Abstract
The Escherichia coli K-12 chromosome encodes at least five proteic toxin-antitoxin (TA) systems. The mazEF and relBE systems have been extensively characterized and were proposed to be general stress response modules. On one hand, mazEF was proposed to act as a programmed cell death system that is triggered by a variety of stresses. On the other hand, relBE and mazEF were proposed to serve as growth modulators that induce a dormancy state during amino acid starvation. These conflicting hypotheses led us to test a possible synergetic effect of the five characterized E. coli TA systems on stress response. We compared the behavior of a wild-type strain and its derivative devoid of the five TA systems under various stress conditions. We were unable to detect TA-dependent programmed cell death under any of these conditions, even under conditions previously reported to induce it. Thus, our results rule out the programmed-cell-death hypothesis. Moreover, the presence of the five TA systems advantaged neither recovery from the different stresses nor cell growth under nutrient-limited conditions in competition experiments. This casts a doubt on whether TA systems significantly influence bacterial fitness and competitiveness during non-steady-state growth conditions.
Most bacterial genomes harbor multiple toxin-antitoxin (TA) systems (for reviews on TA systems, see references 6, 13, 18, and 24). Based on sequence homology, seven families of proteic TA systems have been defined (3, 40). Members of a given family can be found at multiple locations within a genome (e.g., in the genomic core, genomic islands, and mobile genetic elements) and in multiple bacterial species. This ubiquity strongly suggests that TA systems move from one location to another through horizontal gene transfer. The first characterized TA operons were discovered 25 years ago on low-copy-number plasmids, and it became rapidly clear that their role is to maintain the plasmid that carries them in growing bacterial populations. The molecular mechanism underlying plasmid maintenance is called postsegregational killing and relies on the differential stabilities of the two components (antitoxin and toxin). Upon plasmid loss, de novo synthesis of the plasmid-encoded TA components ceases and the unstable antitoxin protein is degraded by an ATP-dependent protease, which results in toxin release. The toxin is then able to interact with its target and interfere with essential cellular processes (e.g., DNA replication or protein synthesis). Ultimately, this leads to the selective killing of the plasmid-free progeny.
In contrast to that of plasmidic systems, the biological function of chromosomal TA systems is still an area of debate. There are at least five proteic TA systems that have been identified so far in the Escherichia coli K-12 (MG1655) chromosome: mazEF, relBE, yefM-yoeB, chpB, and dinJ-yafQ (1, 4, 20, 21, 35, 38, 46). The most prevalent hypothesis is that they are involved in general stress response, as suggested by extensive studies on mazEF and relBE (e.g., references 1 and 10). Interestingly, all five toxins are protein synthesis inhibitors. The molecular mechanism of action is different depending on the toxin. While MazF, ChpBK, and YoeB are endoribonucleases that cleave RNAs directly, RelE cleaves only mRNAs that are being translated (8, 9, 29, 39, 42, 53, 54). How YafQ inhibits translation is still unknown. Activation of mazEF and relBE, reflected by the activities of the MazF and RelE toxins, respectively, appears to have quite different effects on bacterial physiology. On one hand, mazEF was shown to be activated by a variety of short-term stresses (e.g., amino acid starvation, antibiotic treatment, DNA-damaging agents, or oxidative stress) and to provoke cell death (the programmed-cell-death [PCD] hypothesis) (1, 16, 26, 31, 44, 45). On the other hand, Gerdes and colleagues showed that activation of relBE and mazEF under amino acid starvation inhibits protein synthesis without affecting viability (10, 11, 41). It was therefore proposed that relBE, mazEF, and probably other chromosomal TA systems are growth modulators that induce a reversible dormancy state upon amino acid starvation that enhance fitness and competitiveness (18).
If the five chromosomal TA systems participate actively in the general stress response of E. coli, they should provide a selective advantage under stress conditions and/or during the poststress recovery phase. As this hypothesis was never tested directly, we analyzed the behavior of an E. coli mutant strain devoid of all five TA systems cited above (the Δ5 strain) during and after a variety of stresses as well as in competition experiments with the parental wild-type strain under nutrient- limited conditions. Our results rule out TA-dependent PCD under the stress conditions that we tested. They also bring into question whether TA systems significantly influence bacterial fitness during non-steady-state growth.
MATERIALS AND METHODS
Bacterial strains and strain constructions.
The E. coli strains used in this work were MG1655 (50) and its derivatives LVM100 (MG1655 Δ5 [ΔmazEF ΔrelBEF ΔchpB ΔyefM-yoeB ΔdinJ-yafQ]), LVM101 (MG1655 ΔmazEF), LVM102 (MG1655 Nalr), LVM103 (MG1655 Δ5 Nalr), and LVM104 (MG1655 Δ5 ΔyefM-yoeB::kan) (this work) as well as four derivatives of the MC4100 strain (7), MC4100 relA+ and MC4100 ΔmazEF relA+, obtained from H. Engelberg-Kulka (16), and LVM 200 (MC4100 relA+) and LVM201 (MC4100 relA+ ΔmazEF), constructed during this work (Table 1).
TABLE 1.
Bacterial strains
| Bacterial strain | Description | Source or reference |
|---|---|---|
| MG1655 | Wild-type E. coli K-12 | 50 |
| LVM100 (MG1655 Δ5) | MG1655 ΔmazEF ΔrelBEF ΔchpB ΔyefM-yoeB ΔdinJ-yafQ | This work |
| LVM101 | MG1655 ΔmazEF | This work |
| LVM102 | MG1655 Nalr | This work |
| LVM103 | MG1655 Δ5 Nalr | This work |
| LVM104 | MG1655 Δ5 ΔyefM-yoeB::Kan | This work |
| SC314678 | MG1655 relA ΔmazEF ΔrelBEF ΔchpB ΔyefM-yoeB ΔdinJ-yafQ | 9 |
| SC38 | MG1655 relA ΔmazEF | 11 |
| LVM200 | MC4100 relA+ | This work |
| LVM201 | LVM200 ΔmazEF | This work |
| MC4100 relA+ | MC4100 relA mazG::Kan | 16; this work |
| MC4100 relA+ ΔmazEF | MC4100 relA ΔmazEFG::Kan | 16; this work |
The LVM100 strain is a relA+ derivative of SC314678 (MG1655 Δ5 relA) in which the five TA systems are entirely deleted (promoter, antitoxin, and toxin genes) (9). SC314678 was constructed by P1 transduction with SC38 (MG1655 ΔmazEF) (11) as the donor strain. The SC38 strain was reported to behave like a relA strain for unknown reasons (11). We found that the RelA− phenotypes of both SC38 and SC314678 were due to the deletion of 18 nucleotides at the 3′ end of the relA gene including the relA stop codon. This deletion leads to a truncated form of the RelA protein in which the 5 carboxy-terminal amino acids (aa) are replaced by 9 extra aa encoded by the pKD3 (14) sequence replacing the mazEF operon in the ΔmazEF strain. To restore a wild-type relA gene, the missing 18 nucleotides were introduced by homologous recombination (14) in SC314678 and SC38 to obtain LVM100 (MG1655 Δ5) and LVM101 (MG1655 ΔmazEF), respectively. The primers used were chpA1relA+ (5′-ATG TTA TCG ACG CGC GTC GGT TGC ACG GGA GTT AGG TAG GCT GG-3′; the stop codon of relA is in bold) and chpA2 (5′-TAG TGA GCA AAC GGT CGA TTT GAT TCA TTG AAT TGT CCT G-3′). The relA+ recombinants were selected on SMG medium (minimal medium containing serine, methionine, and glycine), on which relA mutants are not able to grow (47). The wild-type sequence of relA was verified in both strains by sequencing.
The MC4100 relA+ (resistant to kanamycin) and MC4100 relA+ ΔmazEF strains received from H. Engelberg-Kulka and described as relA+ derivatives of MC4100 relA1 and MC4100 ΔmazEF::kan, respectively (16), were unable to grow on SMG medium, indicating a RelA− phenotype. As detailed descriptions of the constructions of these strains were not available in the literature, we further characterized their genotypes. Sequencing of the relA region revealed that the so-called MC4100 relA+ strain harbored a frameshift mutation at the 5′ end of the relA gene that rendered it relA (deletion of cytosine 623 modifies the amino acid sequence from codon 115 and generates a stop at codon 118). In the so-called MC4100 ΔmazEF::kan relA+ strain, we found an IS1 element inserted 10 bp downstream of the relA start codon. It should be noted that Aizenman and colleagues reported that the RelA+ phenotype was necessary for mazEF-dependent PCD, since release of the toxin MazF is ppGpp dependent (1). Indeed, ppGpp, the signal molecule produced during the stringent response by the RelA protein, was shown to inhibit transcription of the mazEF promoter (1). We also found that the so-called MC4100 relA+ strain harbors a kanamycin resistance cassette replacing mazG (the gene downstream of mazF) from the mazF stop codon to the promoter region of pyrG (the gene downstream of mazG). In the so-called MC4100 relA+ ΔmazEF::kan strain, the insertion of the kanamycin resistance cassette causes a partial deletion of the mazG gene (to nucleotide 557). It is important to note that the MazG protein was shown to be a regulator of ppGpp- and MazF-dependent PCD (23). We therefore constructed a relA+ derivative of the MC4100 (LVM200) strain by inserting the wild-type relA region of MG1655 spanning the relA1 mutation (which is an IS2 element inserted at nucleotide 534 in the relA gene [36]) by homologous recombination (14). The primers used were oli89 (5′-CAT ATC GGG TAC GTA TCG GCT TAC CAT TAC-3′) and relA5 (5′-CGA TAA AGT GTT GCT GGA C-3′). We selected recombinants that were able to grow on SMG medium. The relA+ genotype was verified by sequencing. We then deleted the mazEF operon by recombineering (14). A linear kanamycin resistance cassette was PCR amplified using the pKD3 plasmid as a template and primers delchpA3′ (5′-GGG TCT GTC AGG TGG AAA CCT GTG ACC AGA ATA GAA GTG AGT TAG TAA CAC ATA TGA ATA TCC TCC TTA G-3′) and delchpA5′2 (5′-ATG TTA TCG ACG CGC GTC GGT TGC ACG GGA GTT AGG TAG GCT GGA GCT GCT TC-3′) and recombineered into LVM200. After the insertion was confirmed, the resistance cassette was eliminated as described in reference 14, yielding strain LVM 201.
The LVM102 (MG1655 Nalr) and LVM103 (MG1655 Δ5Nalr) strains are spontaneous nalidixic acid-resistant mutants selected on LB plates containing 20 μg/ml of nalidixic acid. Nalr mutations were reported to not affect bacterial growth (52).
The LVM104 strain (MG1655 Δ5 ΔyefM-yoeB::kan) was constructed by introducing a kanamycin resistance cassette at the chromosomal location of the yefM-yoeB system in LVM100. This was done by homologous recombination as described in reference 14, using the oli106 (5′-GCT AGC GTA TCA AAA CTG ACA ATT CAT TCT ATG AAT GAA TAA AGC CAC GTT GTG TCT CAA-3′) and oli107 (5′-CCT CTT AGC TGA TGG CAC CAA AAA TAG CGA AGC CTG GAT TTT CGT CTC GCG CTG AGG TCT GCC TCG TGA AG-3′) primers.
Media and growth conditions.
Bacteria were grown in Luria-Bertani (LB) broth (purchased from Invitrogen). Ceria medium (phosphate-based minimal medium containing ammonium and magnesium sulfate as well as oligo-elements) supplemented with 0.4% glucose was used for the nutritional downshift experiments (9). Serine hydroxamate (SHT; a competitive inhibitor of the seryl-tRNA synthetase) and rifampin were purchased from Sigma. LB medium was acidified by addition of HCl (Merck) to the desired pH, measured with a Sartorius pHmeter.
Experiments testing the effects of rifampin, chloramphenicol, spectinomycin, and high temperatures on the viability of MC4100, MG1655, and their relA or relA+ ΔmazEF derivatives (data not shown in Results) were performed with LB medium, with M9 supplemented with a mixture of 20 μg/ml of each amino acid and 1% glucose (1), and with Ceria supplemented with 0.4% Casamino Acids and 0.4% glucose (9) by following the growth and treatment conditions described in reference 44. Antibiotics were tested at the following concentrations: 20 to 200 μg/ml chloramphenicol, 5 to 500 μg/ml rifampin, and 100 to 200 μg/ml spectinomycin.
Bacterial survival assays.
Overnight cultures of individual wild-type and Δ5 strains in LB medium were diluted 200-fold to an optical density at 600 nm of ∼0.01 in 50 ml of LB medium. Cultures were grown at 37°C with shaking at 180 rpm. At early log phase (∼107 bacteria/ml), half of each culture was treated with the different stress agents (e.g., addition of 2.5 mg/ml SHT or 100 μg/ml rifampin or resuspension in acidified LB). The viability of treated and untreated strains was measured by plating serial dilutions of each culture on LB plates. After each stress treatment, bacteria were washed twice by centrifugation and resuspended in fresh LB medium. Viability was then assayed until entry to stationary phase. In the nutritional-downshift experiment, the early-log-phase wild-type and Δ5 cultures were shifted to Ceria medium after two rounds of centrifugation and resuspension in Ceria medium. Viability was measured as described above. In long-term starvation experiments, individual wild-type and Δ5 cultures were tested as described above and the viability of each strain was measured daily for 7 days.
Competition experiments.
Mixed cultures of wild-type and Δ5 strains were performed by mixing equivalent cell numbers of overnight cultures of the wild-type and Δ5 Nalr strains to an optical density at 600 nm of ∼0.01 in 50 ml of LB medium. Competition experiments (SHT-induced amino acid starvation, nutritional downshift, and long-term starvation) were carried out as described for individual cultures. Proportions of each strain were deduced by plating serial dilutions of the mixed cultures on LB plates with or without nalidixic acid (20 μg/ml). For each experiment, the wild type and its Nalr derivative, the Δ5 and its Nalr derivative, and the wild-type Nalr and Δ5 mixed cultures were also tested in order to rule out any effects of the Nalr mutation on bacterial growth (data not shown).
RESULTS
Deletion of the five chromosomal TA systems does not affect several E. coli stress responses.
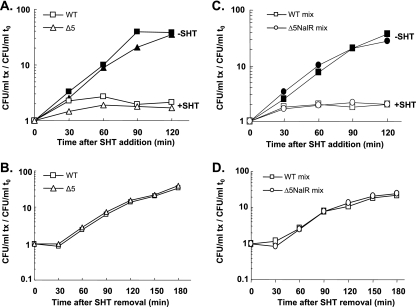
The effect of amino acid starvation induced by SHT on cell viability is quite controversial. On one hand, SHT was reported to rapidly activate mRNA cleavage by RelE and MazF without affecting viability (10, 11), while on the other hand, SHT was said to cause an important mazEF-dependent loss of viability (∼90%) (2, 16). In order to investigate whether the deletion of the five TA systems modifies the response to amino acid starvation, we compared the effects of SHT-induced starvation on the viability of MG1655 and its Δ5 derivative. Figure 1A shows that the growth of the wild-type and Δ5 strains slowed down rapidly after the addition of SHT and ceased completely after 60 min. The plating efficiencies of the two strains were comparable and remained similar for 120 min. Since no significant differences were observed during the SHT treatment, we then tested whether the five TA systems could confer an advantage during the post-SHT recovery phase. Figure 1B shows that 30 min after the SHT washout, growth resumed and viability increased similarly for the two strains for the next 150 min. This suggests that SHT does not cause cell death but rather induces a reversible growth inhibition that is independent of the five TA systems.
FIG. 1.
The five TA systems are not involved in the growth inhibition mediated by amino acid starvation and do not advantage poststress recovery. Individual cultures of the wild-type (WT) (square) and Δ5 (triangle) strains (A and B) and mixed cultures of the wild-type strain and the nalidixic acid-resistant derivative of the Δ5 strain (Δ5 Nalr) (circle) (C and D) were challenged with SHT (2.5 mg/ml) (see Materials and Methods). The viability of treated (open symbols) and untreated (filled symbols) cells was measured for 120 min upon SHT addition (A and C). After 120 min of starvation, SHT was removed and the viability of individual (B) and mixed (D) cultures was measured for 180 min (until early stationary phase). For each graph, the number of CFU/ml at each time point (tx) was normalized to that at time zero (t0). The graphs represent the averages from three (A and B) and two (C and D) independent experiments. Similar results were obtained in experiments in which the wild-type Nalr strain was mixed with the Δ5 strain (data not shown).
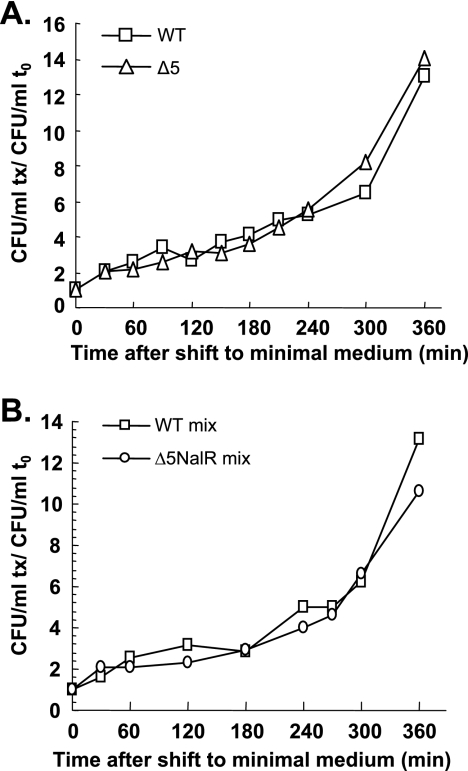
The effects of a nutritional downshift (a shift from nutrient-rich to nutrient-poor medium) on the viability of the MG1655 strain and its Δ5 derivative were also tested. In E. coli, polyphosphate is synthesized in response to nutritional downshift and modulates the specificity of the Lon ATP-dependent protease toward free ribosomal proteins (for a review, see reference 33). Since Lon is known to be responsible for antitoxin degradation (10, 48), we tested whether a nutritional downshift could affect MG1655 viability in a TA-dependent manner. Growth of the wild-type and Δ5 strains shifted from LB to minimal medium slowed down during the first 240 min and then increased in a similar manner (Fig. 2A), indicating that the five TA systems do not contribute to the adaptation of E. coli to a nutritional downshift.
FIG. 2.
The five TA systems are not involved in the adaptation to a nutritional downshift. Individual cultures of the wild-type (WT) (square) and Δ5 (triangle) strains (A) and mixed cultures of the wild-type and Δ5 Nalr (circle) strains (B) were challenged by a nutritional downshift (see Materials and Methods). The number of CFU/ml at each time point (tx) was normalized to that at time zero (t0) for each strain. The graphs represent the averages from three independent experiments. Similar results were obtained when a wild-type Nalr strain was mixed with the Δ5 strain (data not shown).
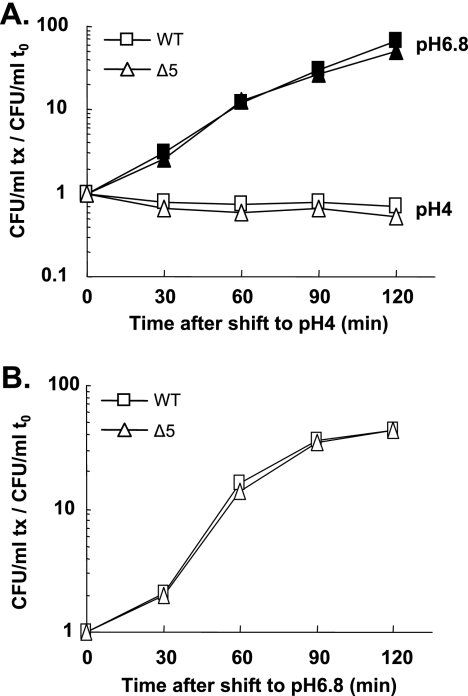
Acidic pH has been shown to down-regulate transcription of the relBE system in Streptococcus mutans (34). Moreover, the double deletion of relBE and mazEF increases resistance to acidic pH when S. mutans is grown in biofilm (34). To test whether TA systems are involved in acid tolerance in E. coli, we analyzed the effect of an acidic pH shift on the viability of MG1655 and its Δ5 derivative. As shown in Fig. 3A, when shifted from pH 6.8 to pH 4, the two strains stopped growing immediately. After 120 min at pH 4, the cultures were shifted back to pH 6.8 and within 30 min growth resumed equally (Fig. 3B) for both strains. Shifts from pH 6.8 to different acidic pHs (5 to 3) had similar effects on growth for both strains (data not shown). These results indicate that the five TA systems are not involved in acidic-shift tolerance.
FIG. 3.
The five TA systems are not involved in the growth inhibition mediated by an acidic shock and do not advantage poststress recovery. Cultures of the wild-type (WT) (square) and Δ5 (triangle) strains were shifted from pH 6.8 to pH 4 medium (see Materials and Methods). The viability of treated (open symbols) and untreated (filled symbols) cells was measured for 120 min (A). After 120 min, the wild-type and Δ5 cultures were shifted back to pH 6.8. Viability was measured for 120 min (until early stationary phase) (B). The number of CFU/ml at each time point (tx) was normalized to that at time zero (t0) for each strain. The graphs represent the averages from three independent experiments.
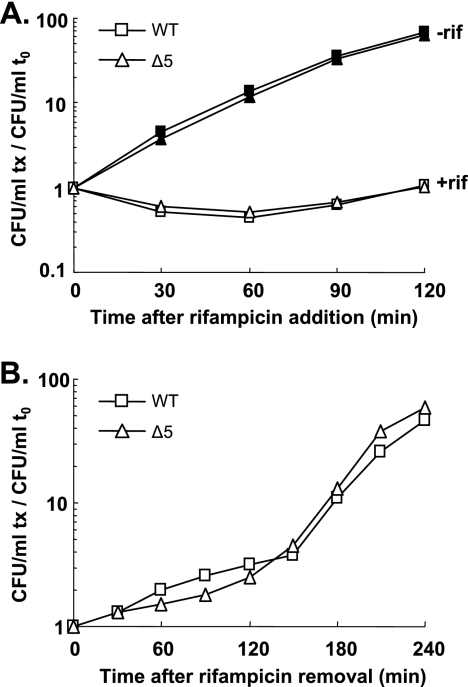
Sat et al. have shown that rifampin causes a drastic loss of viability of an MC4100 wild-type strain within 10 min of addition, in contrast to what occurs with a ΔmazEF MC4100 derivative (44). To test whether other TA systems are involved in the response to rifampin treatment, we compared the behavior of the MG1655 strain and its Δ5 derivative during a 120-min treatment. The viabilities of the wild-type and the Δ5 strains decreased similarly (twofold) during the first 30 min in the presence of rifampin and increased after 60 min to reach their pretreatment plating efficiencies at 120 min (Fig. 4A). After the rifampin was washed out, growth resumed rapidly and the viabilities of both strains increased similarly for the next 240 min (Fig. 4B). Thus, growth inhibition and the ability to recover from a rifampin treatment are independent of the five TA systems.
FIG. 4.
The five TA systems are not involved in the growth inhibition mediated by rifampin (rif) and do not advantage poststress recovery. Individual cultures of the wild-type (WT) (square) and Δ5 (triangle) strains were challenged with rifampin (100 μg/ml) (see Materials and Methods). The viability of treated (open symbols) and untreated (filled symbols) cells was measured for 120 min (A). After 120 min of treatment, rifampin was removed and the viability of both strains was measured for 240 min (until early stationary phase) (B). The graphs represent the averages for three independent experiments. The number of CFU/ml at each time point (tx) was normalized to that at time zero (t0) for each strain.
As we were not able to detect any cell death in the MG1655 wild-type strain with a 100 μg/ml rifampin treatment, we tested a variety of conditions (short-time treatments as described by Sat et al. [44] and higher rifampin concentrations [up to 500 μg] for long exposure times [up to 120 min] in both rich and minimal media [see Materials and Methods]). We also tested different genetic backgrounds: LVM 100 (MC4100 relA+), LVM101 (MC4100 relA+ ΔmazEF), and the so-called MC4100 relA+ and MC4100 relA+ ΔmazEF strains received from H. Engelberg-Kulka (which are phenotypically RelA−; for strain characterization, see Materials and Methods). None of these conditions induced cell death in any genetic background tested here (data not shown). Chloramphenicol and spectinomycin treatments as well as heat shock (48, 50, and 52°C) were also reported to induce mazEF-dependent PCD in a relA+-dependent manner (26, 44). However, in our hands, all of these treatments inhibited the growth of all the strains that we tested in a similar manner (data not shown). Our results show that rifampin, chloramphenicol, spectinomycin, and heat shock do not induce PCD (relA+ dependent or not) but cause growth inhibition independent of the five TA systems.
Deletion of the five chromosomal TA systems does not reduce E. coli competitiveness in a nutrient-limited environment.
Our results showed that the absence of the five TA systems did not prevent bacteria from coping with a variety of stresses. However, in situations in which bacteria are in competition with each other for limited nutrients during long periods of time, the presence of TA systems could be advantageous and enhance bacterial fitness and competitiveness. Moreover, as suggested by Gerdes and colleagues, TA system activity might enhance competitiveness by reducing translational errors (18). The effects of nutrient-limited conditions, such as amino acid starvation and nutritional downshift, were tested with mixed cultures containing equivalent proportions of the MG1655 strain and its nalidixic acid-resistant Δ5 derivative (Δ5 Nalr).
Figure 1C shows that the growth of both strains in mixed cultures was rapidly and similarly slowed down upon SHT addition and ceased completely after 60 min, indicating that neither strain overcame the other. As in individual cultures, the post-SHT recovery phases were similar for both strains in the mixed culture, i.e., growth inhibition resumed after 30 min and viability increased for the next 150 min (Fig. 1D). In the nutritional downshift experiments, following the shift to the nutrient-poor medium, the growth of the wild-type and Δ5 Nalr strains was slowed down for 270 min (Fig. 4B). After this lag, growth resumed similarly for both strains during the next 90 min. Therefore, when in competition under nutrient-limited conditions, the Δ5 Nalr strain is not at a disadvantage compared to the wild-type strain.
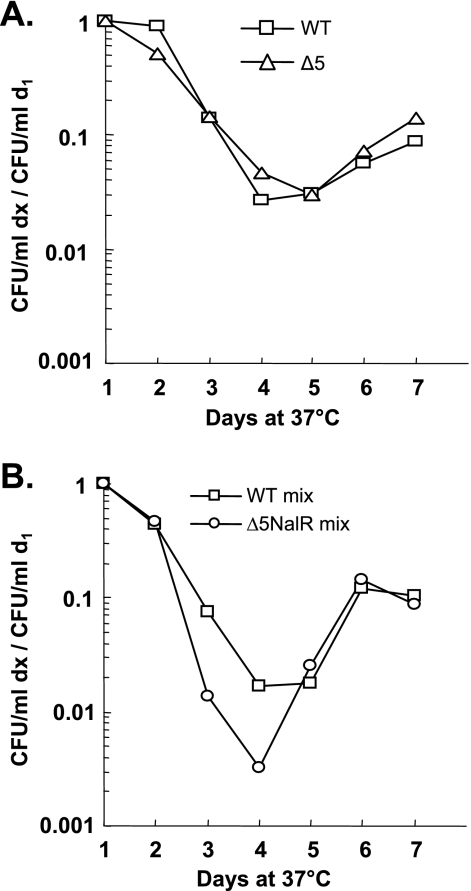
Finally, we tested the effects of long-term starvation (7 days) on the viability of individual and competing MG1655 and Δ5 Nalr strains. All the cultures tested (individual and mixed) presented the classical long-term-starvation growth curve (for a review, see reference 17), i.e., an initial decrease of viability between day 2 and days 4 and 5 (the death phase) followed by a growth phase that reached a plateau at day 6 (Fig. 5A and B). Figure 5A shows that the individual cultures of the wild-type and Δ5 strains displayed similar profiles of viability and growth, indicating that the five TA systems are not responsible for the death phase, in contrast to what was previously suggested (17). In mixed wild-type and Δ5 Nalr cultures, the Δ5 Nalr strain was at a slight disadvantage against the wild-type strain from day 2 to day 4 (∼1 log) (Fig. 5B). The same result was observed when a wild-type Nalr and Δ5 mixed culture was tested, therefore excluding an effect of the Nalr mutation. This result suggests that the absence of the five TA systems reduces competitiveness during the death phase. However, by day 5, both strains were present in equivalent proportions, indicating that the Δ5 strain rapidly overcame this disadvantage. Therefore, it is unlikely that the absence of the five TA systems affects competitiveness in the long term.
FIG. 5.
The five TA systems do not enhance competitiveness in long-term stationary-phase cultures. The viability of individual cultures of the wild-type (WT) (open square) and the Δ5 (open triangle) strains (A) and of mixed cultures of the wild-type and Δ5 Nalr (open circle) cultures (B) was measured daily for 7 days (see Materials and Methods). The graphs represent the averages from three independent experiments. The number of CFU/ml at each time point (dx) was normalized to that at day 1 (d1) for each strain. Similar results were obtained with the symmetrical mixed culture of the wild-type Nalr and Δ5 strains and with a mixed culture of wild-type and Δ5 yefM-yoeB::Kan strains (data not shown; see Materials and Methods).
DISCUSSION
In nature, bacteria are constantly faced with many different environmental stresses. As a result, they have evolved responses that allow them to deal with specific stresses (e.g., acid stress, heat shock, or DNA damages) by expressing proteins that eliminate the stress agents or repair the cellular damages. In general, specific responses allow the cell to survive under specific conditions but do not provide protection against other types of stresses. In contrast, general stress responses that are induced by multiple stresses are also present in bacteria, e.g., the σS regulon in E. coli (for reviews, see references 27 and 28). The σS-controlled genes encode proteins that often provide cross- and broad protection against additional stresses not yet encountered. Induction of specific and general stress responses usually leads to a temporary and reversible reduction or cessation of growth. Moreover, since availability of nutrients is limited in most environments, slow growth is probably the natural bacterial lifestyle. Chromosomally encoded TA systems were therefore good candidates for participating in a general stress response since toxin overexpression down-regulates macromolecular synthesis and thus affects growth rate (9-11, 38).
Numerous and various short-term stresses (e.g., SHT- induced amino acid starvation, antibiotic treatment, DNA-damaging agents, and heat shock) were reported to rapidly and dramatically decrease the viabilities of wild-type strains in a mazEF-dependent manner (1, 2, 16, 25, 26, 31, 44, 45). The mazEF system was thus proposed to be a PCD system. Although expression of the MazF toxin does not seem to induce cell lysis, it was proposed by Engelberg-Kulka and colleagues that the altruistic death of a portion of the bacterial population could supply nutrients and therefore enable the survivors to grow (15, 45). Another well-studied chromosomal TA system is relBE. This TA system was also shown to be involved in the amino acid starvation response induced by SHT (10). Although tested with three different genetic backgrounds (among which the MC4100 background was used by the Engelberg-Kulka group), amino acid starvation did not cause cell death but rather induced a reversible cessation of growth (11). relBE has been reported to be involved in this phenomenon since inhibition of protein synthesis caused by amino acid starvation was reduced in the relBE-deleted mutant compared to what was observed in the wild-type strain (10). This led Gerdes and his collaborators to propose that TA systems are stress response elements that modulate growth by reducing protein synthesis. A decrease of the rate of translation could spare energy and amino acids under stress conditions, which could in turn reduce translational errors and therefore enhance fitness and competitiveness (18).
To gain insights on possible synergetic effects of the five TA systems, we analyzed the behavior of an E. coli mutant strain deleted for all five known TA systems (the Δ5 strain) under a variety of stress conditions (amino acid starvation, rifampin treatment, acidic stress, and nutritional downshift) and during the poststress recovery phase. Under all the conditions tested, the wild-type and mutant strains showed similar reversible growth inhibitions, indicating that neither the growth inhibition nor the poststress recovery phase was dependent on any of the five TA systems. Our observations are in agreement with data from several groups that have tested single-TA-deletion mutants and could not observe TA-dependent PCD under various stress conditions and in different bacterial species, e.g., amino acid starvation in E. coli (11), antibiotic treatments (chloramphenicol and kanamycin) in Vibrio cholerae (5), and long-term starvation, antibiotic treatments (spectinomycin and rifampin), and acidic treatment in S. mutans (34). Conflicting results about the involvement of the mazEF system in thymineless death (TLD) were also recently reported. mazEF was first reported by Sat et al. as being the sole element responsible for TLD (45). However, recent data showed that relBE is also involved in this pathway but that the contribution of both systems to death is far lower than what was previously reported (19). Moreover, as discussed by Morganroth and Hanawalt, relBE and mazEF cannot account solely for TLD and other types of molecular controls are certainly involved (37).
We also performed competition experiments in which the wild-type and Δ5 strains were mixed in equal proportions. Under nutrient-limited conditions (amino acid starvation, nutritional downshift, and long-term cultures), the Δ5 strain was not disadvantaged compared to the wild-type strain, except during the death phase for long-term-starvation cultures, though in a transient way. These results indicate that the absence of the five TA systems does not reduce competitiveness.
Altogether, our results show that the five TA systems are dispensable for the physiological responses to a variety of stresses applied under different culture conditions, including stresses that were never tested before for TA system activation. Nevertheless, we cannot rule out that other TA systems in the E. coli chromosome might contribute to such stress responses. A certain level of redundancy might exist, as an increasing number of chromosomally encoded TA systems have been reported, notably TA systems in which the antitoxin is an antisense RNA (for a review, see reference 18).
At the molecular level, activation of TA systems has been defined by the detection of the toxin's enzymatic activity (i.e., mRNA cleavage) and by an increase of TA operon transcription (10). While the mazEF and relBE systems have been shown to be activated by SHT and chloramphenicol treatments (10, 11), we showed that their deletion did not give any particular phenotype under these conditions. Therefore, it seems that the correlation between the molecular activation of a TA system under a particular stress condition and its involvement in the stress response has to be taken with caution. Similarly, transcription of dinJ-yafQ, yefM-yoeB, relBE, and mazEF is increased in persister cells while single deletions of each system do not affect persister formation frequency (30). On the contrary, the hipBA module (which accounts most likely for a sixth TA system in the E. coli chromosome) is involved in persister cell formation since its deletion significantly reduces the formation frequencies of these cells (30). HipA is a kinase that inhibits macromolecular synthesis (32). Transcription and/or translation inhibition mediated by HipA might cause subsequent activation of the four TA systems cited above. Thus, we propose that TA molecular activation under certain circumstances might be a side effect of the transcriptional/translational arrest that is mediated by other specific pathways.
However, it is possible that conditions under which specific factors directly modulate the regulation (at the transcriptional and/or posttranscriptional level) of TA systems could efficiently trigger them and lead to a detectable modification of bacterial physiology. For instance, the existence of a cofactor modulating the specificity of the Lon ATP-dependent protease was proposed by Gerdes and colleagues and might provide a molecular explanation for the specific activation of yefM-yoeB under Lon overproduction conditions (9, 10). The identification of such factors will certainly clarify how chromosomally encoded TA systems function.
Another hypothesis that has been neglected in the field is that TA systems might have different functions depending on their genomic locations. Recent studies indicate that TA systems located in the superintegron of Vibrionaceae promote genomic stability either by stabilizing the superintegron structure (12, 46) or by preserving the integrity of an entire chromosome (chromosome II of V. cholerae) (51). In E. coli, relBE is actually located in the cryptic Qin prophage while the other four systems seem to be part of the genomic core (43). Interestingly, when cloned in an unstable plasmid, relBE is able to stabilize it (22). In contrast, other systems that are settled in the chromosome, like the ccdO157 gene of E. coli O157:H7, are unable to stabilize a plasmid (49). Moreover, indirect experiments indicate that dinJ-yafQ is unable to promote genomic stabilization (46). Thus, the ability to stabilize genomes might be characteristic of TA systems resident on or recently originated from mobile genetic elements. However, the stabilization potential of some of these systems might fade and eventually be lost once the systems are integrated in the genomic core. Such TA systems could have adapted to their new chromosomal environments by evolving toward a new function that could provide benefit to their host under specific conditions. In the case of the five TA systems considered in this work, these conditions seem not to be linked to general stress responses and remain to be discovered. One might also consider that during adaptation, the activity of the chromosomally encoded toxin becomes neutralized by appropriate regulation and/or repair functions in such a way that fortuitous activation of TA systems (e.g., by transcription and/or translation inhibition) has no effect on bacterial physiology. In such a view, TA systems might not be beneficial to bacteria on a short-time life scale but might rather be elements of long-term evolution.
Acknowledgments
We thank all the members of the laboratory for constant and stringent discussions and Régis Hallez, Myriam Wilbaux, Pierre Smeesters, and especially Nadim Majdalani for critical reading of the manuscript. We also thank the members of the scientific community who encouraged us to pursue and to publish this work. We thank Kenn Gerdes and Hanna Engelberg-Kulka for the generous gift of strains.
This work was supported by the Fonds National de la Recherche Scientifique et Médicale (3.4530.04), by the EU (CRAB LSHM-CT-2005-019023), and by the Fonds Van Buuren. V.T. is supported by the FRIA. L.V.M. is chercheur qualifié du FNRS.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anantharaman, V., and L. Aravind. 2003. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 4:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bech, F. W., S. T. Jorgensen, B. Diderichsen, and O. H. Karlstrom. 1985. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 4:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budde, P. P., B. M. Davis, J. Yuan, and M. K. Waldor. 2007. Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae. J. Bacteriol. 189:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672-679. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705-1717. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 12.Christensen-Dalsgaard, M., and K. Gerdes. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397-411. [DOI] [PubMed] [Google Scholar]
- 13.Condon, C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61:573-583. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 16.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 95:15481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkel, S. E. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113-120. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 19.Godoy, V. G., D. F. Jarosz, F. L. Walker, L. A. Simmons, and G. C. Walker. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 25:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 21.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 22.Gronlund, H., and K. Gerdes. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285:1401-1415. [DOI] [PubMed] [Google Scholar]
- 23.Gross, M., I. Marianovsky, and G. Glaser. 2006. MazG—a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59:590-601. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 25.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227-234. [DOI] [PubMed] [Google Scholar]
- 26.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 28.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamada, K., and F. Hanaoka. 2005. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19:497-509. [DOI] [PubMed] [Google Scholar]
- 30.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda, A. 2006. A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci. Biotechnol. Biochem. 70:325-331. [DOI] [PubMed] [Google Scholar]
- 34.Lemos, J. A., T. A. Brown, Jr., J. Abranches, and R. A. Burne. 2005. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol. Lett. 253:251-257. [DOI] [PubMed] [Google Scholar]
- 35.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 263:15699-15704. [PubMed] [Google Scholar]
- 37.Morganroth, P. A., and P. C. Hanawalt. 2006. Role of DNA replication and repair in thymineless death in Escherichia coli. J. Bacteriol. 188:5286-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Motiejunaite, R., J. Armalyte, A. Markuckas, and E. Suziedeliene. 2007. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 268:112-119. [DOI] [PubMed] [Google Scholar]
- 39.Munoz-Gomez, A. J., S. Santos-Sierra, A. Berzal-Herranz, M. Lemonnier, and R. Diaz-Orejas. 2004. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567:316-320. [DOI] [PubMed] [Google Scholar]
- 40.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 43.Perna, N. T., J. D. Glasner, V. Burland, and G. Plunkett III. 2002. The genomes of Escherichia coli K-12 and pathogenic E. coli, p. 3-53-117-26. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, San Diego, CA.
- 44.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szekeres, S., M. Dauti, C. Wilde, D. Mazel, and D. A. Rowe-Magnus. 2007. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63:1588-1605. [DOI] [PubMed] [Google Scholar]
- 47.Uzan, M., and A. Danchin. 1976. A rapid test for the rel A mutation in E. coli. Biochem. Biophys. Res. Commun. 69:751-758. [DOI] [PubMed] [Google Scholar]
- 48.Van Melderen, L., P. Bernard, and M. Couturier. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11:1151-1157. [DOI] [PubMed] [Google Scholar]
- 49.Wilbaux, M., N. Mine, A. M. Guerout, D. Mazel, and L. Van Melderen. 2007. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J. Bacteriol. 189:2712-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 51.Yamaichi, Y., M. A. Fogel, and M. K. Waldor. 2007. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 104:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zambrano, M. M., and R. Kolter. 1993. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J. Bacteriol. 175:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280:3143-3150. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]