several RND family multidrug efflux systems have been described in P. aeruginosa (34, 35) although the major system contributing to intrinsic multidrug resistance is encoded by the mexAB-oprM operon (19). This system also contributes to acquired multidrug resistance as a result of its hyperexpression in nalB (i.e., mexR) (45, 53), nalC (6, 21), and nalD (42) mutants. The MexAB-OprM efflux system, like other tripartite RND family pumps, consists of an inner membrane drug-proton antiporter (the RND component; MexB), an outer membrane (OM) channel-forming component (also called outer membrane factor [OMF]; OprM) and a periplasmic membrane fusion protein ([MFP]; MexA) (32, 52).
Crystal structures have now been reported for MexA (2, 11) and OprM (1), with the OprM structure reminiscent of that of TolC, the homologous OM component of the Escherichia coli AcrAB-TolC multidrug efflux system (18). The OprM channel is trimeric and is comprised of an OM-spanning β-barrel and a periplasmic α-helical barrel, with an overall length of 135Å (1). The crystal structure of the MexA monomer (residues 29 to 259 only of the 360-residue mature protein) reveals the protein to be elongated and comprised of three linearly arranged subdomains—an α/β domain, a lipoyl domain, and an α-helical hairpin domain—with monomers predicted, in one model, to assemble into a nine-member sheath around the proximal and distal ends, respectively, of OprM and MexB, which are predicted to be in close apposition in the periplasm (2, 11). Still, other models of MFP-RND-OMF pumps suggest one (22) or two (46) MFP monomers per RND and OMF monomer. Although the MexB crystal structure is as yet unavailable, modeling on the available structure of the highly homologous AcrB protein (28) revealed that it also exists as a trimer composed of a 50-Å thick transmembrane (inner membrane) region and a 70-Å headpiece that protrudes into the periplasm (24). Although the original AcrB (and so MexB) structure described a symmetrical trimeric protein with three vestibules linking the periplasm to a central cavity (where substrates were bound) that exited the protein via a funnel-like opening at the top (28), more recent data reveal AcrB to be comprised of asymmetric monomers whose conformations represent different stages in an export process that sees individual monomers transporting substrates from the periplasmic vestibules through channels within each monomer that bypass the central cavity but exit at the previously described funnel-like pore (27, 39, 40).
Despite the available structures of individual components, however, the details of assembly of this tripartite pump remain largely unknown. In vivo interactions between MexA and MexB (25, 29) and between MexA and OprM (25) have been confirmed, and the MexAB-OprM tripartite complex has been recovered from P. aeruginosa in the absence of cross-linking (25). Interestingly, MexA association with MexB is dependent upon the presence of OprM (25, 29) although MexA-OprM association may be independent of MexB (25). Similarly, genetic (8) and biochemical (14, 48, 49) studies have confirmed in vivo interactions between AcrA, AcrB, and TolC in E. coli, and an AcrAB-TolC complex is also recoverable from E. coli without prior cross-linking (48). A C-terminal domain of AcrA is implicated in the binding of this MFP to its cognate RND component, AcrB (7, 49), and while mutations in the corresponding region of MexA have been isolated and shown to abrogate MexA function (29), the importance of this region vis-à-vis MexB binding has not been established. Still, an inactivating mutation in a groove in the MexB structure that in the homologous AcrB is implicated in AcrA binding (27, 28) is suppressed by mutations in the C-terminal half of MexA (30), consistent with the idea that this region of MexA binds MexB. The N-terminal helical hairpin of AcrA has been shown to interact with TolC (22), suggesting that this structure is responsible for MFP-OMF interactions in MFP-RND-OMF pumps. Consistent with this, substituting the hairpin region of AcrA with that of MexA allowed the hybrid AcrA protein to function with MexB and OprM (46). AcrB and TolC have recently been reported to interact in the absence of AcrA (47), although likely weakly (49), with the AcrA adapter responsible for stabilizing the interaction. In the current report we have examined the interaction of the MexAB-OprM constituents through the isolation and characterization of extragenic mexA and oprM suppressors of various pump mutants. Our results highlight the importance of MexA-OprM and MexA-MexB interactions for pump assembly, stability, and function.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were cultivated at 37°C in Luria broth (LB) as described previously (29), supplemented with antibiotics to maintain plasmids as needed (for plasmids pRK415, pEX18Tc, and their derivatives, tetracycline at a concentration of 10 μg/ml for E. coli and 30 μg/ml for P. aeruginosa; for pMMB206 and its derivatives, chloramphenicol at a concentration of 10 μg/ml). Plasmid pDN43, encoding MexB(G220S), was constructed by cloning a 4.5-kb EcoRI-HindIII mexB(G220S)-containing fragment from pJKM14 into pMMB206. Plasmid pDN40, encoding OprM, was constructed by amplifying the oprM gene from the chromosome of P. aeruginosa K870 via PCR and cloning it (as a SacI-HindIII fragment) into pMMB206. Amplification was achieved using the primers Fragment B-forward (5′-GAGCTCGAGCTCTCCAACGACGTGTTCTTCCAGGT-3′; tandem SacI sites are underlined) and OprMR-HindIII [5′-AAGCTTAAGCTTAGGCCGA-GCGGGTCCGTGACGC-3′; tandem HindIII sites are underlined) and reaction conditions and parameters described previously (extension time increased to 2 min) for the amplification of the EcoRI-tagged mexA gene (30). Plasmid pDN42, encoding MexA(V129M), was constructed by amplifying the mexA(V129M) gene from plasmid pDN7 using the primers pMMB-MexA-For (5′-CCCGGGCCCGGGTGAATGTAAGTATTTTGCCTGC-3′; tandem SmaI sites are underlined) and pMMB-MexA-Rev (5′-GGATCCGGATCCGATCACCCACGCGAAAATGG-3′; tandem BamHI sites are underlined) and cloning it (as a SmaI-BamHI fragment) into pMMB206. Amplification conditions were the same as described below for the random mutagenesis of the mexA gene, except for the use of Vent DNA polymerase (New England Biolabs) and 5% (vol/vol) dimethyl sulfoxide.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties or genotypea | Reference |
|---|---|---|
| P. aeruginosa strains | ||
| K767 | Wild-type strain PAO1 | 23 |
| K870 | Spontaneous Smr derivative of wild-type strain PAO1 | 36 |
| K337 | P. aeruginosa ML5087 ilv-220 thr-9001 leu-9001 met-9011 pur-67 aphA | 31 |
| K1110 | K337 ΔoprM | 20 |
| K1113 | K870 ΔmexR ΔoprM | 20 |
| K1589 | K870 ΔmexR ΔmexB | 12 |
| K2274 | K870 ΔmexR ΔmexA | 29 |
| K2275 | K870 ΔmexR ΔmexA ΔmexB | 29 |
| K2547 | K870 ΔmexR ΔmexA ΔoprM | This study |
| K1491 | K767 ΔmexR | 45 |
| K2553 | K1491 ΔmexB ΔoprM | This study |
| E. coli strains | ||
| DH5α | λ− Φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−)supE44 thi-1 gyrA relA1 | 3 |
| S17-1 | thi pro hsdR recA Tra+ | 41 |
| Plasmids | ||
| pRK415 | P. aeruginosa-E. coli shuttle cloning vector; Tcr | 17 |
| pDN3 | pRK415::mexA | 29 |
| pJKM14 | pRK415::mexB(G220S) | 24 |
| pDN52 | pRK415::oprM | This study |
| pDN50 | pRK415::oprM(F439I) | This study |
| pDN51 | pRK415::oprM(T198I) | This study |
| pDN6 | pRK415::mexA(A108T) | 29 |
| pDN7 | pRK415::mexA(V129M) | 29 |
| pDN47 | pRK415::mexA(V66M) | This study |
| pDN48 | pRK415::mexA(A227S) | This study |
| pDN49 | pRK415::mexA(V259F) | This study |
| pDN54 | pRK415::mexA(A263V) | This study |
| pDN55 | pRK415::mexA(V278I) | This study |
| pDN56 | pRK415::mexA(L282M) | This study |
| pDN30 | pRK415::mexA(R221M) | 30 |
| pDN31 | pRK415::mexA(L245F) | 30 |
| pDN32 | pRK415::mexA(E254K) | 30 |
| pDN33 | pRK415::mexA(V259I) | 30 |
| pEX18Tc | Gene-replacement vector; sacB Tcr | 13 |
| pDN53 | pEX18Tc::ΔoprM | This study |
| pMMB206 | P. aeruginosa-E. coli shuttle cloning vector; Cmr | 26 |
| pDN40 | pMMB206::oprM | This study |
| pDN57 | pMMB206::oprM(T209A) | This study |
| pDN58 | pMMB206::oprM(V215A) | This study |
| pDN59 | pMMB206::oprM(V215T) | This study |
| pDN60 | pMMB206::oprM(G216A) | This study |
| pDN61 | pMMB206::oprM(V217T) | This study |
| pDN62 | pMMB206::oprM(T423A) | This study |
| pDN63 | pMMB206::oprM(G424A) | This study |
| pDN64 | pMMB206::oprM(D426A) | This study |
| pDN70 | pMMB206::oprM(T198I) | This study |
| pDN65 | pMMB206::mexA | This study |
| pDN41 | pMMB206::mexA(A108T) | This study |
| pDN42 | pMMB206::mexA(V129M) | This study |
| pDN43 | pMMB206::mexB(G220S) | This study |
| pDN34 | pMMB206::mexB(T578I) | 30 |
| pDN39 | pMMB206::mexB(E864K) | 30 |
| pDN25 | pMMB206::mexB-Hisb | 29 |
| pDN38 | pMMB206::mexA-mexB-His | 30 |
| pDN71 | pMMB206::mexB(G220S)-His | This study |
| pDN44 | pMMB206::mexA-mexB(G220S)-His | This study |
| pDN45 | pMMB206::mexA(V66M)-mexB(G220S)-His | This study |
| pDN46 | pMMB206::mexA(V259F)-mexB(G220S)-His | This study |
| pDN69 | pMMB206::mexA-His | This study |
Mutations in the MexA proteins encoded by the indicated genes are shown in parentheses. Smr, streptomycin resistance; Tcr, tetracycline resistance; Cmr; chloramphenicol resistance.
The indicated genes have been engineered to encode a protein with six C-terminal histidine residues.
DNA manipulations.
Standard protocols were used for restriction endonuclease digestions, ligations, transformation, plasmid isolation, preparation of electrocompetent cells, and agarose gel electrophoresis, as described by Sambrook and Russell (38). Genomic DNA of P. aeruginosa was extracted according to the protocol of Barcak et al. (4). E. coli cells were made competent using the method of Inoue et al. (15). DNA sequencing was performed by ACGT Corporation (Toronto, Ontario, Canada) with universal and custom primers.
Construction of a ΔmexR ΔmexA ΔoprM mutant.
To construct a P. aeruginosa strain lacking mexR and containing an in-frame deletion of mexA and oprM, an in-frame deletion of oprM was engineered into the available ΔmexR ΔmexA strain K2274 (29). The oprM deletion was constructed in the gene replacement vector pEX18Tc following amplification of ca. 1-kb portions upstream and downstream of the oprM sequences being deleted. The upstream region was amplified off the P. aeruginosa K870 chromosome using the primers Fragment B-forward (5′-GAGCTCGAGCTCTCCAACGACGTGTTCTTCCAGGT-3′; tandem SacI sites are underlined) and DeltaMUpstreamReverse (5′-GGATCCGGATCCCGCCGACGTCGTAGCTGCGC-3′; tandem BamHI sites are underlined) and Vent DNA polymerase (New England Biolabs) in a reaction mixture formulated as described previously (43). The reaction mixture was subjected to an initial 2-min denaturation step at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C before a final 10-min elongation at 72°C. The PCR product was purified as described previously (43) and cloned into SacI-BamHI-restricted pEX18Tc. The downstream region was also amplified off the K870 chromosome as above for the upstream fragment, using the primers deltaBM-downstream-Forward (5′-GGATCCGGATCCGGTCCGCGTCACGGACC3′; tandem BamHI sites are underlined) and deltaBM-Downstream-Reverse (5′-AAGCTTAAGCTTGGTGGCGACCGACATCGC-3′; tandem HindIII sites are underlined). The PCR product was purified and cloned into BamHI-HindIII-restricted pEX18Tc carrying the upstream fragment to yield pDN53. The construct was sequenced to ensure that no mutations had been introduced during PCR, and the construct was subsequently mobilized into P. aeruginosa strain K2274 via conjugation with pDN53-carrying E. coli S17-1 as described previously (45). Transconjugants were selected on LB agar containing tetracycline (30 μg/ml) and imipenem (0.5 μg/ml) (to counterselect donor E. coli), and those harboring a chromosomal deletion of oprM were subsequently recovered on sucrose plates (LB agar containing 10% [vol/vol] sucrose) and screened for loss of OprM and oprM using immunoblotting (see below) and colony PCR (37) with primers Fragment B-forward and deltaBM-downstream-reverse.
Random mutagenesis of mexA and isolation of suppressors of MexB(G220S).
Mutagenic PCR of the mexA gene was conducted by PCR amplification of the mexA gene from plasmid pDN3, exactly as described previously (30), using the primers JT-28 (5′-AAGCTTAAGCTTTGAATGTAAGTATTTTGCCTGC-3′; tandem HindIII sites are underlined) and JT-27 (5′-GAGCTCGAGCTCGATCACCCACGCGAAAATGG-3′; tandem SacI sites are underlined). The resultant mutagenized mexA-containing PCR products were digested with HindIII and SacI and cloned en masse into pRK415, with the recombinant plasmids electroporated into E. coli S17-1 and mobilized into P. aeruginosa K2275 (ΔmexR ΔmexA ΔmexB) harboring plasmid pDN43 [pMMB206::mexB(G220S)] via conjugation as described previously (29). Selection of suppressor mutations was performed by spreading the conjugation mixture on LB agar containing tetracycline (10 μg/ml), carbenicillin (20 μg/ml), imipenem (0.5 μg/ml; to counterselect the donor E. coli), and 1 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) [to induce transcription of mexB(G220S) from pDN43]. P. aeruginosa K2275 expressing the pDN43-encoded MexB(G220S) and chromosomal MexA [i.e., a MexA-MexB(G220S)-OprM pump] is unable to grow in the presence of 20 μg/ml carbenicillin, while P. aeruginosa expressing a wild-type, functional MexAB-OprM system can. Therefore, potential MexA suppressors would restore growth of pDN43-carrying K2275 on 20 μg/ml carbenicillin. Plasmid pRK415 derivatives carrying mutagenized mexA were recovered from carbenicillin-resistant transconjugants and reintroduced into P. aeruginosa K2275 harboring pDN43 to confirm that restored carbenicillin resistance was dependent upon the mutagenized plasmid-borne mexA gene in each instance before sequencing to identify the suppressor mutations.
Random mutagenesis of oprM and isolation of suppressors of MexA(V129M).
Random PCR-based mutagenesis of the oprM gene was carried out as described for mexA except for the use of P. aeruginosa K870 chromosomal DNA as a template, an extension time of 2 min, and primers Fragment B-forward (5′-GAGCTCGAGCTCTCCAACGACGTGTTC-TTCCAGGT-3′; tandem SacI sites are underlined) and pRK-OprM-Rev [5′-GAATTCGAATTCAGGCCGAGCGGGTCCGTGACGC-3′; tandem EcoRI sites are underlined). The resultant mutagenized oprM-containing PCR products were digested with SacI and EcoRI and cloned en masse into pRK415, with the recombinant plasmids electroporated into E. coli S17-1 and mobilized into P. aeruginosa strain K2547 (ΔmexR ΔmexA ΔoprM) harboring plasmid pDN42 [pMMB206:: mexA(V129M)]. Potential suppressors were selected and processed as above except that 12 μg/ml carbenicillin was used.
Site-directed mutagenesis of oprM.
The oprM gene of plasmid pDN40 was subjected to site-directed PCR mutagenesis using mutagenic primers and KOD HiFi DNA polymerase (Novagen, Mississauga, Ontario) in the presence of dimethyl sulfoxide and using an annealing temperature of 65°C as described by the manufacturer. The PCR-mutated plasmids were digested with DpnI to eliminate the methylated (i.e., wild-type) template copies and introduced into E. coli DH5α via transformation, with plasmid-carrying E. coli selected on LB agar containing 10 μg/ml chloramphenicol. Plasmid DNA was isolated from pDN40-carrying E. coli and sequenced to confirm both the introduction of the engineered mutation and the absence of any unwanted changes to the nucleotide sequence of the oprM gene.
MexA-MexB interaction assay.
To evaluate an interaction between MexB(G220S) and wild-type and suppressor mutant MexA proteins, a C-terminal polyhistidine tag was first engineered onto MexB(G220S) encoded by pDN43 (to yield pDN57) using an approach described elsewhere (30), and corecovery of MexA with MexB(G220S)-His on Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) was assessed as described previously without cross-linker (29). Following construction of pDN57, genes for wild-type and suppressor mutant (V66M and V259F) MexA proteins were amplified from plasmids pDN3, pDN47, and pDN49, respectively, and individually cloned upstream of mexB(G220S)-His exactly as described previously (30) to yield plasmids pDN44, pDN45, and pDN46. These plasmids, on which the mexA and mexB(G220S)-His genes are coexpressed from the same IPTG-inducible vector-borne promoter, were then introduced into P. aeruginosa K2275 (ΔmexR ΔmexA ΔmexB). Triton X-100-soluble membrane fractions were prepared from overnight cultures and incubated with Ni-NTA agarose (QIAGEN) to recover MexB(G220S)-His as described previously (29). Corecovery of wild-type or mutant MexA proteins was assessed using immunoblotting and used as a measure of in vivo MexA-MexB(G220S)-His interaction (29).
MexA-OprM interaction assay.
To assess an in vivo interaction between MexA and OprM, a polyhistidine tag was engineered into MexA to permit assessment of corecovery of MexA-His and OprM on Ni-NTA as above. His tagging of mexA was achieved by amplifying mexA from the chromosome of P. aeruginosa PAO1 using the primer pMMB-MexA-forward (5′-CCCGGGCCCGGGTGAATGTAAGTATTTTGC-3′; tandem SmaI sites are underlined) and the primer MexA-His reverse (5′-AAGCTTAAGCTTTCAGTGGTGGTGGTGGTGGTGGCCCTTGCTGTCGGTTTTCGCCGGAGC-3′; tandem HindIII sites are underlined; six-histidine-encoding codons are in bold, and the stop codon is italicized) in reaction mixtures formulated and processed as above for the PCR amplification of oprM. The mexA-His-containing PCR product was digested with SmaI and HindIII and cloned into pMMB206 to yield pDN69, which was mobilized into P. aeruginosa strains K2274 (ΔmexR ΔmexA) and K2275 (ΔmexR ΔmexA ΔmexB). Triton X-100-soluble membrane extracts were again prepared from overnight cultures as above and incubated with Ni-NTA agarose (QIAGEN) (29) to recover MexA-His. Corecovery of the chromosomally encoded OprM (assessed using immunoblotting [29]) was used as an indicator of MexA-OprM association in vivo.
MexB-OprM interaction assay.
The in vivo interaction of OprM with MexB was examined using MexB-His exactly as described for MexA-MexB-His (29), except that corecovery of OprM with MexB-His on Ni-NTA agarose was assessed using immunoblotting (29).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
The protocols for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting have been described previously (44). The preparation of antibodies used to detect MexA (29), MexB (44), and OprM (9) has also been described elsewhere. To confirm expression of wild-type and mutant MexA or OprM in P. aeruginosa strains K2275 and K2547, immunoblotting was carried out on whole-cell extracts prepared from overnight LB cultures as described previously (37). When equal loading of protein samples was critical for the immunoblots, whole-cell extracts were electrophoresed on duplicate gels, one of which was stained with Coomassie Brilliant Blue G-250 (37).
Susceptibility testing.
The antimicrobial susceptibilities of P. aeruginosa strains carrying pRK415 and pMMB206 and their derivatives encoding wild-type and mutant MexA, MexB, and OprM were assessed using a twofold serial microtiter broth dilution method described previously (16) with an inoculum of 5 × 105 cells per ml and a final IPTG concentration of 1 mM (to enhance expression of genes cloned onto these plasmids). MICs were recorded as the lowest concentration of antibiotic inhibiting visible growth after an 18-h incubation at 37°C.
RESULTS
Isolation of mexA extragenic suppressors of mexB(G220S).
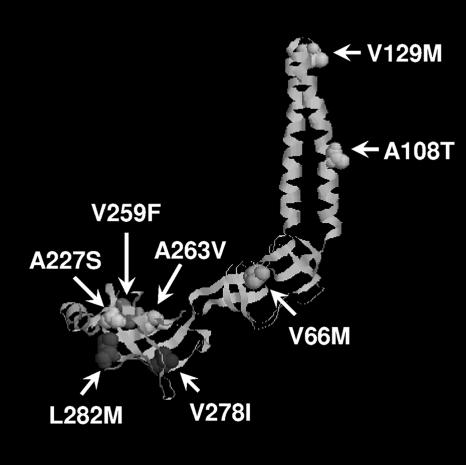
The G220S mutation on the “thumb” region of MexB monomers predicted to facilitate trimerization of this RND pump component has been shown to compromise MexB-promoted antimicrobial resistance, presumably owing to disruption/destabilization of the MexB trimer (24). Intragenic suppressors of this mutation were mapped to the distal end of MexB assumed to be in close apposition to OprM (24), suggesting that OprM might play a role in the suppression/stabilization of the mutant MexB trimer. To assess this, attempts were made to isolate oprM suppressor mutations of mexB(G220S) but without success. Still, previous studies showed that other MexB mutations that apparently compromised pump assembly (e.g., T578I) were suppressed by mutations in MexA that enhanced MexA association with the mutant MexB (30), and so attempts were made to isolate mexA suppressors of mexB(G220S). Plasmid-encoded mexA (pDN3) was, therefore, mutagenized, mobilized into the ΔmexR ΔmexAB P. aeruginosa strain K2275 harboring the MexB(G220S) plasmid (pDN43) and transconjugants expressing a MexAB-OprM system with restored function selected on 20 μg/ml carbenicillin [P. aeruginosa expressing a wild-type MexAB-OprM system is able to grow at this level of carbenicillin while one expressing an MexA-MexB(G220S)-OprM pump is not]. Of several transconjugants carrying possible MexA suppressors, six showed increased resistance to antimicrobials known to be substrates for MexAB-OprM (Table 2), consistent with their harboring a MexAB-OprM pump with restored activity. Isolation of the mutagenized pDN3 from each of these transconjugants and their subsequent reintroduction into P. aeruginosa K2275 confirmed that restored multidrug resistance in K2275 was, indeed, dependent upon the mutagenized mexA gene in each instance. Nucleotide sequencing of the mexA genes confirmed single mutations in each of them producing single amino acid changes in MexA (V66M, A227S, V259F, A263V, V278I, and L282M) (Table 2), all but one of which mapped to the α/β domain of the protein (Fig. 1), where previous MexA suppressors of MexB(T578I) also mapped (30). These earlier suppressors (R221H, L245F, E254K, and V259I) also suppressed the MexB(G220S) mutation, restoring wild-type levels of antimicrobial resistance to K2275 harboring the mexB(G220S) vector pDN43 (Table 2). As with the suppressors of MexB(T578I), however, the MexB(G220S) suppressors did not rescue the hypersusceptibility phenotype attributable to a MexB mutation (Table 2, E864K) situated in the predicted vestibule region of the MexB trimer (24). Interestingly, suppressor mutations were never recovered in the C-terminal region of MexA that is missing from the available crystal structure of this protein.
TABLE 2.
Influence of MexA suppressors of MexB(G220S) on antibiotic susceptibility of P. aeruginosa expressing wild-type and mutant MexB proteinsa
| Plasmid(s) | Expressed proteinb
|
MIC (μg/ml)c
|
|||
|---|---|---|---|---|---|
| MexA | MexB | CAR | NOV | NAL | |
| pDN3, pDN25 | WT | WT | 128 | 512 | 128 |
| pDN3, pDN43 | WT | G220S | 4 | 32 | 16 |
| pDN47, pDN43 | V66M | G220S | 128 | 256 | 128 |
| pDN48, pDN43 | A227S | G220S | 128 | 256 | 128 |
| pDN49, pDN43 | V259F | G220S | 128 | 256 | 128 |
| pDN54, pDN43 | A263V | G220S | 128 | 256 | 128 |
| pDN55, pDN43 | V278I | G220S | 128 | 256 | 128 |
| pDN56, pDN43 | L282M | G220S | 128 | 256 | 128 |
| pDN30, pDN43 | R221H | G220S | 128 | 256 | 128 |
| pDN31, pDN43 | L245F | G220S | 128 | 256 | 128 |
| pDN32, pDN43 | E254K | G220S | 128 | 256 | 128 |
| pDN33, pDN43 | V259I | G220S | 128 | 256 | 128 |
| pDN3, pDN39 | WT | E864K | 2 | 16 | 8 |
| pDN47, pDN39 | V66M | E864K | 2 | 16 | 8 |
| pDN49, pDN39 | V259F | E864K | 2 | 16 | 8 |
| pDN38 | WT | WT-His | 128 | 256 | 128 |
| pDN44 | WT | G220S-His | 4 | 32 | 16 |
| pDN45 | V66M | G220S-His | 128 | 256 | 128 |
| pDN46 | V259F | G220S-His | 128 | 256 | 128 |
P. aeruginosa K2275 (MexA− MexB− OprM++) harboring the indicated plasmids was used to perform antibiotic susceptibility testing as described in Materials and Methods. IPTG was included in the growth medium to induce MexB expression from plasmid pMMB206 derivatives carrying the mexB gene. OprM++, OprM is hyperexpressed.
WT, wild type. The His-tagged MexB protein was used to assess antibiotic resistance in some instances in order to be consistent with the interaction studies, which utilized MexB-His. Mutations are indicated.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid.
FIG. 1.
Mapping suppressor and inactivating mutations onto the three-dimensional structure of residues 29 to 259 of monomeric MexA (Protein Data Bank identifier 1VF7; http://www.ncbi.nlm.nih.gov). Mutated residues are represented in space-fill.
MexA suppressors show enhanced interaction with MexB(G220S).
A possible explanation for the finding that mutations in mexA suppress the antimicrobial susceptibility resulting from a G220S mutation in MexB is that the purportedly less stable MexB(G220S) trimer is intrinsically less active and/or interacts less effectively with MexA (wild type) and that the suppressor MexA proteins are better able to interact with the mutant MexB trimer, thereby stabilizing the trimer and permitting formation of a functional tripartite efflux system. To assess this directly, P. aeruginosa K2275 expressing plasmid-encoded MexA and either MexB(G220S)-His (from pDN44) or wild-type MexB-His (from plasmid pDN38) was extracted with detergent, and the extracts were incubated with Ni-NTA agarose beads to recover the histidine-tagged MexB(G220S) proteins. Corecovery of MexA (assessed using immunoblotting) was then used as a measure of MexA binding to the corresponding MexB protein in vivo. As seen previously, MexA was readily recovered together with wild-type MexB-His (Fig. 2, lane 2, upper panel), confirming the ability of these proteins to interact in vivo. In contrast, very little MexA was recovered together with MexB(G220S)-His (Fig. 2, lane 4, upper panel), even though levels of MexB(G220S)-His recovered from the Ni-NTA agarose beads in this experiment were comparable to the level of wild-type MexB-His (Fig. 2, lower panel, compare lanes 2 and 4). Clearly, MexB(G220S)-His was less able to bind MexA than its wild-type counterpart. Significantly, when the MexA V66M and V259F suppressors of MexB(G220S) were individually coexpressed with MexB(G220S)-His off the same vector (pDN45 and pDN46, respectively) in K2275, both showed substantially increased corecovery with MexB(G220S)-His (relative to wild-type MexA) on the Ni-NTA agarose beads (Fig. 2, upper panel, compare lanes 6 and 8 with lane 4). The levels of MexA(V66M) and MexA(V259F) bound to MexB(G220S) were, in fact, enhanced relative to the levels of wild-type MexA bound to wild-type MexB (Fig. 2, upper panel, compare lanes 6 and 8 with lane 2). These data clearly show that the V66M and V259F mutations in MexA restore the protein's ability to interact with the MexB(G220S) mutant protein and in so doing restore pump function (Table 2).
FIG. 2.
Western immunoblotting assessment of in vivo binding of wild-type and mutant MexA proteins to MexB(G220S)-His. Cell envelopes from P. aeruginosa strain K2275 carrying plasmid pDN38 (pMMB206::mexA-mexB-His) (lanes 1 and 2), pDN44 [pMMB206::mexA-mexB(G220S)-His] (lanes 3 and 4), pDN45 [pMMB206::mexA(V66M)-mexB(G220S)-His] (lanes 5 and 6), or pDN46 [pMMB206::mexA(V259F)-mexB(G220S)-His] (lanes 7 and 8) were extracted as described in Materials and Methods. Triton X-100-soluble extracts of cell envelope preparations were incubated with Ni-NTA agarose, and Triton X-100-soluble cell envelope extracts (odd-numbered lanes) and elution fractions off Ni-NTA (even-numbered lanes) were immunoblotted and developed with antibodies to MexA and MexB.
Isolation of oprM suppressors of mexA(V129M).
Several mexA mutations that compromise MexA function have been reported (29) including one, V129M, that occurs within the α-helical hairpin region (Fig. 1) that has been implicated in various MFP components in interactions with cognate OMF components (22, 46). To assess whether this residue or region of MexA might be important for interaction with OprM, attempts were made to isolate extragenic suppressors of mexA(V129M) in oprM. Thus, vector-borne oprM (pDN40) was randomly mutagenized and introduced into P. aeruginosa strain K2547 (ΔmexR ΔmexA ΔoprM) expressing MexA(V129M) from plasmid pDN42, and transconjugants expressing a now functional MexAB-OprM efflux system were selected on 12 μg/ml carbenicillin. A number of carbenicillin-resistant transconjugants were recovered and were shown to exhibit increased resistance to several MexAB-OprM antimicrobial substrates. The oprM-carrying vectors from these transconjugants invariably carried one of two mutations, yielding a T198I or F439I change in OprM, and their reintroduction into pDN42-carrying strain K2547 restored resistance to all tested antimicrobials (Table 3), consistent with the finding that these OprM changes suppress the V129M mutation in MexA. This suppression was, however, specific, inasmuch as these oprM mutations did not restore antimicrobial resistance in P. aeruginosa K2547 expressing a MexA variant carrying an inactivating mutation in helix 2 of the helical hairpin α-domain (A108T) (Fig. 1; Table 3). T198I and F439I both mapped to the proximal end of the periplasmic α-helical barrel of OprM, in helices 3 (T198I) and 8 (F439I), very near to each other in the three-dimensional structure (Fig. 3). Our data therefore support idea that the helical hairpin region of MexA interacts with the distal periplasmic portion of OprM.
TABLE 3.
Influence of OprM suppressors of MexA(V129M) on antibiotic susceptibility of P. aeruginosa expressing mutant or wild-type MexA proteinsa
| Plasmids | Expressed proteinb
|
MIC (μg/ml)c
|
|||
|---|---|---|---|---|---|
| MexA | OprM | CAR | NOV | NAL | |
| pDN3, pDN52 | WT | WT | 64 | 256 | 64 |
| pDN42, pDN52 | V129M | WT | 2 | 16 | 16 |
| pDN41, pDN52 | A108T | WT | 2 | 16 | 16 |
| pDN42, pDN51 | V129M | T198I | 128 | 256 | 128 |
| pDN42, pDN50 | V129M | F439I | 128 | 256 | 128 |
| pDN41, pDN51 | A108T | T198I | 2 | 16 | 16 |
| pDN41, pDN50 | A108T | F439I | 2 | 16 | 16 |
| pDN65, pDN51 | WT | T198I | 128 | 256 | 128 |
| pDN65, pDN50 | WT | F439I | 128 | 256 | 128 |
P. aeruginosa K2547 (MexA− MexB++ OprM−) harboring the indicated plasmids was used to perform antibiotic susceptibility testing as described in Materials and Methods. IPTG was included in the growth medium to induce MexA expression from pMMB206-derived plasmids. MexB++, MexB is hyperexpressed.
WT, wild type. Mutations are indicated.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid.
FIG. 3.
Mapping suppressor and inactivating mutations onto the three-dimensional structure of the OprM trimer (Protein Data Bank identifier 1WP1; http://www.ncbi.nlm.nih.gov). Mutated residues are represented in space-fill and labeled on one monomer only. The in vivo orientation of the structure with respect to the OM and periplasm (PP) is indicated. Residues are numbered according to the precursor form of the protein where the initiator methionine is residue number 1.
OprM suppressors show evidence of enhanced interaction with MexA(V129M).
To assess whether the V129M mutation in MexA compromises an interaction with OprM that is subsequently restored by the T198I and F439I suppressor mutations in OprM, attempts were made to develop a MexA-OprM interaction assay with His-tagged MexA, using corecovery of OprM on Ni-NTA agarose as an indicator of in vivo interaction, as above for MexA-MexB. An OprM-MexA interaction was confirmed using this assay (data not shown), in agreement with Trepout et al. (50), who recently demonstrated MexA binding to periplasmic OprM in vitro. Still in order to assess the impact of the V129M mutation on MexA's association with OprM and the subsequent impact of the T198I and F439I mutations in OprM on this association, it was necessary to express His-tagged versions of wild-type MexA or MexA(V129M) together with wild-type or suppressor OprM proteins off the same vector (30) (see above for MexA and MexB) since experience has shown that expression of any of MexA, MexB, or OprM from different vectors failed to provide evidence of an interaction using this assay (D. Nehme, unpublished results). Unfortunately, P. aeruginosa carrying plasmids with mexA-His and oprM engineered in tandem routinely failed to show any evidence of OprM production, precluding the use of the Ni-NTA agarose approach to look at the impact of the OprM suppressor mutations on binding to MexA(V129M). Subsequent examination of whole-cell immunoblots of P. aeruginosa, however, revealed a correlation between the presence of MexA and yields of OprM (Fig. 4) that could provide an indirect measure of in vivo OprM-MexA interaction. Thus, while substantial OprM was detectable in P. aeruginosa expressing MexA (Fig. 4A, lane 2), OprM was barely detectable in a ΔmexA mutant (Fig. 4A, lane 1). OprM levels were not, however, adversely impacted by loss of mexB (Fig. 4A, lane 5) unless mexA was also absent (Fig. 5A, lane 6), indicating that this OprM “instability” was specific to MexA and was, therefore, a reasonable measure of MexA-OprM interaction in vivo. As expected, the A108T and V129M mutations in MexA yielded reduced levels of OprM (Fig. 4A, compare lanes 3 and 4 with lane 2, and B, compare lanes 1 and 2), consistent with these MexA proteins interacting less well with OprM. This was reversed when MexA(V129M) was expressed with OprM(T198I), where OprM(T198I) levels were comparable to those of wild-type OprM expressed with wild-type MexA (Fig. 4B, compare lanes 1 and 3). This increased yield of OprM(T198I) was not an intrinsic feature of the mutant protein, however, inasmuch as it was MexA dependent [i.e., OprM(T198I) was not detectable in immunoblots of P. aeruginosa lacking MexA (Fig. 4B, lane 5)]; this observation is best explained by the capacity of the T198I mutation to restore/promote OprM interaction with the V129M mutant MexA. Strikingly, OprM(T198I) yields were greatly enhanced when this protein was coexpressed with wild-type MexA versus MexA(V129M) (Fig. 4B, compare lanes 3 and 4) and, indeed, exceeded the levels of wild-type OprM seen in P. aeruginosa expressing wild-type MexA (Fig. 4B, lane 1). P. aeruginosa expressing OprM(T198I) and wild-type MexA and MexB also showed increased levels of both MexA (Fig. 4B, upper panel, compare lanes 1 and 2) and MexB (Fig. 4C, compare lanes 1 and 3). This increased MexB, however, was lost upon deletion of mexA (Fig. 4C, lane 4) even though MexB levels were not compromised upon loss of mexA in a strain producing wild-type OprM (Fig. 4C, compare lanes 1 and 2). Apparently, changes imparted to OprM by the T198I substitution that restore an interaction with MexA somehow stabilize the tripartite complex.
FIG. 4.
Western immunoblotting assessment of OprM yields in whole-cell protein extracts as a measure of MexA-OprM interaction in vivo. (A) Whole-cell extracts prepared from P. aeruginosa K2274 (MexA−) harboring plasmids pMMB206 (no MexA; lane 1), pDN65 (MexA wild type [WT]; lane 2), pDN41 [MexA(A108T); lane 3] or pDN42 [MexA(V129M); lane 4] and from P. aeruginosa strains K1589 (MexB−; lane 5) and K2275 (MexA− MexB−; lane 6). Extracts were immunoblotted with antibodies to MexA and OprM. (B) Whole-cell extracts prepared from P. aeruginosa strain K1113 (OprM−) harboring plasmids pDN40 (OprM wild type [WT]; lane 1) or pDN70 [OprM(T198I); lane 2] and strain K2547 (MexA− OprM−) harboring plasmids pDN40 and pDN7 [OprM wild type (WT) and MexA(V129M); lane 3], pDN70 and pDN7 [OprM(T198I) and MexA(V129M); lane 4] or pDN70 alone [OprM(T198I); lane 5]. Extracts were immunoblotted with antibodies to MexA and OprM. (C) Anti-MexB immunoblotting of whole-cell extracts prepared from P. aeruginosa strains K1113 (OprM−; lanes 1 and 3) and K2547 (MexA− OprM −; lanes 2 and 4) harboring plasmids pDN40 (OprM wild type [WT]; lanes 1 and 2) or pDN70 [OprM(T198I); lanes 3 and 4]. MexA detectable in lanes 1 and 6 of panel A and lane 5 of panel B represents overflow from an adjacent well. The status (wild type or mutant) of relevant MexAB-OprM components is indicated above each lane.
FIG. 5.
Anti-OprM immunoblotting of whole-cell extracts prepared from P. aeruginosa strain K1110 (OprM−) harboring plasmids pDN57 [OprM(T209A); lane 1], pDN58 [OprM(V215A); lane 2], pDN59 [OprM(V215T); lane 3], pDN61 [OprM(V217T); lane 4], pDN62 [OprM(T423A); lane 5], pDN64 [OprM(D426A); lane 6], pDN60 [OprM(G216A); lane 7], pDN63 [OprM(G424A); lane 8], pMMB206 (no OprM; lane 9), or pDN40 (OprM wild type; lane 10).
oprM suppressors of mexA(V129M) suppress mutations in mexB.
In light of our data showing that different pump-destabilizing mutations in mexB could be suppressed by the same suppressor mutations in mexA, it was of interest to see if the oprM suppressors of mexA(V129M) might also be able to suppress the mexB mutations. Thus, the plasmid-borne oprM(T198I) and oprM(F439I) genes were introduced into P. aeruginosa K2553 expressing MexB(G220S) or MexB(T578I). While P. aeruginosa K2553 expressing wild-type OprM and the mutant MexB proteins were, as noted previously (24), multidrug hypersusceptible (Table 4), consistent with the mexB mutations compromising MexAB-OprM pump activity, both oprM suppressors restored resistance of the mutant MexB-producing P. aeruginosa to wild-type levels (i.e., restored pump activity) (Table 4). As with mexA suppressors of these mexB mutations, however, the oprM suppressors did not restore resistance to P. aeruginosa expressing MexB with a mutation in the vestibule region of the protein (E864K) (Table 4).
TABLE 4.
Influence of OprM suppressors of MexA(V129A) on antibiotic susceptibility of P. aeruginosa expressing mutant MexB proteinsa
| Plasmids | Expressed proteinb
|
MIC (μg/ml)c
|
|||
|---|---|---|---|---|---|
| MexB | OprM | CAR | NOV | NAL | |
| pDN25, pDN52 | WT | WT | 128 | 256 | 128 |
| pDN43, pDN52 | G220S | WT | 4 | 32 | 16 |
| pDN34, pDN52 | T578I | WT | 4 | 32 | 16 |
| pDN43, pDN51 | G220S | T198I | 128 | 256 | 128 |
| pDN43, pDN50 | G220S | F439I | 128 | 256 | 128 |
| pDN34, pDN51 | T578I | T198I | 128 | 256 | 128 |
| pDN34, pDN50 | T578I | F439I | 128 | 256 | 128 |
| pDN39, pDN51 | E864K | T198I | 2 | 16 | 16 |
| pDN39, pDN50 | E864K | F439I | 2 | 16 | 16 |
P. aeruginosa K2553 (MexA++ MexB− OprM−) harboring the indicated plasmids was used to perform antibiotic susceptibility testing as described in Materials and Methods. IPTG was included in the growth medium to induce MexB expression from pMMB206-derived plasmids. MexA++, MexA is hyperexpressed.
WT, wild type. Mutations are indicated.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid.
oprM mutations compromising OprM function.
Two glycine residues (G147 and G365) that occur at the proximal (periplasmic) end of TolC have been shown to directly contact AcrB (47) and so might be of functional importance. It was of interest, therefore, to determine if the corresponding residues (Fig. 3, G216 and G424) and others found at the proximal end of OprM (T209, V215, V217, T423, and D426) were important for MexAB-OprM function. Thus, alanine (or threonine) substitutions were engineered at each site, the mutant proteins were expressed in P. aeruginosa K1110 (ΔoprM) (Fig. 5), and the impact on antimicrobial resistance was assessed (Table 5) as a measure of MexAB-OprM pump function. The T209A substitution abrogated OprM production (Fig. 5, lane 1) and therefore resistance (Table 5), while all other substitutions yielded wild-type levels of OprM (Fig. 5). Of these latter, only the G216A and G424A substitutions had a detrimental impact on antibiotic resistance (Table 5). Interestingly, this resistance defect was reversed when MexA was replaced with representative MexA suppressors of MexB(G220S) [MexA(V66M) and MexA(V259F)] in OprM(G216A)- or OprM(G424A)-expressing P. aeruginosa (Table 6). In trying to assess whether the OprM G216A and G424A substitutions did, indeed, compromise an OprM-MexB interaction, an OprM-MexB association was first established using corecovery of OprM and MexB-His on Ni-NTA as above (data not shown). Again, however, attempts at expressing MexB-His and OprM(G216A) or OprM(G424A) from genes cloned in tandem failed, precluding assessment of the impact of these mutations on MexB association.
TABLE 5.
Antibiotic susceptibility of P. aeruginosa expressing mutant OprM proteinsa
| Plasmid | OprM protein expressedb | MIC (μg/ml)c
|
||
|---|---|---|---|---|
| CAR | NOV | NAL | ||
| pDN40 | WT | 64 | 256 | 64 |
| pMMB206 | None | 1 | 8 | 8 |
| pDN57 | T209Ad | 2 | 8 | 8 |
| pDN58 | V215A | 64 | 256 | 64 |
| pDN59 | V215T | 64 | 256 | 64 |
| pDN61 | V217T | 64 | 256 | 64 |
| pDN62 | T423A | 64 | 256 | 64 |
| pDN64 | D426A | 64 | 256 | 64 |
| pDN60 | G216A | 2 | 16 | 16 |
| pDN63 | G424A | 2 | 16 | 16 |
P. aeruginosa K1110 (MexA+ MexB++ OprM−) harboring the indicated plasmid was used to perform antibiotic susceptibility testing as described in Materials and Methods. IPTG was included in the growth medium to induce OprM expression from pMMB206-derived plasmids. MexB++, MexB is hyperexpressed.
WT, wild type.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid.
OprM(T209A) was not stably expressed as assessed by Western immunoblotting (Fig. 5).
TABLE 6.
Influence of MexA suppressors of MexB(G220S) on antibiotic susceptibility of P. aeruginosa expressing mutant OprM proteinsa
| Plasmids | Expressed proteinb
|
MIC (μg/ml)c
|
|||
|---|---|---|---|---|---|
| MexA | OprM | CAR | NOV | NAL | |
| pDN3, pDN40 | WT | WT | 64 | 256 | 64 |
| pDN3, pDN60 | WT | G216A | 2 | 16 | 16 |
| pDN3, pDN63 | WT | G424A | 2 | 16 | 16 |
| pDN47, pDN60 | V66M | G216A | 128 | 256 | 128 |
| pDN47, pDN63 | V66M | G424A | 128 | 256 | 128 |
| pDN49, pDN60 | V259F | G216A | 128 | 256 | 128 |
| pDN49, pDN63 | V259F | G424A | 128 | 256 | 128 |
P. aeruginosa K2547 (MexA− MexB++ OprM−) harboring the indicated plasmids was used to perform antibiotic susceptibility testing as described in Materials and Methods. IPTG was included in the growth medium to induce OprM expression from pMMB206-derived plasmids. MexB++, MexB is hyperexpressed.
WT, wild type.
CAR, carbenicillin; NOV, novobiocin; NAL, nalidixic acid.
DISCUSSION
Previous studies have implicated the helical hairpin α-domain of MFPs like AcrA and MexA in these proteins' interaction with their cognate OMF components (22, 46, 49). Consistent with this, we have shown here that a mutation in the helical hairpin region of MexA (V129M) could be suppressed by mutations in OprM (T198I and F439I). Significantly, these mutations mapped to helices 3 (T198I) and 8 (F439I) of the equatorial domain of OprM in a region that in TolC has been defined as the site of interaction with its MFP, AcrA (22, 46). Moreover, gain-of-function mutations within the VceC component of the Vibrio cholerae VceABC efflux system that allow it to operate with E. coli AcrAB also map to helices 3 and 8 (51) as do mutations in TolC that permit this OMF to function with the P. aeruginosa MexAB components (5). One possibility is that these mutations somehow alter the MFP binding domain to allow the OMFs to accommodate noncognate or mutant MFPs although one cannot rule out that they simply render OMF function independent of a functional MFP interaction. Still, the observation that the OprM suppressors of MexA(V129M) reported here are unstable without MexA and do not promote antibiotic resistance in a MexA− mutant (D. Nehme, unpublished results) argue for the former possibility. It is interesting, too, that these gain-of-function and suppressor mutations do not adversely affect the ability of the mutant OMF components to interact with their cognate or wild-type MFP components. Thus, mutant TolC and VceC that can functionally associate with MexAB and AcrAB, respectively, retain the ability to function with AcrAB and HlyAB (TolC) (5) and VceAB (VceC) (51) while OprM(T198I) and OprM(F439I) operate with wild-type MexAB (Table 3). This suggests that these OMF mutations increase flexibility and/or reduce specificity as regards their interaction with MFP components. If so, the fact that wild-type VceC can interact with AcrAB (51) and wild-type TolC can interact with MexAB, despite not forming a functional complex with these noncognate efflux components, argues that the aforementioned gain-of-function and suppressor mutations must somehow promote a functional association between the OMF and noncognate or mutant MFP.
A surprising finding of this study is that the same mutations within the α/β domain of MexA suppress a variety of inactivating MexB and OprM mutations. Since both of the MexB T158I and G220S mutations were shown to interfere with pump assembly, inasmuch as much as their interactions with MexA in vivo were reduced relative to wild-type MexB (30) (Fig. 2), the recovery of common MexA suppressors of these mutations is understandable since they simply overcome the assembly defect by restoring an interaction with the mutant MexB proteins. Consistent with this interpretation, these suppressors did not restore functionality to a pump with a mutation in the vestibule region of MexB (E864K), expected to be defective in substrate acquisition and not assembly (24). Why these MexA mutants would also suppress mutations at the proximal (periplasmic) tip of OprM is less clear. In light of recent data showing that the proximal tip of TolC interacts with its cognate RND component, AcrB (47), it may be that these mutations in OprM compromise an OprM-MexB interaction. Perhaps, then, the suppressor mutations in MexA simply enhance MexA interaction with both of these proteins and stabilize them or force a functional interaction. It is worth noting, for example, that previously reported intragenic suppressors of the MexB(G220S) mutant all mapped to the distal (periplasmic) tip of MexB where, in light of the TolC-MexB interaction study (24), MexB is expected to interact with OprM. The implication is that the G220S mutation compromises MexB's interaction with OprM as well as MexA, and so the MexA suppressors of MexB(G220S) must overcome both defects. As such, these suppressors would also be expected to overcome defects in OprM-MexB association caused by mutations in the periplasmic tip of OprM. Extragenic suppressors of a TolC mutant that map predominantly to the α/β domain of AcrA have also been reported (8), suggesting that changes to this domain in particular positively impact MFP association with and/or stabilization of the other pump components, perhaps by influencing the disposition/conformation of the OMF- and/or RND-binding domains of these MFPs.
While technical difficulties precluded a direct examination of the impact of a MexA(V129M) mutation on this protein's interaction with OprM and the effect of OprM suppressor mutations on a MexA(V129M)-OprM interaction, MexA-dependent OprM stability proved a useful surrogate for assessing this. Thus, mutations in the helical hairpin α-domain of MexA expected to adversely impact an interaction with OprM yielded reduced levels of OprM that were, however, restored by the T198I or F439I suppressor mutations in OprM. Importantly, the increased apparent stability of OprM(T198I) and OprM(F439I) was MexA dependent, consistent with the increased stability reflecting a restored interaction with MexA(V129M). This is reminiscent of AcrA suppressors of mutations in TolC, where the assembly defect and instability of a mutant TolC protein were partially reversed by the MexA suppressors; i.e., mutant TolC was stabilized by a direct interaction with AcrA (8). The observations here that the OprM(T198I) suppressor was functional with wild-type MexA and MexB and that its expression in P. aeruginosa produced noticeably enhanced yields of all three pump components are consistent with the idea that this mutation stabilizes the entire tripartite complex. This has potentially important implications vis-à-vis complex crystallization and determination of pump structure since a stable complex might be easier to crystallize.
Acknowledgments
We thank N. Gotoh for providing monoclonal antibodies to OprM.
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation. D.N. was the recipient of a CCFF studentship.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Akama, H., M. Kanemaki, M. Yoshimura, T. Tsukihara, T. Kashiwagi, H. Yoneyama, S. I. Narita, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the drug-discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 17:52816-52819. [DOI] [PubMed] [Google Scholar]
- 2.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 279:25939-25942. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 4.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 5.Bokma, E., E. Koronakis, S. Lobedanz, C. Hughes, and V. Koronakis. 2006. Directed evolution of a bacterial efflux pump: adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 580:5339-5343. [DOI] [PubMed] [Google Scholar]
- 6.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 7.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 185:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerken, H., and R. Misra. 2004. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol. Microbiol. 54:620-631. [DOI] [PubMed] [Google Scholar]
- 9.Gotoh, N., H. Tsujimoto, M. Tsuda, K. Okamoto, A. Nomura, T. Wada, M. Nakahashi, and T. Nishino. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3:247-255. [DOI] [PubMed] [Google Scholar]
- 11.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 101:9994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.Husain, F., M. Humbard, and R. Misra. 2004. Interaction between the TolC and AcrA proteins of a multidrug efflux system of Escherichia coli. J. Bacteriol. 186:8533-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue, H., H. Nojima, and H. Okayama. 1991. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 16.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 19.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, X. Z., and K. Poole. 2001. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 183:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobedanz, S., E. Bokma, M. F. Symmons, E. Koronakis, C. Hughes, and V. Koronakis. 2007. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA 104:4612-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 186:1258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokhonov, V. V., E. I. Mokhonova, H. Akama, and T. Nakae. 2004. Role of the membrane fusion protein in the assembly of resistance-nodulation-cell division multidrug efflux pump in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 322:483-489. [DOI] [PubMed] [Google Scholar]
- 26.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 27.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 143:173-179. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 29.Nehme, D., X. Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 186:2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nehme, D., and K. Poole. 2005. Interaction of the MexA and MexB components of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification of MexA extragenic suppressors of a T578I mutation in MexB. Antimicrob. Agents Chemother. 49:4375-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okii, M., S. Iyobe, and S. Mitsuhashi. 1983. Mapping of the gene specifying aminoglycoside 3′-phosphotransferase II on the Pseudomonas aeruginosa chromosome. J. Bacteriol. 155:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 33.Poole, K. 2003. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms, p. 273-298. In I. T. Paulsen and K. Lewis (ed.), Microbial multidrug efflux. Horizon Scientific Press, Norfolk, United Kingdom.
- 34.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 35.Poole, K. 2004. Efflux pumps, p. 635-674. In J.-L. Ramos (ed.), Pseudomonas, vol. I. Genomics, lifestyle and molecular architecture. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 36.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redly, G. A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable extracytoplasmic-function sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295-1298. [DOI] [PubMed] [Google Scholar]
- 40.Sennhauser, G., P. Amstutz, C. Briand, O. Storchenegger, and M. G. Grutter. 2006. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLOS Biol. 5:0106-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 42.Sobel, M. L., D. Hocquet, L. Cao, P. Plesiat, and K. Poole. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stegmeier, J. F., G. Polleichtner, N. Brandes, C. Hotz, and C. Andersen. 2006. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry 45:10303-10312. [DOI] [PubMed] [Google Scholar]
- 47.Tamura, N., S. Murakami, Y. Oyama, M. Ishiguro, and A. Yamaguchi. 2005. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 44:11115-11121. [DOI] [PubMed] [Google Scholar]
- 48.Tikhonova, E. B., and H. I. Zgurskaya. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279:32116-32124. [DOI] [PubMed] [Google Scholar]
- 49.Touze, T., J. Eswaran, E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53:697-706. [DOI] [PubMed] [Google Scholar]
- 50.Trepout, S., S. Mornet, H. Benabdelhak, A. Ducruix, A. R. Brisson, and O. Lambert. 2007. Membrane protein selectively oriented on solid support and reconstituted into a lipid membrane. Langmuir 23:2647-2654. [DOI] [PubMed] [Google Scholar]
- 51.Vediyappan, G., T. Borisova, and J. A. Fralick. 2006. Isolation and characterization of VceC gain-of-function mutants that can function with the AcrAB multiple-drug-resistant efflux pump of Escherichia coli. J. Bacteriol. 188:3757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 53.Ziha-Zarifi, I., C. Llanes, T. Koehler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]