Toxin-antitoxin systems are very commonly found both on large, low-copy plasmids, where they increase effective stability (35), and on bacterial chromosomes, where their function has been the subject of considerable speculation. In this issue of the Journal of Bacteriology, Virginie Tsilibaris and colleagues (65) in the laboratory of Laurence Van Melderen ask “What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome?” To answer this question, the authors compared the fitness levels of wild-type Escherichia coli and an isogenic strain simultaneously deleted for five chromosomal toxin-antitoxin systems. Relative fitness was assessed by head-to-head competition in liquid culture with reciprocal markers. By this rigorous test, the benefit, under the conditions tested, is little or none. This is a stunning bit of news that poses a serious challenge to the heretofore preeminent stress response/growth control hypothesis.
DEFINING CHARACTERISTICS
A proteic toxin-antitoxin system (35) can be defined an “addictive” genetic element (Fig. 1) that encodes two intracellular proteins, a stable toxin and an unstable antitoxin (Fig. 2). Transcription of the two genes is typically autoregulated by the protein products. This homeostatic negative-feedback loop may help to maintain steady levels of toxin and antitoxin (43).
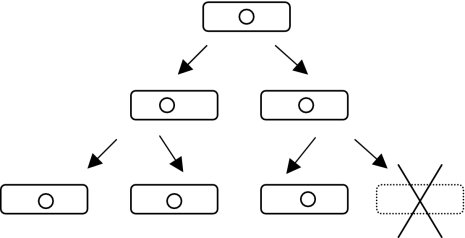
FIG. 1.
Toxin-antitoxin systems are addictive. An addictive genetic element (small circle) can increase effective plasmid stability approximately 10- to 1,000-fold (35) by the arrest or elimination of element-free daughter cells (lower right). Potentially, the loss of the addictive genetic element may occur by (mis-) segregration, exclusion, mutation, or recombination. If the environment is sufficiently structured and has limited resources, then the elimination of element-free cells will leave more food for element-containing cells. Since plasmid-borne toxin-antitoxin systems can be found on chromosomes (66) and since chromosomal toxin-antitoxin systems can stabilize plasmids (12, 26), there is at present little evidence of any obligatory location-specific specializations. Restriction modification systems (48), hok-sok-type postsegregational killing systems (25), and eukaryotic meiotic drive systems (6) possess similar addictive properties but are mechanistically distinct.
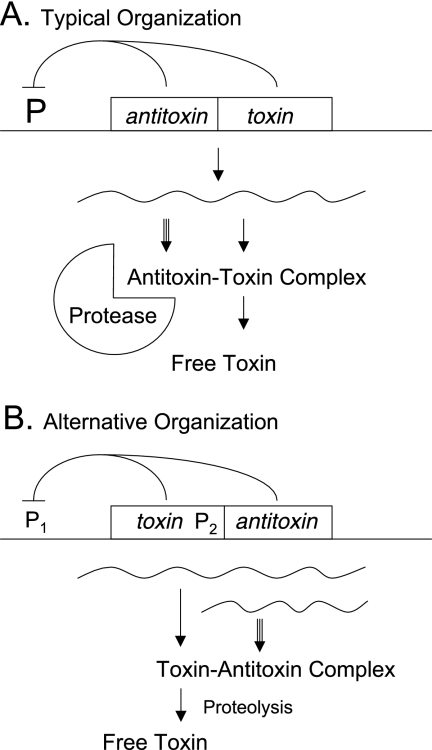
FIG. 2.
Organization of toxin-antitoxin systems. (A) Typically, the antitoxin gene precedes the toxin gene. (B) In some families, the usual order is reversed and a second promoter is observed. In both cases, antitoxin and toxin genes are usually small and often have overlapping stop and start codons, indicating that translation may be coupled. Transcription of the two genes is generally autoregulated by the protein products. The antitoxin is synthesized at a greater rate than the toxin but is also degraded at greater rate, due to the action of a specific host-encoded ATP-dependent protease, such as Lon, ClpXP, or ClpAP. At steady state, there is sufficient antitoxin to bind and neutralize the toxin. If the genes are lost or if their synthesis is sufficiently disrupted, the continuing proteolysis of the unstable antitoxin may liberate the stable toxin and produce biological effects. In at least a few atypical instances, a third protein product is implicated in the functioning of the system or the toxin is unusually large. Numerous reviews are available (19, 23, 29, 31, 35, 56, 57, 68, 70).
Multiple mechanisms for activation.
If the homeostatic capacity of the system is exceeded by transcriptional inhibitors, translational inhibitors (61), severe starvation, or addition of serine hydroxamate (a convenient if severe facsimile of starvation) (10), then the toxin may be unveiled and the cell arrested. Potentially, the specific transcriptional or translational downregulation of the operon (2), the activation of another toxin-antitoxin system (34), or the specific upregulation of the appropriate protease (9) might also be sufficient or helpful in revealing the toxin. Additionally, the actual loss of the toxin-antitoxin genes, by segregation, exclusion, mutation, or recombination, can liberate the toxin and arrest the cell.
Multiple mechanisms for action.
The toxins so far characterized include gyrase inhibitors, phosphotransferases, site-specific ribonucleases, ribosome-dependent ribonucleases, and a possible riboexonuclease (5, 7, 70). In principle, any essential structure or process could be targeted by these toxins. In practice, mRNA-targeting ribonucleases are particularly common (13) (Fig. 3). Interestingly, the immediate effects of these RNase toxins are bacteriostatic rather than bactericidal (3, 53) and can be counteracted by the action of tmRNA (11). Importantly, no toxin has been reported to be directly bacteriolytic.
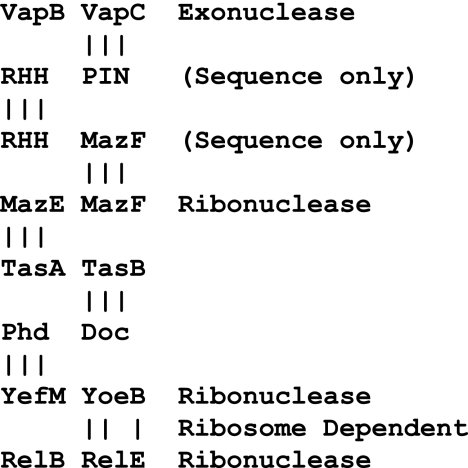
FIG. 3.
Phylogenetic network of chimeric toxin-antitoxin systems. The known toxins belong to a diverse and polyphyletic set of at least four major families, and the antitoxins are equally heterogeneous (4, 51). The N-terminal moiety of the antitoxin typically contains a DNA-binding domain belonging to any one of four of more repressor families, while the C-terminal moiety contains a flexible antitoxin domain that is typically too short and too diverse to be of any significant bioinformatic utility. The major toxin-antitoxin families can be loosely connected in an evolutionary sense only by their propensities to swap operator-repressor and antitoxin-toxin modules, thereby generating a network of chimeric systems (4, 22, 27, 30, 44, 63).
Multiple and polymorphic toxin-antitoxin systems.
Bioinformatic surveys indicate that prokaryotic genomes may contain zero, one, or many toxin-antitoxin systems (51). Escherichia coli, with at least a half a dozen toxin-antitoxin systems, is fairly typical. It is increasingly apparent that toxin-antitoxin systems are frequently polymorphic, meaning that they are found in some but not all isolates of a given species (51, 66, 67) (Table 1). Stable polymorphisms may be attributed to niche-specific adaptations or to frequency-dependent selection. Multiplicity is generally regarded as strong evidence of functional specialization. Other selfish genetic elements, such as restriction-modification systems (49) and colicins (59), share these distinctive bioinformatic characteristics.
TABLE 1.
Polymorphism of toxin-antitoxin systems in Escherichia coli
| Escherichia coli strain | Presence of indicated K-12 toxina
|
|||||
|---|---|---|---|---|---|---|
| yoeB | mazF | relE | chpB | yafQ | hipA | |
| K-12 | + | + | + | + | + | + |
| 0157:H7 | − | + | + | + | + | + |
| UT189 | + | + | + | − | − | + |
| CFT073 | + | − | − | + | − | + |
Results indicate the presence and absence of K-12 toxins in the genomes of Escherichia coli strains. +, a very good match for the K-12 toxin; −, no match or only a distant match for the K-12 toxin.
NINE POSSIBLE FUNCTIONS OF CHROMOSOMAL TOXIN-ANTITOXIN SYSTEMS
(i) Junk.
Chromosomal toxin-antitoxin systems are genomic junk, acquired from plasmids or other sources and lost in due course, albeit at an unusually low rate, due to their addictive qualities. This is an important null hypothesis against which any other hypothesis must be compared (41, 50).
(ii) Stabilization of genomic parasites.
The presence of toxin-antitoxin systems on larger genomic parasites, such as conjugative transposons (17) and temperate bacteriophage (16), may benefit the parasite by reducing the effective deletion rate. Many “chromosomal” toxin-antitoxin systems may be found, upon closer examination, to map to chromosomal parasites or to the remnants of such parasites (58).
(iii) Selfish alleles.
The recombinational (28) or segregational (14, 15, 37, 45, 46) loss of a toxin-antitoxin system following a sexual encounter results in addictive arrest. Thus, nonaddictive alleles cannot replace addictive alleles, but the reciprocal event is permitted (Fig. 4). In a limited and structured environment (8), this asymmetry generates a slight benefit for the addictive element and a slight detriment to any nonaddictive allele. Since different toxin-antitoxin systems at different locations occupy different ecological niches, the multiplicity of the toxin-antitoxin systems is well explained. Polymorphisms may reflect a balance between infected and uninfected loci and are thus also expected. There are serious quantitative difficulties with this hypothesis, but they may not be insuperable (64).
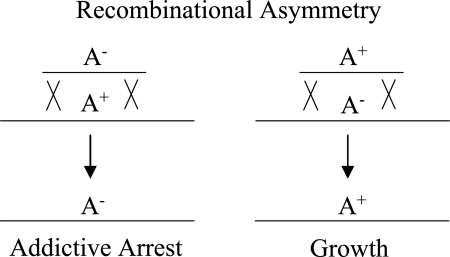
FIG. 4.
Recombinational asymmetry. The replacement of an addictive allele (A+) with a nonaddictive allele (A−) leads to addictive arrest, but the reciprocal process gives a viable recombinant. Thus, toxin-antitoxin systems and other addictive genetic elements can introduce an asymmetry into an otherwise symmetric gene transfer process, such as transduction, conjugation, or transformation. If the environment is structured and has limited resources, then the elimination of A− cells may leave more food for A+ cells and thus generate a slight benefit for the addictive allele as well as a slight detriment to the nonaddictive allele.
(iv) Gene regulation.
Since many of the toxins encode site-specific ribonucleases, they can shift gene expression away from site-rich messages and toward site-poor messages. This effect may be used for autoregulation (60, 69), for specific regulation (47, 54), and possibly for global regulation (18, 20).
(v) Growth control.
Toxin-antitoxin systems can be activated by starvation or other stresses (1, 7, 10, 33, 38). Since many of the RNase toxins are bacteriostatic rather than bactericidal in nature, their effects can potentially be reversed (53) by the action of tmRNA (11). The quick arrest of growth, in response to starvation or other stresses, may permit improved survival during starvation or a quicker resumption of growth when conditions improve or both (23, 24). At present, however, there is no evidence that toxin-accentuated arrest actually creates an advantage later on, and the first real attempt to test this prediction (65) has failed to reveal any substantial benefit during head-to-head competition in liquid culture. Additionally, this hypothesis cannot predict the existence of multiple and polymorphic toxin-antitoxin systems (Table 1) without invoking accessory hypotheses (such as those regarding functional specializations and niche-specific adaptations) for which there is little or no independent evidence.
(vi) Persisters.
At least one toxin-antitoxin system is implicated in the generation of “persisters,” a subfraction of the population that is characterized by low growth and high resistance to β-lactam antibiotics (36, 39). By adapting a mixed strategy in which some cells are specialized for growth while others are specialized for persistence, the culture can insure itself against a sudden and catastrophic loss (40).
(vii) Programmed cell arrest and the preservation of the commons.
The toxin-mediated early arrest of growth by the culture may leave more resources in the medium and therefore may improve long-time survival. This scenario requires a reasonably well-structured environment (such as an agar plate) or some other mechanism to ensure that the benefits do not accrue without limit to the cheaters (42). Polymorphisms might be interpreted as a balance between good citizens and selfish cheaters. A multiplicity of toxin-antitoxin systems might be useful in reducing the rate at which cheaters are generated. This hypothesis and the following ones predict that toxin-antitoxin systems will be beneficial in pure culture or in a structured habitat, such as an agar plate, but may fail to show any benefit in a mixed-liquid culture.
(viii) Programmed cell death.
Programmed cell death is similar to the process for the preceding hypothesis, except that it requires a heterogeneous response to stress (since no one can benefit if everyone dies) (2, 18, 42). The prediction of heterogeneity might be tested experimentally using green fluorescent protein fusions or other single-cell techniques. Currently, there is no uncontested and unambiguous evidence of toxin-mediated cell death (65).
(ix) Antiphage.
Since bacteriophage can interfere with host transcription and translation (55), they may activate addictive systems, which would then limit phage production (32, 52), unless the phage could inhibit host proteases (21, 62), neutralize the toxins, or exit the cell before the toxins were activated. Toxin-antitoxin diversity and multiplicity arise as a natural consequence of the host-parasite conflict.
Multiple hypotheses may apply.
Multiple hypotheses may apply to a single toxin-antitoxin system. In principle, one toxin-antitoxin system could be a parasite (hypothesis iii), could stabilize a larger parasite (hypothesis ii), could benefit individual cells (hypotheses iv, v, and vi), and could benefit the colony, by mediating cooperative (hypothesis vii) and altruistic (hypotheses viii or ix) strategies, all at the same time. At present, however, there is no particularly compelling case for even one of these possibilities. Progress will require interdisciplinary studies and a combination of creativity, collegiality, and critical thinking.
Acknowledgments
I acknowledge Ry Young for helpful discussions and key intellectual contributions regarding the possible role of toxin-antitoxin systems as a defense against bacteriophages.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Aguirre-Ramirez, M., J. Ramirez-Santos, L. Van Melderen, and M. C. Gomez-Eichelmann. 2006. Expression of the F plasmid ccd toxin-antitoxin system in Escherichia coli cells under nutritional stress. Can. J. Microbiol. 52:24-30. [DOI] [PubMed] [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantharaman, V., and L. Aravind. 2003. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 4:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcus, V. L., K. Backbro, A. Roos, E. L. Daniel, and E. N. Baker. 2004. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 279:16471-16478. [DOI] [PubMed] [Google Scholar]
- 6.Bull, J. J., I. J. Molineux, and J. H. Werren. 1992. Selfish genes. Science 256:65. [DOI] [PubMed] [Google Scholar]
- 7.Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672-679. [DOI] [PubMed] [Google Scholar]
- 8.Chao, L., and B. R. Levin. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78:6324-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705-1717. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 12.Christensen-Dalsgaard, M., and K. Gerdes. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397-411. [DOI] [PubMed] [Google Scholar]
- 13.Condon, C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61:573-583. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper, T. F., and J. A. Heinemann. 2005. Selection for plasmid post-segregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc. Biol Sci. 272:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeShazer, D. 2004. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 186:3938-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dziewit, L., M. Jazurek, L. Drewniak, J. Baj, and D. Bartosik. 2007. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J. Bacteriol. 189:1983-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelberg-Kulka, H., S. Amitai, I. Kolodkin-Gal, and R. Hazan. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 20.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 118:4327-4332. [DOI] [PubMed] [Google Scholar]
- 21.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 95:15481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fico, S., and J. Mahillon. 2006. TasA-tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system. BMC Genomics 7:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 25.Gerdes, K., and E. G. Wagner. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10:117-124. [DOI] [PubMed] [Google Scholar]
- 26.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 27.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 28.Handa, N., Y. Nakayama, M. Sadykov, and I. Kobayashi. 2001. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol. Microbiol. 40:932-940. [DOI] [PubMed] [Google Scholar]
- 29.Hayes, C. S., and R. T. Sauer. 2003. Toxin-antitoxin pairs in bacteria: killers or stress regulators? Cell 112:2-4. [DOI] [PubMed] [Google Scholar]
- 30.Hayes, F. 1998. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180:6415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 32.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227-234. [DOI] [PubMed] [Google Scholar]
- 33.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 36.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi, I. 2004. Genetic addiction: a principle of gene symbiosis in a genome, p.105-144. In B. E. Funnel and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 38.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 40.Kussell, E., R. Kishony, N. Q. Balaban, and S. Leibler. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerat, E., and H. Ochman. 2005. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 33:3125-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magnuson, R., and M. B. Yarmolinsky. 1998. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180:6342-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinley, J. E., and R. D. Magnuson. 2005. Characterization of the Phd repressor-antitoxin boundary. J. Bacteriol. 187:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochizuki, A., K. Yahara, I. Kobayashi, and Y. Iwasa. 2006. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics 172:1309-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mongold, J. A. 1992. Theoretical implications for the evolution of postsegregational killing by bacterial plasmids. Am. Nat. 139:677-689. [Google Scholar]
- 47.Muñoz-Gómez, A. J., M. Lemonnier, S. Santos-Sierra, A. Berzal-Herranz, and R. Díaz-Orejas. 2005. RNase/anti-RNase activities of the bacterial parD toxin-antitoxin system. J. Bacteriol. 187:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 49.Nobusato, A., I. Uchiyama, and I. Kobayashi. 2000. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene 259:89-98. [DOI] [PubMed] [Google Scholar]
- 50.Ochman, H., and L. M. Davalos. 2006. The nature and dynamics of bacterial genomes. Science 311:1730-1733. [DOI] [PubMed] [Google Scholar]
- 51.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pecota, D. C., and T. K. Wood. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 178:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 54.Pimentel, B., M. A. Madine, and G. de la Cueva-Mendez. 2005. Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 24:3459-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pineda, M., B. D. Gregory, B. Szczypinski, K. R. Baxter, A. Hochschild, E. S. Miller, and D. M. Hinton. 2004. A family of anti-sigma70 proteins in T4-type phages and bacteria that are similar to AsiA, a transcription inhibitor and co-activator of bacteriophage T4. J. Mol. Biol. 344:1183-1197. [DOI] [PubMed] [Google Scholar]
- 56.Rawlings, D. E. 1999. Proteic toxin-antitoxin, bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol. Lett. 176:269-277. [DOI] [PubMed] [Google Scholar]
- 57.Rice, K. C., and K. W. Bayles. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729-738. [DOI] [PubMed] [Google Scholar]
- 58.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 34:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley, M. A., L. Cadavid, M. S. Collett, M. N. Neely, M. D. Adams, C. M. Phillips, J. V. Neel, and D. Friedman. 2000. The newly characterized colicin Y provides evidence of positive selection in pore-former colicin diversification. Microbiology 146:1671-1677. [DOI] [PubMed] [Google Scholar]
- 60.Ruiz-Echevarría, M. J., G. de la Cueva, and R. Díaz-Orejas. 1995. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol. Gen. Genet. 248:599-609. [DOI] [PubMed] [Google Scholar]
- 61.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skorupski, K., J. Tomaschewski, W. Ruger, and L. D. Simon. 1988. A bacteriophage T4 gene which functions to inhibit Escherichia coli Lon protease. J. Bacteriol. 170:3016-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, J. A., and R. D. Magnuson. 2004. Modular organization of the Phd repressor/antitoxin protein. J. Bacteriol. 186:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spratt, B. G., W. P. Hanage, and E. J. Feil. 2001. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 4:602-606. [DOI] [PubMed] [Google Scholar]
- 65.Tsilibaris, V., G. Maenhaut-Michel, N. Mine, and L. Van Melderen. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyndall, C., H. Lehnherr, U. Sandmeier, E. Kulik, and T. A. Bickle. 1997. The type IC hsd loci of the enterobacteria are flanked by DNA with high homology to the phage P1 genome: implications for the evolution and spread of DNA restriction systems. Mol. Microbiol. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 67.Wilbaux, M., N. Mine, A. M. Guerout, D. Mazel, and L. Van Melderen. 2007. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J. Bacteriol. 189:2712-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]
- 70.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]