Abstract
Repetitive DNA sequences have been demonstrated to play an important role for centromere function of eukaryotic chromosomes, including those from fission yeast, Drosophila melanogaster, and humans. Here we report on the isolation of a repetitive DNA element located in the centromeric regions of cereal chromosomes. A 745-bp repetitive DNA clone, pSau3A9, was isolated from sorghum (Sorghum bicolor). This DNA element is located in the centromeric regions of all sorghum chromosomes, as demonstrated by fluorescence in situ hybridization. Repetitive DNA sequences homologous to pSau3A9 also are present in the centromeric regions of chromosomes from other cereal species, including rice, maize, wheat, barley, rye, and oats. Probe pSau3A9 also hybridized to the centromeric region of B chromosomes from rye and maize. The repetitive nature and its conservation in distantly related plant species indicate that the pSau3A9 family may be associated with centromere function of cereal chromosomes. The absence of DNA sequences homologous to pSau3A9 in dicot species suggests a faster divergence of centromere-related sequences compared with the telomere-related sequences in plants.
Among the most distinguishing and characteristic landmarks of chromosomes of higher eukaryotes is the location of the centromere. The centromere plays an essential role in the proper segregation of chromosomes during mitosis and meiosis, thus ensuring equal distribution of genetic information to the next generation. The centromeric region of higher eukaryotic chromosomes is structurally specified by the primary constriction at which the sister chromatids associate and a pair of kinetochores to which microtubules of the mitotic and meiotic spindle attach.
The centromeres from budding yeast (Saccharomyces cerevisiae), known as point centromeres, have been well characterized. The genetic information specifying full centromere function in these species is contained within a 125-bp DNA segment (1). Such centromeres bind to a single microtubule and can move chromosomes of 0.26–3 megabases in size. Extensive studies also have been carried on centromeres from fission yeast (Schizosaccharomyces pombe), Drosophila melanogaster, and mammalian species. The centromeres from these species are much more complex compared with those from budding yeast. These centromeres, called regional centromeres, encompass kilobases or megabases of DNA and include both unique and repetitive DNA sequences. Several different repetitive DNA elements were identified in the centromeres of fission yeast. It is well established that these repetitive elements are essential for full centromere function (1). Full function of centromeres in Drosophila also requires the presence of satellite DNA (2). The α-satellite DNA, the major DNA component in the centromeric region of human chromosomes, has long been regarded as junk DNA. However, the current evidence indicates that the α-satellite DNA plays an important role in centromere function (3, 4, 5).
Thus far, no plant DNA sequences essential for centromere function have been identified. Alfenito and Birchler (6) reported on the cloning and characterization of a repetitive DNA element located in the centromeric region of maize B chromosomes. Sequence analysis of this repeat shows homology to motifs of the maize knob sequence. The maize knob has a neocentromere function in certain genetic backgrounds. However, this B chromosome-specific repeat is not located in the centromeres of maize A chromosomes and is, therefore, unlikely to be related to centromere function of the A chromosomes. Here we report on the cloning and characterization of a repetitive DNA element conserved in the centromeric region of cereal chromosomes.
MATERIALS AND METHODS
Materials.
Bacterial artificial chromosome (BAC) clone 52A4 was randomly selected from a sorghum BAC library for chimerism analysis (7). This BAC was found to be located in the centromeric region of all sorghum chromosomes. The following lines or varieties of the cereal species were used in both fluorescence in situ hybridization (FISH) and DNA analysis: 607E (sorghum), B73 (maize), DV85 (rice), Chinese Spring (wheat), Chilton (barley), Hancock (rye), and Ogle (oats). Rye and maize lines with B chromosomes were provided by B. Friebe at Kansas State University and S. M. Kaeppler at the University of Wisconsin–Madison, respectively.
Plasmid Cloning and Sequencing.
DNA fragments to be subcloned were isolated by cutting out the agarose containing the fragments and purified using the Geneclean II kit (Bio 101). The fragments then were ligated to a linear pUC18 plasmid. The ligation mixture was used to transform Escherichia coli strain DH5α. White recombinant clones on 5-bromo-4-chloro-3-indolyl β-d-galactoside/isopropyl β-d-thiogalactoside plates were picked and analyzed for the presence of insert DNA using plasmid minipreparation and restriction digestion. Cycle sequencing reactions were performed with an Applied Biosystems AmpliTaq DNA polymerase, an FS Dye Terminator Ready Reactions kit, and a Perkin–Elmer Thermocycler (model 9600). The reaction products were purified with MicroSpin G-50 Columns (Pharmacia Biotech) and analyzed on an Applied Biosystems Automated DNA Sequencer (model 373).
FISH.
Detailed protocols for chromosome preparation, probe labeling, in situ hybridization, and signal detection were described previously (8). Slides were examined using an Olympus BX60 fluorescence microscope. Gray scale images were obtained using a SenSys CCD camera (Photometrics) and then pseudo-colored and merged using IPLab spectrum software.
Southern Blotting.
Genomic DNA was isolated from young leaf tissue using a hexadecyltrimethylamonium bromide method (9). DNA samples were digested with restriction enzymes and blotted to Hybond N+ (Amersham) membrane. Prehybridization and hybridization were done at 65°C in 5× SSC, 0.1% N-lauroylsarcosine, 0.02% SDS, 0.02% denatured salmon sperm DNA, and 1% Blocking Reagent (Boehringer Mannheim). After hybridization, the membranes were washed at different stringencies by controlling the concentrations of SSC, and then the membranes were exposed to x-ray films.
RESULTS
During an attempt to cytologically map random BAC clones from sorghum, we found that BAC 52A4 hybridized to the centromeric region of all 20 sorghum chromosomes. Under the same hybridization stringency (50% formamide in 2× SSC at 37°C), this clone hybridized strongly to the centromeres of all maize chromosomes. At a lower stringency (30% formamide in 2× SSC at 37°C), 52A4 also hybridized with different signal intensities to the centromeres of chromosomes from different cereal species, including rice, wheat, barley, rye, and oats. However, it did not hybridize to any specific chromosomal regions of the several dicot species analyzed, including Vicia faba, tomato, tobacco, soybean, and Arabidopsis thaliana.
The BAC clone 52A4 was digested with various restriction enzymes. A blot with digested DNA was hybridized to genomic DNA from wheat, maize, and rice. Several fragments were hybridized to DNA from all three species. A Sau3AI fragment (Fig. 1) was subcloned in plasmid pUC18. When this subclone, pSau3A9, was analyzed by in situ hybridization to chromosomes of various cereal species, the result was identical as when 52A4 was used as the probe. The subclone hybridized with different signal intensities to centromeric regions of chromosomes from all of the cereal species analyzed (Fig. 2). In several species, some chromosomes showed stronger signals than others. For example, about one-half of the chromosomes in sorghum line “607E” had relatively large and strong FISH sites at the centromere. In “Chilton” barley, at least one pair of chromosomes consistently showed weaker signals than the other chromosomes. In the oats line “Ogle,” about 14 chromosomes had very weak signals in the centromeric region. However, wheat and rye chromosomes had a relatively uniform signal intensity. At present, it is not known whether such differences in the intensity of FISH signals is species-specific or genotype-specific. FISH signals were detected in the centromeres of rye B chromosomes with pSau3A9 as a probe. The signal intensities in rye B chromosomes are similar to those on A chromosomes (Figs. 2, 3, 4, 5). Similarly, pSau3A9 also hybridized to the centromeric regions of maize B chromosomes (data not shown). No specific FISH signals were detected when pSau3A9 was hybridized to chromosomes from the several dicot species analyzed (data not shown). This result was also confirmed by Southern blot hybridization analysis by probing the pSau3A9 subclone to a restriction enzyme digested DNA from cereal and several dicot species (Fig. 3).
Figure 1.
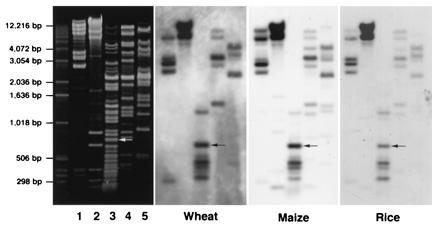
DNA of sorghum BAC clone 52A4 was digested with restriction enzymes DraI (lane 1), SalI (lane 2), Sau3AI (lane 3), AccI (lane 4), and HindIII (lane 5). The same blot was probed with genomic DNA from wheat, maize, and rice, respectively. A 745 Sau3AI fragment (arrow) hybridized to the genomic DNA from all the three species. This fragment was subcloned as plasmid pSau3A9.
Figure 2.
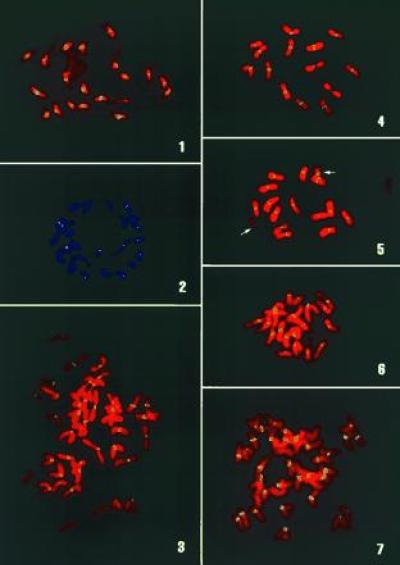
FISH analysis using pSau3A9 as a probe: 1, sorghum; 2, rice; 3, wheat; 4, barley; 5, rye, 14 A and 2 B (arrows) chromosomes have similar FISH signals at centromeres; 6, maize; 7, oat.
Figure 3.
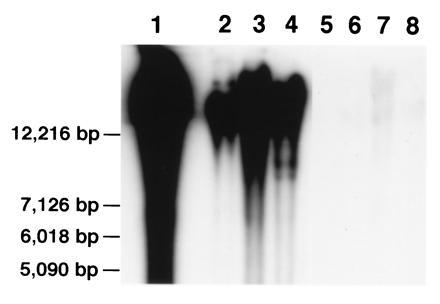
Genomic DNA from sorghum (lane 1), maize (lane 2), wheat (lane 3), rice (lane 4), A. thaliana (lane 5), tobacco (lane 6), tomato (lane 7), and soybean (lane 8) was digested with SalI and probed with pSau3A9. Strong hybridization signals were detected in the four cereal species, and very weak or no signals were detected in the four dicot species.
Figure 4.

Nucleotide sequence of pSau3A9.
Clone pSau3A9 was sequenced (Fig. 4). No significant homologies were found with any sequences in the EMBL/GenBank data base. The clone contains 745-bp sequences with 44% GC and 56% AT. Sequence analysis did not reveal any special characteristics that often are associated with repetitive DNA families, such as internal short repeats, inverted repeats, and palindromes.
DISCUSSION
Constitutive heterochromatin is often located at the centromeric regions of plant chromosomes as demonstrated by pachytene chromosome analysis and C-banding analysis in many plant species. Repetitive DNA sequences are the major components of the centromeric heterochromatin. Repetitive DNA elements, located mainly or exclusively in the centromeric region, have been cloned from various plant species (10, 11, 12, 13, 14). However, all of the reported elements are species- or genome-specific. Some of them are not present on all of the chromosomes in the same species or in species with related genomes. Clone pSau3A9 is the first centromeric repetitive DNA element conserved in distantly related plant species. Rice, maize, and the Triticeae species, including wheat, barley and rye, diverged from a common ancestor about 60 to 100 million years ago (15, 16). No repetitive DNA elements, except the telomeric DNA sequences, have been reported to be conserved among all of these species. The high conservation of the Sau3A9 sequence among cereal species suggests that this element may play a role in centromeric function.
Thus far, the telomeric DNA sequence, first isolated from A. thaliana (17), is the only reported repetitive DNA element conserved among both monocot and dicot species. The lack of conservation of the Sau3A9 sequence in dicot species is not surprising. The α-satellite DNA family is present on centromeric regions of chromosomes of all primates (18, 19) but absent in other mammalian species. Budding and fission yeasts have completely different centromeric DNA sequences. Therefore, it is well documented that the DNA sequences related to centromere function are not conserved to the same degree as the telomeric DNA sequences. Even the α-satellite itself has been modified significantly during primate evolution. The α-satellite from New and Old World primates share only 64% sequence identity (19). The present work indicates that the Sau3A9 family also has been modified during the divergence of cereal species. Based on the signal intensities of FISH using pSau3A9 as a probe under the same stringency, the homology between Sau3A9 and its related sequences in maize is higher than those between Sau3A9 and its related sequences in other cereal species. The order of such homology is sorghum/maize > sorghum/wheat > sorghum/oats > sorghum/rice.
The α-satellite in primates and the minor satellite in mouse (18, 20) are the only repetitive DNA elements that were demonstrated to be related to centromeric function. However, the centromeric region of fission yeast chromosomes contains several classes of repetitive DNA sequences (21, 22, 23, 24, 25). At least four different repetitive DNA elements seem to be responsible for full centromere function (1). At present, it is not known whether repetitive DNA elements different from Sau3A9 are present in the cereal genomes. This question could be answered by molecular analysis of BAC clone 52A4 and other large insert genomic DNA clones isolated using pSau3A9.
Clone pSau3A9 can be possibly applied in a number of related research projects. Sequences homologous to Sau3A9 can be isolated from other cereal species. Comparison of such sequences may provide useful information about the evolution of cereal species. The immediate application of the Sau3A9 sequence is to locate the centromeres on the genetic maps of the cereal species either by directly mapping this sequence or by isolating and mapping the adjacent unique sequences of Sau3A9. Probe pSau3A9 can be used to analyze the structure and molecular organization of plant centromeres. Cloning and characterization of centromeres of plant chromosomes is a prerequisite for constructing plant artificial chromosomes. Such artificial chromosomes may be the ultimate vector for functional analysis of large complex plant genes. Plant artificial chromosomes will also provide a vehicle to move complex genes among different plant species in the future.
Acknowledgments
This research is supported in part by start-up funds from the Graduate School (101-0503) and the College of Agriculture and Life Sciences (150–2689), University of Wisconsin-Madison (to J.J.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
References
- 1.Clarke L. Trends Genet. 1990;6:150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- 2.Murphy T D, Karpen G H. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haaf T, Warburton P E, Willard H F. Cell. 1992;70:681–696. doi: 10.1016/0092-8674(92)90436-g. [DOI] [PubMed] [Google Scholar]
- 4.Tyler-Smith C, Oakey R, Larin Z, Fisher R B, Crocker M, Affara N A, Ferguson-Smith M A, Muenke M, Zuffardi O, Jobling M A. Nat Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- 5.Larin Z, Fricker M D, Tyler-Smith C. Hum Mol Genet. 1994;3:689–695. doi: 10.1093/hmg/3.5.689. [DOI] [PubMed] [Google Scholar]
- 6.Alfenito M R, Birchler J A. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo S-S, Jiang J, Gill B S, Paterson A H, Wing R A. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang, J., Hulbert, S. H., Gill, B. S. & Ward, D. C. (1996) Mol. Gen. Genet. in press. [DOI] [PubMed]
- 9.Gill K S, Lubbers E L, Gill B S, Raupp W J, Cox T S. Genome. 1991;34:362–374. [Google Scholar]
- 10.Martinez-Zapater J M, Estalle M A, Somerville C R. Mol Gen Genet. 1986;204:417–423. [Google Scholar]
- 11.Ganal M W, Lapitan N L V, Tanksley S D. Mol Gen Genet. 1988;213:262–268. [Google Scholar]
- 12.Iwabuchi M, Itoh K, Shimamoto K. Theor Appl Genet. 1991;81:349–355. doi: 10.1007/BF00228675. [DOI] [PubMed] [Google Scholar]
- 13.Calasso I, Schmidt T, Pignone D, Heslop-Harrison J S. Theor Appl Genet. 1995;91:928–935. doi: 10.1007/BF00223902. [DOI] [PubMed] [Google Scholar]
- 14.Kamm A, Galasso I, Schmidt T, Heslop-Harrison J S. Plant Mol Biol. 1995;27:853–862. doi: 10.1007/BF00037014. [DOI] [PubMed] [Google Scholar]
- 15.Martin W, Gierl A, Saedler H. Nature (London) 1989;339:46–48. [Google Scholar]
- 16.Wolfe K H, Gouy M, Yang Y-W, Sharp P M, Li W-H. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards E J, Ausubel F M. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 18.Willard H F. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- 19.Alves G, Seuánez H N, Fanning T. Chromosoma. 1994;103:262–267. doi: 10.1007/BF00352250. [DOI] [PubMed] [Google Scholar]
- 20.Wong A K C, Rattner J B. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke L, Amstutz H, Fishel B, Carbon J. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakaseko Y, Kinoshita N, Yanagida M. Nucleic Acids Res. 1987;15:4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]


