Abstract
We report here functional and topological analyses of TraG and Eex, the donor and recipient cell inner membrane proteins that mediate entry exclusion in the SXT/R391 family of integrative conjugative elements. We found that the exclusion-determining regions of the Eex variants EexS (SXT) and EexR (R391) are located in distinct yet overlapping regions of the proteins. Unexpectedly, the carboxyl-terminal regions of TraG and Eex, which contain the residues essential for exclusion activity and specificity, were found to localize in the cell cytoplasm. These observations suggest that complex topological rearrangements of conjugative proteins must occur during mating to enable these domains to interact.
Conjugative DNA transfer among bacteria plays a prominent role in horizontal gene flow and bacterial evolution (7). Conjugative elements typically encode proteins that mediate three processes: (i) mating pair and bridge formation, (ii) DNA processing and delivery to the mating pore, and (iii) exclusion functions that inhibit redundant conjugative DNA transfer. Two types of exclusion, surface and entry, have been described. Surface exclusion, which has been found only in IncF plasmids, is mediated by an outer membrane protein (TraT) that is thought to inhibit mating pair formation (1). In contrast, a wide variety of plasmids produce entry exclusion proteins, which are thought to inhibit DNA transfer after mating pairs have formed (11, 20). Entry exclusion genes found in different plasmids ordinarily lack similarity, yet they all encode proteins that localize to the inner membrane (8-10, 26, 27). The mechanism by which this diverse group of proteins inhibits conjugative DNA transfer is not well understood.
We are investigating entry exclusion in SXT and R391, two closely related integrative conjugative elements (ICEs) derived from Vibrio cholerae and Providencia rettgeri, respectively (4, 6). These ICEs have nearly identical genes that mediate integration into and excision from the host chromosome (3). The conjugation genes of SXT and R391 are also almost identical; they are distantly related to those found in the F plasmid (3). However, despite their nearly identical conjugative machinery, we found that SXT and R391 do not exclude each other (17). As with conjugative plasmids, entry exclusion of these ICEs is mediated by small inner membrane proteins expressed in recipient cells. These proteins, EexS (SXT) and EexR (R391), are 87% identical within their first 86 amino acids (aa), while the remaining 56 carboxyl-terminal residues are only 41% identical (18). We generated chimeric Eex proteins and demonstrated that the specificity of the Eex variants is dictated by residues found in their respective carboxyl termini (17). Furthermore, recent DNA sequence analyses of many SXT/R391 family ICEs revealed that there are only two exclusion groups in this large family of ICEs. These elements harbor Eex variants that are almost identical to EexS or EexR (18).
Additional genetic analyses identified TraG, another inner membrane protein, as the Eex target in donor cells (17). TraG is essential for ICE transfer, and it is thought to be a component of the donor cell mating pair formation apparatus (4, 16). TraGS and TraGR, derived from SXT and R391, respectively, are 98% identical, and a stretch of only 3 aa (606-607-608) determines the exclusion specificity of the TraG variants (17). For example, R391 encoding a chimeric TraG with aa 606 to 608 switched to the residues found in TraGS was excluded from a recipient producing EexS and not from a recipient producing EexR. Our findings that ICE entry exclusion is mediated by particular amino acid sequences in both TraG and Eex suggests that direct interactions between element-specific forms of TraG in donor cells and Eex in recipient cells mediate exclusion.
It is difficult to imagine how interactions between inner membrane proteins in donor and recipient cells can interact to mediate exclusion. To begin to address how Eex and TraG mediate exclusion, we mapped the Eex residues important for exclusion specificity and determined the subcellular localization of the TraG and Eex residues that are critical for exclusion specificity.
Distinct but overlapping regions of EexS and EexR determine exclusion specificity.
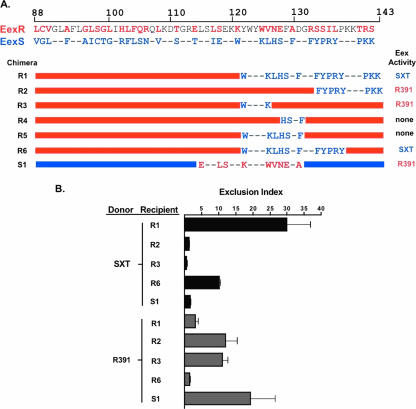
Strains and plasmids used in this study are listed in Table 1. We created chimeric Eex proteins to further define which Eex carboxyl residues determine SXT and R391 exclusion specificity. These chimeras were constructed by swapping portions of eexR and eexS as indicated in Fig. 1A. For example, chimera R1 is composed of the first 120 aa residues from EexR, with the remaining residues (aa 121 to 143) derived from EexS. This chimera displayed SXT exclusion specificity; a recipient expressing chimera R1 excluded SXT transfer (exclusion index for SXT [EIS], 30) but not R391 transfer (exclusion index for R391 [EIR], 1) (Fig. 1B). Additional chimeras were constructed to explore whether all of the EexS residues in chimera R1 were necessary for SXT exclusion specificity. EexS residues 133 to 143, in chimera R2, were not sufficient to dictate SXT exclusion specificity, since a recipient expressing this chimera excluded R391 and not SXT (EIR, 15; EIS, 0.50) (Fig. 1). Similarly, a chimeric Eex protein with aa 121 to 132 derived from SXT (chimera R5) also did not exclude SXT; however, unlike chimera R2, this protein also lacked exclusion activity for R391 (Fig. 1) (EIS, 0.50; EIR, 0.95), despite being expressed at approximately the same level as the wild-type exclusion proteins and the other chimeras (data not shown). These observations suggest that at least some of aa 121 to 132, but not aa 133 to 143, are required for EexR activity and that additional residues besides aa 121 to 132 are necessary for EexS exclusion activity. In agreement with the latter idea, we found that chimera R6, a protein that contained EexR sequences except for EexS residues 121 to 137, selectively excluded SXT transfer (EIS,10.2; EIR, 1.53) (Fig. 1). Thus, EexS residues 121 to 137 appear sufficient to mediate EexS exclusion activity when these amino acids are expressed within an EexR-based chimeric protein.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype/description | Reference |
|---|---|---|
| E. coli K-12 strains | ||
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 | 21 |
| Bi533 | MG1655 Nalr | 13 |
| Jo214 | Bi533 SXT ΔfloR::kan | 17 |
| Jo100 | Bi533 R391 | 12 |
| Jm655 | MG1655 R391 traGR (Thr604-Cys) | This study |
| Jm676 | BW25113 SXT EexS59::GFP | This study |
| Jm559 | BW25113 SXT EexS36::PhoA | This study |
| Plasmids | ||
| pR1 | pBAD-Topo derivative expressing Eex chimera R1 | This study |
| pR2 | pBAD-Topo derivative expressing Eex chimera R2 | This study |
| pR3 | pBAD-Topo derivative expressing Eex chimera R3 | This study |
| pR4 | pBAD-Topo derivative expressing Eex chimera R4 | This study |
| pR5 | pBAD-Topo derivative expressing Eex chimera R5 | This study |
| pR6 | pBAD-Topo derivative expressing Eex chimera R6 | This study |
| pS1 | pBAD-Topo derivative expressing Eex chimera S1 | This study |
| pEexS59-GFP | pBAD-Myc/His derivative encoding EexS with GFP fused at residue 59 | This study |
| pEexS143-GFP | pBAD-Myc/His derivative encoding EexS with GFP fused at residue 143 | This study |
| pEexS143-Cys | pBAD-Topo derivative encoding EexS containing a cysteine residue inserted at the carboxyl terminus | This study |
| pTraGR1109-GFP | pBAD-Myc/His derivative encoding TraGR with GFP fused at residue 604 | This study |
| pTraGR604-Cys | pBAD-Myc/His derivative encoding TraGR containing the Thr604-Cys substitution | This study |
Bacterial strains were grown and maintained as described previously (4). See the supplemental material for information regarding strain and plasmid construction.
FIG. 1.
The amino acid residues that determine EexS and EexR exclusion specificities are located in distinct regions of the proteins. (A) Schematic depiction of the Eex chimeric constructs and their exclusion activities. The top two lines show the last 56 aa of EexR and EexS. Blue and red letters represent residues specific to EexS or EexR, respectively. Dashes represent conserved residues. In the maps of the chimeras, red and blue bars represent EexR and EexS sequences, respectively. (B) Exclusion activities of recipients expressing the indicated chimeric Eex proteins are expressed as exclusion index scores as previously described (17). Bacterial conjugation assays were performed as previously described (17). All recipients were E. coli CAG18439 derivatives expressing the indicated chimeric exclusion protein. The E. coli donors were MG1655 SXT ΔfloR::kan and MG1655 R391. Each bar represents the mean from three experiments. Error bars, standard deviations.
We also identified amino acids within EexR that appear to be responsible for its exclusion activity. An EexS-based chimera containing aa 114 to 130 of EexR (chimera S1) excluded R391 transfer, suggesting that these 17 aa are sufficient (within the context of full-length EexS protein) for dictating EexR exclusion activity (EIS, 1.6; EIR, 19.3) (Fig. 1). However, based on findings from EexR-based chimeras containing sequences derived from EexS, not all 17 of the EexR amino acid residues in this region seem to be required for R391 exclusion. For example, chimera R3, which lacked EexR residues 121 to 125, retained R391 exclusion specificity (EIR, 12; EIS, 1.4) (Fig. 1). On the other hand, chimera R4, which contains all EexR residues except for residues 127 to 130, which are derived from EexS, did not exclude R391 or SXT (EIS, 0.57; EIR, 1.04) (Fig. 1), even though this chimera was expressed as well as the other chimeras. Thus, these amino acids appear to be required for the exclusion activities of both EexS and EexR. In aggregate, our observations suggest that overlapping yet distinct regions of EexR and EexS determine their respective exclusion specificities. These findings are in contrast to our previous findings for TraG exclusion specificity, where only 3 aa at the same location dictate SXT or R391 exclusion specificity (17). Nonetheless, since amino acids at certain positions (e.g., aa 127 to 130) seem to be required for the activities of both EexR and EexS, it appears that it is not possible to generate a variant of Eex that can exclude both SXT and R391.
The exclusion-determining residues of EexS are cytoplasmic.
To gain insight into the process by which EexS excludes SXT transfer, we performed analyses of the topology of this protein. Initially, we used bioinformatics to predict the number and locations of the transmembrane domains in EexS. The results of these analyses were incongruent. Two programs, TMHMM (23) and SPLIT (15), predicted four transmembrane domains, whereas TMPred (14) and HMTOP (24) predicted three transmembrane domains. All four predictions agreed on the positions of the first two transmembrane domains (residues 12 to 30 and 41 to 59), but they differed as to whether residues 63 to 104 contained one or two transmembrane domains. Thus, these predictions also differ over whether the N terminus and C terminus of EexS are in the same or distinct cellular compartments.
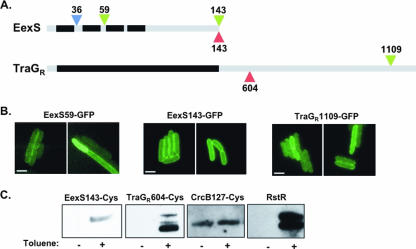
To begin to discriminate between these two models, we investigated the localization of the amino-terminal end of EexS. We engineered a fusion of PhoA to EexS aa 36 in SXT (Fig. 2A); alkaline phosphatase (PhoA) activity is detectable only when the enzyme is located in the periplasm. This EexS-PhoA translational fusion was expressed, and cell fractionation analysis indicated that the fusion protein was membrane bound (data not shown). Furthermore, the alkaline phosphatase activity detected for the strain harboring this chromosomal EexS::PhoA fusion was significantly greater than that for the isogenic strain lacking this fusion (36.8 ± 6.6 versus 9.4 ± 2.4 Miller units), suggesting that aa 36 of EexS is located in the periplasm. This finding, coupled with the prediction that the amino acid stretch from 12 to 30 contains a transmembrane domain, suggests that the amino-terminal end of EexS is localized in the cytoplasm. These data also suggest that aa 59 to 62 should be cytoplasmic. The latter supposition was confirmed using a hybrid protein consisting of green fluorescent protein (GFP) fused to aa 1 to 59 of EexS. This protein was found to be fluorescent, which occurs only if GFP resides within the cytoplasm (Fig. 2B).
FIG. 2.
TraGR and EexS exclusion residues are localized in the cytoplasm. (A) Schematic depiction of the EexS and TraGR proteins with the predicted transmembrane (black) and soluble (gray) regions. Residue numbers and colors indicate the sites where GFP (green), PhoA (blue), and cysteine (red) insertions were placed. (B) Fluorescence images of cells expressing the indicated GFP fusion constructs. Bars represent 2 μM scales. (C) Detection of biotinylation (see the discussion of methods in the supplemental material) of the indicated proteins from cells either left untreated or treated with toluene to permeabilize the membrane. Estimated protein sizes are as follows: EexS143-Cys, 18 kDa; TraGR604-Cys, 128 kDa; CrcB127-Cys, 13 kDa; RstR, 10 kDa. Residue 127 of CrcB is known to be periplasmic, while RstR is a known cytoplasmic protein.
We then determined the localization of the EexS carboxyl terminus. First, we generated a fusion protein consisting of GFP fused to EexS aa 143 (EexS143-GFP) (Fig. 2A). This fusion protein exhibited wild-type exclusion activity (data not shown), and cells expressing EexS143-GFP were fluorescent (Fig. 2B), with the fluorescence observed predominantly at the cell periphery (Fig. 2B). Furthermore, only full-length EexS143-GFP was detected in immunoblot analyses with anti-GFP antisera (data not shown), indicating that the observed fluorescence was not due to GFP cleaved from the fusion protein. Together, these results suggest that the EexS143-GFP fusion protein is membrane bound, and they indicate that the EexS carboxyl terminus must lie in the cytoplasm. The latter finding is consistent with the positive-inside rule (25), since 13 of the 43 putatively soluble amino acids at the EexS carboxyl terminus are positively charged. Our observations that both the amino and carboxyl termini of EexS are in the cytoplasm are consistent with the prediction of TMHMM and SPLIT that EexS contains four membrane-spanning regions (Fig. 2A).
We used cysteine accessibility analysis (5) to corroborate the GFP data suggesting that the EexS carboxyl terminus is located in the cytoplasm. This approach takes advantage of the impermeability of the bacterial inner membrane to MPB [N-(3-maleimidopropionyl)biocytin], a reagent that specifically couples biotin to cysteine thiols. When MPB is added to intact cells, it preferentially reacts with cysteines found in the periplasm. However, when cells are gently treated with toluene, MPB can cross the inner membrane and gain access to cytoplasmic cysteines. Using this system, we never observed biotinylation of wild-type EexS, which contains four native cysteines (data not shown), consistent with the prediction that these residues are found within membrane domains, which are not accessible to MPB even in toluene-permeabilized cells. However, we found that a Cys residue inserted at the carboxyl-terminal end of EexS could be biotinylated, but only after the inner membrane was permeabilized with toluene (Fig. 2C), indicating that this domain is cytoplasmic. Since EexS143-Cys is functional for exclusion (data not shown), its structure is likely representative of the native protein structure. In control experiments, we found that a native cysteine in the cytoplasmic protein RstR was labeled only after toluene permeabilization, while a cysteine engineered into the known periplasmic region of CrcB (CrcB127Cys), was labeled in unpermeabilized cells (Fig. 2C). These experiments demonstrated the reliability of this approach, strengthening our confidence in the conclusion that the EexS residues that determine exclusion activity and specificity are located in the cytoplasm.
The exclusion specificity residues in TraG are also cytoplasmic.
Our previous work indicated that a stretch of only 3 aa at TraG positions 606 to 608 determines its exclusion specificity and activity (17). All four computer programs mentioned above predicted different numbers of TraG membrane domains, yet all predictions agreed that the carboxyl-terminal half (aa 510 to 1189) of TraGR, containing the exclusion specificity residues, is soluble. To examine the cellular localization of the TraGR carboxyl terminus, we used GFP fusion and cysteine accessibility analysis as described above. When GFP was fused to TraGR residue 1109, the resulting protein was fluorescent (Fig. 2B) and detectable only as a full-length fusion protein (data not shown). These observations suggest that the TraGR carboxyl terminus is localized in the cytoplasm. Cysteine accessibility analysis yielded the same conclusion. TraGR contains two native cysteines that were not accessible to MPB, as evidenced by the fact that we observed no biotinylation of the wild-type protein (data not shown). When we replaced Thr604 with a cysteine residue in TraGR, we detected biotinylation of this construct only in permeabilized cells (Fig. 2C), confirming that the carboxyl-terminal region of TraGR is cytoplasmic. R391 harboring TraGR604-Cys was proficient for transfer, yet interestingly, this protein did not have exclusion activity (data not shown). The latter observation suggests that residues outside of the TraG exclusion specificity region may also influence TraG-Eex interactions. In summary, as with its counterpart Eex, the TraG exclusion activity and specificity region is localized in the cytoplasm of the cell.
Conclusions.
The structural and functional investigations of the entry exclusion partners Eex and TraG presented here yielded several unexpected observations that challenge our previous model that interactions between periplasmic regions of these proteins mediate exclusion (17). Given our new finding that the TraG and Eex regions that mediate exclusion specificity are cytoplasmic, we propose that TraG-Eex interactions occur in the cytoplasm. However, the mechanism by which cytoplasmic proteins in mating cells can come in contact is as yet unknown. It is possible that an intact protein or a fragment of either TraG or Eex might be translocated into the mating partner. Such intercellular translocation of membrane proteins has been reported for myxobacteria (19). Eex or TraG translocation could occur either through the lumen of the mating channel or outside of the mating channel, perhaps concurrently with the formation of the mating bridge. Several scenarios can be imagined. In one scenario, only a proteolytic fragment of one of the proteins moves. In a second scenario, TraG or Eex, as part of the mating channel, undergoes a conformational change that flips its cytoplasmic domain to contact the recipient cell cytoplasm, while its N terminus remains anchored in the donor cell inner membrane. In a third scenario, the entire TraG or Eex protein is transported between the donor and the recipient cell, with its topology retained. Finally, since our topological analyses were not performed on mating cells, it is formally possible that the Eex and/or TraG domains of interest do not reside within the cytoplasm during the mating process and thus that entirely different means of interaction are possible.
Like Eex, the other characterized entry exclusion proteins have also been shown to localize to the inner membrane (8-10, 26, 27). However, in these other exclusion proteins, the cellular localization of the residues critical for exclusion activity remains to be determined. The donor cell targets of entry exclusion proteins are known only for Eex and TraS. As additional entry exclusion systems are defined, it will be interesting to learn whether cytoplasmic interactions between exclusion determinants are a conserved feature in a mechanism for inhibiting DNA transfer. A recent study of the F plasmid suggests this may not be the case, since the F TraG exclusion-determining region was reported to be periplasmic (2). A fundamental question remaining is how interactions between TraG and Eex abolish DNA transfer. Some ideas for exploring this question can be derived from studies of superinfection immunity in some phage systems (e.g., T4 [22]), where interactions between proteins in the cytoplasm have been shown to block DNA import. Ultimately, understanding how entry exclusion inhibits conjugative DNA transfer should provide insight into the mechanics of the conjugative process. Finally, further studies on entry exclusion will also highlight the importance of the (heretofore neglected) recipient cell in the conjugative process.
Supplementary Material
Acknowledgments
We are grateful for support from NIH grant AI-42347, HHMI, and the Tufts-NEMC GRASP center.
We thank Brigid Davis for insightful comments on the manuscript.
Footnotes
Published ahead of print on 15 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M., N. Kennedy, and R. Skurray. 1977. Cell-cell interactions in conjugating Escherichia coli: role of TraT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74:5104-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audette, G. F., J. Manchak, P. Beatty, W. A. Klimke, and L. S. Frost. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153:442-451. [DOI] [PubMed] [Google Scholar]
- 3.Beaber, J. W., V. Burrus, B. Hochhut, and M. K. Waldor. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 59:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanov, M., W. Zhang, J. Xie, and W. Dowhan. 2005. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods 36:148-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 8.Finlay, B. B., and W. Paranchych. 1986. Nucleotide sequence of the surface exclusion genes traS and traT from the IncF0 lac plasmid pED208. J. Bacteriol. 166:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase, J., M. Kalkum, and E. Lanka. 1996. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncPα plasmid RP4. J. Bacteriol. 178:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartskeerl, R. A., J. E. Bergmans, M. C. Kamp, and W. P. Hoekstra. 1983. Cloning of an exclusion-determining fragment of the IncI plasmid, R144. Plasmid 10:11-20. [DOI] [PubMed] [Google Scholar]
- 11.Hartskeerl, R. A., and W. P. Hoekstra. 1984. Exclusion in IncI-type Escherichia coli conjugations: the stage of conjugation at which exclusion operates. Antonie Leeuwenhoek 50:113-124. [DOI] [PubMed] [Google Scholar]
- 12.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 15.Juretic, D., L. Zoranic, and D. Zucic. 2002. Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comput. Sci. 42:620-632. [DOI] [PubMed] [Google Scholar]
- 16.Manning, P. A., G. Morelli, and M. Achtman. 1981. TraG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc. Natl. Acad. Sci. USA 78:7487-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marrero, J., and M. K. Waldor. 2005. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev. Cell 8:963-970. [DOI] [PubMed] [Google Scholar]
- 18.Marrero, J., and M. K. Waldor. 2007. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J. Bacteriol. 189:3302-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nudleman, E., D. Wall, and D. Kaiser. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125-127. [DOI] [PubMed] [Google Scholar]
- 20.Sheehy, R. J., C. Orr, and R. Curtiss III. 1972. Molecular studies on entry exclusion in Escherichia coli minicells. J. Bacteriol. 112:861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder, L., S. Blight, and J. Auchtung. 2003. Regulation of translation of the head protein of T4 bacteriophage by specific binding of EF-Tu to a leader sequence. J. Mol. Biol. 334:349-361. [DOI] [PubMed] [Google Scholar]
- 23.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 24.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 26.Winans, S. C., and G. C. Walker. 1985. Entry exclusion determinant(s) of IncN plasmid pKM101. J. Bacteriol. 161:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada, Y., M. Yamada, and A. Nakazawa. 1995. A ColE1-encoded gene directs entry exclusion of the plasmid. J. Bacteriol. 177:6064-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.