Abstract
Xanthomonas campestris pv. vesicatoria is the causal agent of bacterial spot disease of tomato and pepper. The disease process is interactive and very intricate and involves a plethora of genes in the pathogen and in the host. In the pathogen, different genes are activated in response to the changing environment to enable it to survive, adapt, evade host defenses, propagate, and damage the host. To understand the disease process, it is imperative to broaden our understanding of the gene machinery that participates in it, and the most reliable way is to identify these genes in vivo. Here, we have adapted a recombinase-based in vivo expression technology (RIVET) to study the genes activated in X. campestris pv. vesicatoria during its interaction with one of its hosts, tomato. This is the first study that demonstrates the feasibility of this approach for identifying in vivo induced genes in a plant pathogen. RIVET revealed 61 unique X. campestris pv. vesicatoria genes or operons that delineate a picture of the different processes involved in the pathogen-host interaction. To further explore the role of some of these genes, we generated knockout mutants for 13 genes and characterized their ability to grow in planta and to cause disease symptoms. This analysis revealed several genes that may be important for the interaction of the pathogen with its host, including a citH homologue gene, encoding a citrate transporter, which was shown to be required for wild-type levels of virulence.
Xanthomonas campestris pv. vesicatoria is a biotrophic gram-negative gammaproteobacteria and is the causal agent of bacterial spot disease of tomato (Solanum lycopersicum L.) and pepper (Capsicum annuum) (22). Recently, X. campestris pv. vesicatoria has been reclassified into four different species: Xanthomonas vesicatoria, Xanthomonas euvesicatoria, Xanthomonas gardneri, and Xanthomonas perforans (20, 21). Because the acceptance of the new nomenclature is yet to be determined by the scientific community, here we use the classical nomenclature. Bacterial spot symptoms include defoliation and chlorotic necrotic lesions on leaves, stems, fruits, and flowers, subsequently leading to reduced yield. The disease is present worldwide, with optimal conditions for breakout of the disease being temperatures ranging between 24°C and 30°C and high relative humidity.
In the past 2 decades, progress has been made in elucidating the molecular mechanisms underlying the interactions between this pathogen and its hosts. It has been determined that, like many other gram-negative biotrophic bacteria, X. campestris pv. vesicatoria uses a type III secretion system to secrete effectors into the host cell, where they interact with the host cellular processes to promote disease or to elicit a defense response (3). However, the establishment of bacterial pathogens in their host and their ability to cause disease expand beyond the ability to form this secretion system and transfer effectors through it. The infectious process is very intricate and requires the coordinated activity of a myriad of bacterial genes whose identity and mode of action are still largely unknown.
In attempts to shed light on the different bacterial genes involved in virulence and pathogenicity, investigators have applied various in vitro approaches, mimicking certain aspects of infectious processes. These approaches, however, are limited in their ability to mimic all aspects of the in vivo state. Therefore, in vivo experimental approaches are highly desirable to further our understanding of pathogenesis. In vivo approaches showing high potential to identify genes required for virulence and/or survival of bacterial pathogens have been developed. These include, in vivo expression technology (IVET) (28), differential fluorescence induction (46), signature-tagged mutagenesis (18), differential display (1), selective capture of transcribed sequences (16), and in vivo transposon screens using an effector derivative deprived of its secretion signal as a reporter (17, 39). Here, we report the utilization of an IVET approach to identify novel genes from X. campestris pv. vesicatoria that facilitate the pathogen in its interaction with tomato.
IVET systems are promoter-trap methods designed to identify promoters whose activities are specifically induced or enhanced in a certain environment. Different IVET variations have been developed to study bacterial responses to various ecological niches (such as animal or plant hosts, in the case of bacterial pathogens), leading to the identification of many in vivo upregulated genes (38). Most IVET systems use reporters that are auxotrophic markers, genes essential for pathogenicity, or antibiotic markers for capturing in vivo activated promoters. Since the activation of such reporters by in vivo induced promoters complements an essential function, their activation is required so that each strain within a pooled population can compete and eventually be detected by the screen. Thus, promoters that are only weakly or transiently induced might be missed. Some attempts have been made to circumvent this limitation, for instance, by screening individual clones in an efficient manner (51) or by using reporters that even when weakly induced still complement the essential function (29). Another IVET approach that provides a very elegant solution to this problem is a recombinase (or resolvase)-based IVET (RIVET) (13). RIVET relies on the induction of a promoterless resolvase gene (tnpR), which irreversibly captures activated promoters. TnpR recognizes specific sites (res) inserted within the bacterial chromosome to create a reporter strain, and its action on these sites leads to the excision of a reporter marker flanked by them.
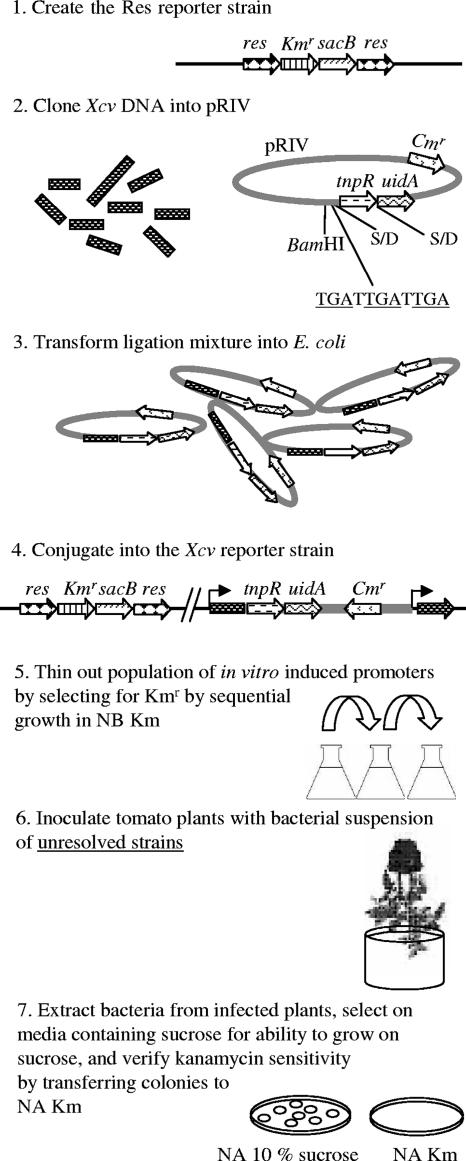
In this study, RIVET was implemented to gain a better understanding of the gene machinery at work during the interaction of X. campestris pv. vesicatoria with its tomato host. To our knowledge, this is the first RIVET screen on a phytopathogenic bacterium. The screen flow and its rationale are detailed in Fig. 1. It was designed to identify promoters that are expressed during infection based on their ability to drive expression of a recombinase, which results in excision (resolution) of a reporter construct from the X. campestris pv. vesicatoria genome. Resolution of the reporter from individual strains is screened for after the bacteria are isolated from the plant tissue. It should be noted that prior to plant inoculation, in vitro induced strains are diluted out of the population since they are propagated in a medium that inhibits growth of resolved strains. Analysis of promoter regions of in vivo induced fusions revealed a list of 61 unique genes (or operons), of which 13 were selected for site-directed mutagenesis to further investigate their contribution to the virulence of X. campestris pv. vesicatoria. Pathogenicity assays with the mutated strains revealed that a citrate transporter is essential for full virulence of X. campestris pv. vesicatoria on tomato.
FIG. 1.
General scheme of the RIVET screen for selection of IVX genes. The screen is set to identify promoters that are activated during infection based on their ability to drive expression of a recombinase (TnpR), which results in excision of a reporter construct from the X. campestris pv. vesicatoria genome. (1) An X. campestris pv. vesicatoria reporter strain was generated that carries res sites flanking a Kmr gene and a sacB gene. The latter inhibits growth in the presence of high sucrose concentrations, allowing positive selection on sucrose of clones that have lost the construct (30). (2) A library of X. campestris pv. vesicatoria DNA fragments was cloned into pRIV into the BamHI site upstream of a promoterless tnpR-uidA transcriptional fusion. Translational stop codons (underlined) in all three reading frames (TGATTGATTGA) were added to the vector downstream of the cloning site. The Shine-Dalgarno (S/D) sequences of the tnpR and uidA genes are indicated. (3) The vectors containing the cloned fragments were first transformed into E. coli. (4) Pools of E. coli transformants were conjugated into the X. campestris pv. vesicatoria reporter strain (Res strain), resulting in their integration into its chromosome. (5) In vitro induced strains were diluted out of the population by sequential growth in NB supplemented with Km prior to plant inoculation. (6) Tomato plants were inoculated with pools of unresolved clones. (7) In vivo induced/resolved strains were selected on NA containing 10% sucrose, and clones that were able to grow on sucrose were verified for their inability to grow in NA with Km.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Wild-type X. campestris pv. vesicatoria 97-2 (race T3; X. perforans, according to the new nomenclature) (21) and mutant strains were grown on nutrient agar (NA; Becton, Dickinson and Co., Sparks, MD) or in XVM2 minimal medium (48) at 28°C. Escherichia coli strains were grown on Luria-Bertani medium at 37°C. A modified M9 minimal medium (potassium phosphate buffer [pH 7.0], NaCl 2.5 mg/ml, NH4Cl 5 mg/ml, 0.3% Casamino Acids, 5 mM MgSO4, 0.1 mM CaCl, 10 mM Na-citrate) was used in several experiments for characterization of the citH deletion mutant as described previously. Carbon sources were added to the medium at a concentration of 10 mM. Antibiotics were added at the indicated concentrations: kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 25 μg/ml; cephalexin, 20 μg/ml; and gentamicin, 30 μg/ml. Oligonucleotide primers (purchased from Hy Laboratories, Rehovot, Israel) used for PCR and sequencing in this study are also listed in Table 1.
TABLE 1.
Strains, plasmids, and DNA primers used in this study
| Strain, plasmid, or primer | Characteristic(s) of strain or plasmid and primer sequences (5′-3′)a | Reference or source |
|---|---|---|
| X. campestris pv. vesicatoria strains | ||
| 97-2 | Wild type; belongs to race T3 | 5 |
| Res strain | Kmr; reporter strain containing the res cassette inserted within the hpaG gene | This study |
| ResPhrpA | Kmr Cmr; PhrpA::tnpR::uidA in the Res strain background | This study |
| ResPhrpG | Kmr Cmr; PhrpG::tnpR::uidA in the Res strain background | This study |
| ResPhrpGrev | Kmr Cmr; PhrpGrev::tnpR::uidA in the Res strain background | This study |
| E. coli S17-1 λ pir | λ lysogenic S17-1 derivative producing π protein for replication of plasmids carrying oriR6K; recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 λ− pir | 43 |
| Plasmids | ||
| pIVET | Kmr; vector carrying the hrcC-uidA transcriptional fusion | 8 |
| pIVETn | Apr; vector containing the tnpR gene | A. Camilli |
| pRes | Apr Kmr; vector containing the res-Kmr-sacB-res | A. Camilli |
| pJP5603 | Kmr; R6K-based suicide vector; requires the pir-encoded π protein for replication | 34 |
| pUCPET | Apr; pUC18 with the uidA gene from pIVET inserted into its multiple cloning site | This study |
| pUC18 | Apr; cloning vector | 52 |
| PJPCm | Cmr; pJP5603 with the Kmr gene replaced by the Cmr gene | This study |
| PJPCmhpaG | Cmr; X. campestris pv. vesicatoria hpaG gene cloned into pJPCm | This study |
| pJPCmhpaGRes | Cmr; res cassette cloned into X. campestris pv. vesicatoria hpaG gene within pJPCm | This study |
| PRIV | Cmr; vector for X. campestris pv. vesicatoria library cloning for the RIVET screen | This study |
| PRIVPhrpA | Cmr; 614-bp hrpA promoter region in correct orientation relative to the tnpR::uidA | This study |
| PRIVPhrpG | Cmr; 551-bp hrpG promoter region in correct orientation relative to the tnpR::uidA | This study |
| pRIVPhrpGrev | Cmr; hrpG promoter region in opposite orientation relative to the tnpR::uidA | This study |
| Primers | ||
| RtnpRIVET | CTTTCCGATCGCATGAACTG | This study |
| FwPASacBam2 | GGGGGAGCTCGGATCCCGACGGTGTTCTGGATCG (SacI, BamHI) | This study |
| RPAXhoBam | GGGGCTCGAGGGATCCGCTAGGCAAGTGAAAAATCG (XhoI, BamHI) | This study |
| FphrpGXho | GGGGCTCGAGACAGTCGGACAACGTCTGC (XhoI) | This study |
| RphrpGXho | GGGGCTCGAGCGTGTCGGCGTACAAGC (XhoI) | This study |
| RtnpRHind | GGGGAAGCTTTTTCCTCCTTAGTTGC (HindIII) | This study |
| FtnpR1353stop | GGGGCATATGCTCGAGTGATTGATTGAGCGGCCGCCTCAATTGTGAGAA TTTGAG (NdeI, XhoI, Not) | This study |
| FhpaGHind | GGGGAAGCTTCGCAAGACAGTCCTTCACG (HindIII) | This study |
| RhpaGKpn | GGGGGGTACCCTAATGGATGCCCCATTCC (KpnI) | This study |
| FRESal | GGGGGTCGACACTACTTAGGGCGAATTGGG (SalI) | This study |
| RRESal | GGGGGTCGACCACTAAAGGGAACAAAAGCTGG (SalI) | This study |
In primers, underlined nucleotides represent restriction sites of enzymes indicated in parentheses. In primer FtnpR135stop, the three stop codons in the different reading frames are in boldface. Kmr, Cmr, and Apr indicate kanamycin, chloramphenicol and ampicillin resistance, respectively.
Plant material and inoculation procedures.
Tomato (Solanum lycopersicum L.) cultivar Hawaii 7998 (H7998) (53) was grown from seeds in the greenhouse (25 to 28°C). Inoculation of plants with X. campestris pv. vesicatoria libraries and assessment of in planta growth were carried out by dipping 4- to 5-week-old plants into bacterial suspensions of 105 CFU/ml containing 0.02% of the surfactant Silwet L-77 (24) and 10 mM MgCl2. When plants were inoculated with X. campestris pv. vesicatoria libraries, vacuum infiltration was applied (50), and the majority of bacteria were recovered after 48 h, although some were recovered 4 and 8 days postinoculation (dpi). When in planta growth was assessed, no vacuum was applied, and the plants were immediately covered with plastic bags for 48 h to keep them in a moist environment. At least three plants per treatment were inoculated in each experiment. Leaf discs were extracted from the first three fully expanded leaves, macerated, and plated to determine bacterial concentration (CFU/g) in inoculated leaves. To evaluate symptom development, inoculations were done by syringe infiltration of bacterial suspensions at 105 CFU/ml (unless otherwise stated) in 1 mM MgCl2 containing 0.02% of the surfactant Silwet L-77. Wild-type and mutant strains were infiltrated several times into leaflets on opposing sides. This step was repeated for each mutant on the leaflets of the first three fully expanded leaves on three different plants. Phenotypes were scored based on comparisons between symptoms at the sites of the inoculations of the wild-type and the mutant strains on opposing leaflets.
Genetic manipulations.
Routine molecular manipulations and cloning procedures were carried out as described by Sambrook et al. (40). All enzymes were purchased from Fermentas (Ontario, Canada). Kits for plasmid and PCR product extraction and purification were purchased from Real Biotech Corporation (Taipei, Taiwan). Genomic DNA from X. campestris pv. vesicatoria was prepared with a GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO). Southern blot hybridization was done using an ECL Direct Nucleic Acid Labeling and Detection System (Amersham Biosciences, Buckinghamshire, United Kingdom) according to the manufacturer's instructions. DNA sequencing was performed by the Big Dye Terminator Cycle Sequencing Kit from ABI, using an automated sequencer at Hy Laboratories.
Generation of an X. campestris pv. vesicatoria reporter strain carrying the res-Kmr-sacB-res cassette.
The hpaG gene was PCR amplified from X. campestris pv. vesicatoria 97-2 with the FhpaGHind and RhpaGKpn primer pair and cloned into pUC18 after digestion of the PCR product and the vector with KpnI and HindIII. The cloned fragment was sequenced to verify that the correct region was amplified and cloned. This vector was then digested with SalI and EcoRI, both cutting within hpaG and resulting in the generation of two hpaG fragments, of 0.7 kb and 0.6 kb. These fragments were ligated in a three-fragment ligation into pJPCm, and the resulting vector was named pJPCmhpaG. This vector was cut with EcoNI, blunt ended with Klenow fragment, and dephosphorylated. The res cassette (res-Kmr-sacB-res) was amplified from pRes (kindly supplied by A. Camilli) with the FRESal and RRESal primer pair and cloned into pJPCmhpaG after digestion of the vector with SalI and blunt ending with Klenow. The resulting vector was named pJPCmhpaGRes, and it was conjugated from E. coli S17 λpir into X. campestris pv. vesicatoria. Transconjugants were selected on NA containing cephalexin and Km and replica plated onto NA with Cm. A Kmr Cms mutant that was unable to grow on NA containing 10% sucrose was further confirmed for a single insertion of the cassette by Southern analysis and named X. campestris pv. vesicatoria Res strain.
Construction of pRIV and cloning of previously characterized promoters.
The tnpR gene was amplified from the pIVETn vector (kindly supplied by A. Camilli) with FtnpR1353stop as the forward primer, containing stop codons in the three reading frames, and RtnpRHind as the reverse primer. The amplified product was ligated into pUCPET (pUC18 carrying the uidA gene) after the insert and vector were cut with NdeI and HindIII. The resulting tnpR-uidA transcriptional fusion was then excised and ligated into the multiple cloning site of pJPCm. The BamHI site for library cloning was added later by cloning the hrpA promoter region, containing BamHI sites internal to the ScaI and XhoI cloning sites, upstream of tnpR-uidA. The hrpA promoter region was PCR amplified from the wild-type X. campestris pv. vesicatoria strain 97-2 with the FWPASacBam2 and RPAXhoBam primer pair. This resulted in pResPhrpA that was used as a control. To generate pResPhrpG and pResPhrpGrev (vectors carrying the tnpR-uidA fusion downstream of a DNA fragment containing the hrpG promoter in the correct and in the opposite orientations, respectively), the hrpA promoter was excised from pResPhrpA by digestion with BamHI. The vector was then self-ligated to create pRIV. The hrpG promoter region was then PCR amplified from X. campestris pv. vesicatoria 97-2 with the FPhrpGXho and RPhrpGXho primer pair and ligated upstream of the tnpR-uidA transcriptional fusion after digestion with XhoI of the vector and insert. The resulting vectors were introduced into E. coli S17 λpir and then conjugated into the X. campestris pv. vesicatoria Res strain to assess the viability of the RIVET approach.
Construction of X. campestris pv. vesicatoria transcriptional fusion libraries.
Chromosomal DNA fragments from X. campestris pv. vesicatoria 97-2 were obtained by partial digestion with Sau3AI and isolation of 1.5- to 2.5-kb fragments. These were cloned into pRIV digested with BamHI and dephosphorylated. The ligation products were transformed into E. coli S17 λpir. The transformants were then grouped into four pools, each containing roughly 1,000 transformants. Each pool was mated with the X. campestris pv. vesicatoria Res reporter strain. Since pRIV does not replicate in X. campestris pv. vesicatoria, the conjugated plasmids had to recombine at the site in the genome with homology to the cloned genomic fragments. Approximately 5,000 transconjugants were selected in rich medium (NA) with Km and Cm and confirmed for their inability to grow in the presence of sucrose by replica plating. To enrich the strains which did not resolve in rich medium, each of the four pools of 5,000 transconjugants was transferred to rich medium with Km and Cm three times before being used for plant inoculations.
Screening of transcriptional libraries for in vivo overexpressed (IVX) genes.
Four X. campestris pv. vesicatoria libraries, each containing 5,000 transconjugants, were grown to saturation three consecutive times in nutrient broth (NB) supplemented with Km and Cm to dilute out strains with in vitro induced promoters. Each pool of transconjugants was then used for plant inoculation. The majority of bacteria were extracted from the plants 48 h after inoculation, although some were extracted 4 and 8 days after inoculation. The extracted clones were plated on NA containing Cm and 10% sucrose. Colonies that were able to grow on sucrose due to the loss of the sacB gene were verified by replica plating for their sensitivity to Km and were also analyzed for glucuronidase (GUS) expression on rich and minimal media, using X-GlcA (5-bromo-4-chloro-3-indoyl-ß-D-glucoside) as a substrate. The mutants were then analyzed by Southern hybridization to eliminate those containing multiple insertions of the plasmid.
Recovery of integrated plasmids from the chromosome of IVX strains.
Recovery of inserted plasmids from the chromosome was carried out by isolating genomic DNA from strains carrying IVX genes (or operons; hereafter, IVX strains), digesting it with SacI, which cuts once within the integrated plasmid upstream of the cloned sequence, and self-ligating the digested DNA. Ligation products were transformed into E. coli S17 λpir, and plasmids were isolated from the transformants that were able to grow on Cm. IVX promoter regions were sequenced using the RtnpRIVET primer matching the 5′ region of the tnpR gene.
Generation of knockout mutants for selected IVX genes.
Primers were designed to amplify fragments internal to a chosen set of IVX genes (such that the amplified fragment did not span both the 5′ and 3′ ends of the gene). Disruption of the putative open reading frames was done by cloning the amplified products into pJP5603 (34) and introducing the vector back into X. campestris pv. vesicatoria 97-2 by electrotransformation (after first sequencing the cloned fragment to verify that the desired sequence was cloned). Interruption of the desired gene was verified by Southern analysis. To ensure that plasmids were not lost after chromosomal insertion, bacteria extracted from plants were constantly monitored for their ability to grow on Km by plating dilutions of macerated tissue both on NA and NA supplemented with Km.
GUS expression analysis.
The qualitative GUS assay was conducted by replicating colonies from growth plates onto filter paper, which was then washed with 3 ml of 10 mg/ml lyzosyme for 30 min at 37°C. After the liquid was drained, 1 ml of 1 mg/ml X-Gluc (5-bromo-4-chloro-3-indoyl-ß-D-glucoronic acid) cyclohexylammonium (Duchefa Biochemie, Haarlem, The Netherlands) resuspended in 50 mM sodium phosphate buffer (pH 7.0), with a K+ ferricyanide/ferrocyanide mixture, was added for overnight incubation at 37°C. For quantitative analysis, the fluorometric substrate 4-methylumbelliferyl-β-d-glucuronide (MUG) (Duchefa Biochemie) was used as described previously (19). Essentially, the assay buffer was prepared by resuspending 1 mM MUG in extraction buffer (50 mM sodium phosphate buffer [pH 7.0], 10 mM β-mercaptoethanol, 10 mM Na2-EDTA, 0.1% sodium lauryl sarcosine, 0.1% Triton X-100). A total of 450 μl of the assay buffer was added to a 50-μl sample (consisting of cells after growth in liquid medium or macerated leaf tissue that had been pelleted and resuspended in 50 μl of double distilled water), and 50-μl aliquots were taken at different times and added to 450 μl of stop buffer (0.2 M Na2CO3). Fluorescence was measured on a Cary Eclipse fluorescence spectrophotometer (Varian Inc., Palo Alto, CA) with 4-methylumbelliferone (Sigma-Aldrich) as the standard. For the in vitro quantitative GUS assay, 500 μl of liquid culture in its mid-exponential to late exponential phase was centrifuged, resuspended in 50 μl of double distilled water, and stored at −80°C until it was used. For the in vivo assay, bacteria at 105 CFU/ml in 1 mM MgCl2 were inoculated into leaves of 5-week-old plants. Each mutant was inoculated into 3 plants, and samples from each plant were collected and macerated 24, 48, and 72 h postinoculation (hpi). The macerated tissue was centrifuged at low speed to get rid of plant material, and the supernatant was centrifuged again to pellet the cells, which were then resuspended in double distilled water. An aliquot of the resuspension from each sample was diluted and plated for CFU counts, and an aliquot was stored at −80°C until it was used for GUS activity measurements.
Nucleotide sequence accession numbers.
Nucleotide sequence data of IVX clones identified following growth on tomato H7998 were deposited in the GenBank database under the accession numbers EF633111 to EF633171.
RESULTS
General overview and rationale of the RIVET screen.
A RIVET approach was developed to identify IVX genes of X. campestris pv. vesicatoria. The screen is delineated in Fig. 1. Basically, we first generated an X. campestris pv. vesicatoria reporter strain, named X. campestris pv. vesicatoria Res strain (see below). This strain carries the res cassette (res-Kmr-sacB-res), in which the res sites are flanking an antibiotic resistance gene (Km) and a second reporter gene, sacB, that allows positive selection of in vivo induced strains (30). The res sites are specifically recognized by a transposase (TnpR) that catalyzes the excision of the res cassette in a process called resolution. Cells that have deleted the res cassette lose their ability to grow in the presence of the antibiotic but are able to grow in the presence of a high sucrose concentration. In a second step, a library of X. campestris pv. vesicatoria DNA fragments was fused upstream of a promoterless tnpR-uidA transcriptional fusion and introduced into the chromosome of the reporter strain, creating cis-merodiploid strains without disrupting the downstream gene (except when the cloned fragment does not span both the 5′ and 3′ ends of the transcriptional unit) and avoiding problems related to the use of plasmids. Next, in vitro induced strains (namely, strains that were resolved in culture) were diluted out of the population prior to plant inoculation, by growing the pools of strains in rich medium (NB) with Km for three cycles. The resulting unresolved strains were then used to inoculate tomato plants and in vivo resolved strains (namely, those strains that lost Km resistance and gained the ability to grow in the presence of a high sucrose concentration) were isolated from leaves. The integrated vectors were then isolated, and the genes of interest were further characterized.
Assessment of the X. campestris pv. vesicatoria Res mutant as a reporter strain for screening for IVX promoters.
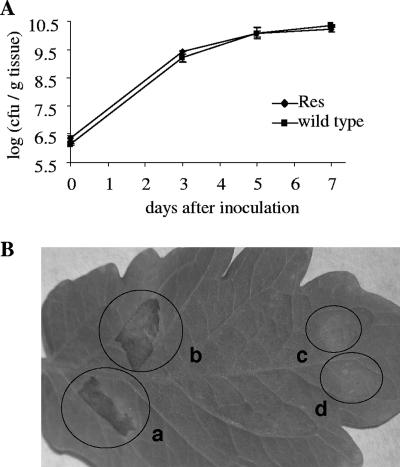
The mutant strain containing the res cassette was evaluated for its suitability as a reporter strain for the screening of promoters that are upregulated during the interaction of the pathogen with the plant. We first assessed the neutrality of the res cassette insertion into the hpaG site. The hpaG gene is located in the region downstream to the hrpF gene and encodes a putative protein of 432 amino acids (31). It was selected as a target for inserting the res cassette because disruption of this gene was shown to have no effect on the virulence of X. campestris pv. vesicatoria on pepper (31). In the interaction of X. campestris pv. vesicatoria with tomato, the virulence of the reporter strain was compared to that of the wild type by comparing symptom development and growth in planta. Our data show that disruption of the hpaG gene by insertion of the res cassette did not affect growth or symptom development under the tested conditions (Fig. 2), thus supporting the idea that this gene is not required for pathogenicity in the interaction of the pathogen with tomato.
FIG. 2.
Growth in planta and symptom induction of the X. campestris pv. vesicatoria Res strain compared to the wild-type strain on tomato. (A) Growth in planta. Plants were inoculated by vacuum infiltration with 105 CFU/ml suspensions. (B) Symptom development following syringe infiltration at a relatively high inoculum concentration (108 CFU/ml). This method allows differentiation between pathogenic and nonpathogenic bacteria a few days after inoculation. Wild-type (a) and X. campestris pv. vesicatoria Res mutant (b) strains did not differ in their ability to induce typical disease symptoms (necrotic lesions). Negative controls were a nonpathogenic hrpA deletion mutant strain (c) and 10 mM MgCl2 (d). The photograph was taken 72 hpi.
The X. campestris pv. vesicatoria Res strain was also evaluated for background resolution of the res cassette in the absence of an active TnpR. Resolution was determined in planta and in culture. Bacteria extracted from the plant 1 h and 2 and 8 dpi had no detectable resolution (defined as the number of Kms CFU compared to the total number of CFU). Also, background resolution could not be detected after growth to saturation in rich (NB) and in minimal (XVM2) media, without antibiotic, two consecutive times (Table 2). XVM2 medium was used since it is a minimal medium that mimics to some extent the plant apoplast environment and has been shown to efficiently induce the expression of hrp genes (48).
TABLE 2.
Resolution by TnpR in different X. campestris pv. vesicatoria strains carrying the res cassette
| Strain | Resolution under the indicated growth conditiona
|
||
|---|---|---|---|
| NB | XVM2 | In planta | |
| Res strain | None | None | Noneb |
| ResPhrpG | 77.4 ± 1.1c | Complete | Completed |
| ResPhrpA | None | Complete | Completed |
| ResPhrpGrev | None | ND | Noned |
None, less than 2 % resolution based on a comparison of growth between NA-Km and NA-10 % sucrose plates with NA-Cm plates; complete resolution, at least 98 % resolution based on a comparison of growth between NA-Km and NA-10 % sucrose plates with NA-Cm plates; ND, not determined. XVM2 is a minimal medium that imitates to some extent the plant environment.
Tested 1 h and 2 and 8 days postinoculation.
Based on replica of NA-Cm plates compared to NA-Km plates.
Tested 4 dpi.
TnpR activity in the X. campestris pv. vesicatoria Res reporter strain.
To gauge the activity of TnpR in the X. campestris pv. vesicatoria Res strain, fragments containing the hrpA and hrpG promoter regions were cloned upstream of the promoterless tnpR-uidA transcriptional fusion in the pRIV vector to create pRIVPhrpA and pRIVPhrpG, respectively. These plasmids were introduced into the X. campestris pv. vesicatoria Res chromosome by single homologous recombination to create strains ResPhrpA and ResPhrpG, which were confirmed by Southern blotting (data not shown). These promoters were chosen since the activity of hrpA and hrpG promoters had previously been characterized in X. campestris pv. vesicatoria strain 85-10 following growth in rich (NB) and in minimal (XVM2) media (49). The hrpA promoter was reported to have a very weak activity in rich medium and about a 1,000-fold induction in XVM2 medium, whereas the hrpG promoter has a relatively strong activity in rich medium and has only about a threefold induction in XVM2 medium (49). A similar pattern of hrpA and hrpG promoter activities in minimal and rich media was also observed in this study (Fig. 3). The ResPhrpG strain showed 80% and 100% resolution after growth in NB and XVM2 media, respectively, while the ResPhrpA strain did not resolve in NB medium and resolved completely in XVM2 medium (Table 2).
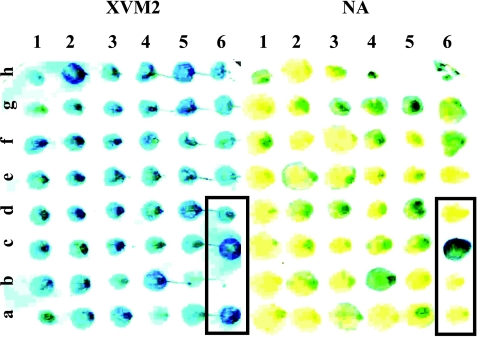
FIG. 3.
GUS analysis of IVX strains after growth in XVM2 medium and in NA. The strains grown on XVM2 medium (left) and on NA (right) are replicates of the same original plate of plant-extracted CFU. The controls (boxed) are as follows (identified by lane and row): 6a, ResPhrpG; 6b, ResPhrpGrev; 6d, ResPhrpA; 6c, an unidentified clone isolated in a preliminary IVET screen, showing strong GUS activity in rich and in minimal media.
To verify that there was no background activity, the hrpG promoter was also cloned in the reverse orientation to create the pRIVPhrpGrev plasmid and, subsequently, the ResPhrpGrev strain. When the plasmid was inserted into the reporter strain, there was no detectable resolution in NB medium or after growth in planta (Table 2). Together, these data demonstrate that tnpR expression can be induced in X. campestris pv. vesicatoria and that TnpR activity is sufficient to target its substrate in the reporter strain, confirming the utility of this system to identify X. campestris pv. vesicatoria upregulated genes during infection of tomato.
The RIVET screen.
To identify genes of X. campestris pv. vesicatoria upregulated during its interaction with tomato, we constructed a library of X. campestris pv. vesicatoria 97-2 genomic fragments in pRIV. A total of approximately 4,000 transformants from two independent ligations and four electroporations were obtained. The resulting transformants from each of the electrotransformations were sampled to verify that they contained a variety of plasmids and that these plasmids were of the expected size. The transformants were then grouped into four pools, each containing roughly 1,000 transformants. Each pool was mated with the X. campestris pv. vesicatoria Res strain. Approximately 5,000 transconjugants for each E. coli pool were obtained.
Before utilization of the pooled strains for plant inoculation, strains that did not resolve in rich medium were enriched. This was done by growing each pool in rich medium supplemented with Km and Cm to saturation and then diluting the culture into fresh medium. This process was repeated two consecutive times. The rationale was that in vitro resolved strains, being Kms, would not be able to propagate under these conditions. The Kmr strains were used for tomato plant inoculations. The majority of mutants were isolated from inoculated plants 2 dpi, but some were also isolated 4 and 8 dpi (Table 3). The isolated mutants were tested for their resolution state. Namely, they were verified for their ability to grow on sucrose and their sensitivity to Km (Fig. 1). To validate the screening method, the isolated strains were analyzed qualitatively for their ability to express GUS on rich (NA) and on minimal (XVM2) media. Indeed, most of IVX strains after growth on NA expressed GUS to lower levels than when grown on XVM2 medium (Fig. 3).
TABLE 3.
IVX clones identified following growth on tomato H7998
| Functiona | Gene no.b | Description (conserved domains) |
|---|---|---|
| Transport | Xcv4089c,d | ABC-type antimicrobial peptide transport system, ATPase component (COG1136, cd03255) |
| Xcv4319c,d | tatC; Sec-independent protein translocase (pfam00902) | |
| Xcv3892 | clcB; voltage gated chloride channels (COG0038, COG2239) | |
| Xcv 4128c | Iron uptake factor c-terminal fragment (COG0369, pfam00175, pfam00258, pfam03929) | |
| Xcv3613d,e | citH; citrate:H+ symporter (pfam03606) | |
| Xcv1541d | mexF; RND multidrug efflux transporter (pfam00873) | |
| Virulence | Xcv0052 | avrBs2 (pfam03009) |
| Regulatory function | Xcv2189c,d | Transcriptional regulator, AraC family (smart00342) |
| Xcv0232c,d | Two-component system, sigma 54-dependent response regulator (cd00009, cd00156, COG2204, pfam02954) | |
| Xcv0115c,d | Two-component system, regulatory protein (cd00156, COG2197, smart00421) | |
| Xcv4439 | Putative DNA binding protein (COG3093) | |
| Xcv1455c | Putative transcriptional regulator, padR- like family (pfam03551) | |
| Xcv3621c | Ribosome-associated GTPase (COG1162, cd01854) | |
| Xcv0645 | Putative signal transduction (cd01949) | |
| Mobile genetic element | Xcv3676 | xerD; tyrosine recombinase (cd00798, COG4974) |
| Metabolism | ||
| Macromolecular metabolism | Xcv4356c,g | bga2; degradation of polysaccharides (pfam02836, pfam02837) |
| Xcv3342c | rluD; ribosomal large subunit pseudouidine synthase D (cd00165, cd02869, COG0564) | |
| Xcv4443c | recC; exodeoxyribonuclease V gamma chain (COG0564) | |
| Xcv4441d | recD, exodeoxyribonuclease V alpha chain (COG0507, pfam01443) | |
| Xcv0477 | nucH; nuclease (COG2374, pfam03372) | |
| Xcv3652 | Putative glycosyltransferase (COG1216) | |
| Xcv4333c | aguA; alpha glucuronidase (COG3661, pfam03648, pfam07477, pfam07488) | |
| Biosynthesis of small molecules | Xcv2047c,f | NUDIX hydrolase family protein (pfam00293) |
| Intermediary metabolism | Xcv4285 | 2-Hydroxyhepta-2,4-diene-1,7-dioate isomerase (COG0179) |
| Xcv1627 | sseA; thiosulfate sulfurtransferase (cd01448, cd01449) | |
| Xcv0399 | Short chain dehydrogenase (pfam00106) | |
| Xcv0264d | mls; putative malate synthase (cd00727) | |
| Xcv4482d | phoN; acid phosphatase (cd03397) | |
| Xcv3051c | Dienelactone hydrolase (COG0412) | |
| Xcv0181c,f,g | Putative hydrolase (COG0429, COG0596) | |
| Xcv3809c | dadA2; d-amino acid dehydrogenase small subunit (COG0665, COG3349) | |
| Xcv3586 | leuC; isopropylmalate isomerase large subunit (cd01583) | |
| Xcv0713 | Putative alcohol dehydrogenase class III (COG1062, pfam00107, pfam08240) | |
| Secondary metabolite | Xcv3302 | cypX; conserved hypothetical cytochrome P-450 (COG2124) |
| Cell structure | Xcv0246 | Ddl; d-alanylalanine synthetase (pfam01820) |
| Hypothetical | ||
| Putative secreted | Xcv1882 | Putative secreted (COG1409) |
| Xcv2059 | Putative secreted (no conserved domain found) | |
| Xcv4456 f | Putative secreted (no conserved domain found) | |
| Xcv2861c,f | Putative secreted protein (COG3794) | |
| Xcv0561c | Putative secreted protein (no conserved domain found) | |
| Xcv1554g | Putative secreted (no conserved domain found) | |
| Xcv0132 | Putative secreted protein (no conserved domain found) | |
| Xcv0168 | Putative secreted protein (COG0520) | |
| Xcv0536 | Putative secreted lipase (pfam03585) | |
| Xcv0048f | Putative secreted protein (COG1748) | |
| Membrane associated | Xcv4437f | Putative aminopeptidase precursor mechanism (cd00989, COG3975, pfam05299) |
| Xcv0050c,f | TonB-dependent outer membrane receptor (cd01347) | |
| Conserved | Xcv4314c | Conserved hypothetical (no conserved domain found) |
| Hypothetical | Xcv3974c | Hypothetical protein (no conserved domain found) |
| Xcv0299 | Hypothetical protein (no conserved domain found) | |
| Xcv0085c,e | Hypothetical protein (no conserved domain found) | |
| Xcv4415c,e | Hypothetical protein (pfam05016) | |
| Xcv4120 | Hypothetical protein (no conserved domain found) | |
| Xcv3588 | Putative sensor protein (cd01949, COG3292 pfam03739) | |
| Xcv4207c,f | Putative phosphatase, possible role in heat shock or osmotic stability (cd00143) | |
| Xcv0586c | Hypothetical protein (COG3660, pfam06258) | |
| Xcv0786 | Hypothetical protein (no conserved domain found) | |
| Xcv1239 | Hypothetical protein (no conserved domain found) | |
| Xcv4050 | Hypothetical protein (no conserved domain found) |
Function assignment is mainly based on the ONSA/FAPESP/Brazil Xanthomonas genome project.
Gene numbers and names are according to the sequence of X. campestris pv. vesicatoria strain 85-10 (GenBank accession no. AM039952) (45).
Gene is part of an operon.
Knockout mutant generated for this gene.
Isolated 4 days after plant inoculation.
Isolated 8 days after plant inoculation.
Imperfect PIP BOX (TTCG-N17-TTCG) identified within a 1-kb region upstream of the putative translation start site.
Results of the screen.
In total, 384 in vivo resolved clones (i.e., Kms and lacking sacB) were obtained. Of these, 120 were eliminated because they contained multiple plasmid insertions as determined by Southern blot analysis (not shown). The rest were analyzed to determine the sequence of the region cloned upstream of the tnpR gene by excising the integrated plasmids from the chromosome of each clone as described above. Sequences of the fragments fused to the tnpR gene were determined using the RtnpRIVET primer matching the 5′ region of the tnpR gene and analyzed by BLAST (4). We were able to determine the sequence of 219 fragments, of which 80 were siblings and 80 were fragments whose annotated genes are not predicted to be transcribed in the same orientation as in the tnpR-uidA transcriptional fusion. Sixty-one fragments were unique and were inserted in the orientation expected from the X. campestris pv. vesicatoria genome annotation. These unique sequences were assigned to functional categories, based largely on the gene assignments made by the ONSA/FAPESP/Brazil Xanthomonas Genome Project (http://cancer.lbi.ic.unicamp.br/xanthomonas). Based on these categories, 21 of the sequences matched hypothetical proteins, 22 were homologous to proteins with metabolic functions, 7 were homologous to regulatory proteins, 1 was homologous to a previously identified virulence protein, 6 were homologous to transport proteins, 1 was homologous to a mobile genetic element, and 1 matched a cell structure protein (Table 3). Two sequences had no matches in the public database. However, since it is not known yet how these genes function in the X. campestris pv. vesicatoria-tomato interaction, the category assignment is merely speculative, at least for some of the genes. For instance, several genes assigned to different functional categories were found to contribute to the virulence of other animal or plant pathogenic bacteria and could have been placed in the virulence category. Two such genes, categorized under transport, encode for two types of pumps that efflux antimicrobial compounds: Xcv4089 and Xcv1541 encode an ATP-binding cassette (ABC) transporter of antimicrobial peptides and an inner membrane component of the tripartite resistance-nodulation-cell division (RND) type multidrug efflux transporter similar to mexF, respectively. While multidrug efflux ABC transporters were shown to contribute to virulence of fungal phytopathogens (15, 41), RND multidrug transporters were shown to contribute to the virulence of several phytopathogens, such as Erwinia chrysanthemi in chicory, and Erwinia amylovora in apple rootstock (6, 12). Furthermore, genes categorized under general metabolism could also be considered for other functionalities. For example, recC (Xcv4443) and recD (Xcv4441), both encoding exoribonucleases, could be induced for nutritional purposes (breakdown of macromolecules). Otherwise, these genes could be induced in response to stress as a means of DNA repair. Also, sseA (Xcv1627) encoding a thiosulfate sulfotransferase may be involved in the formation of thiosulfate and/or cyanide detoxification. However, since sulfated molecules are involved in intercellular interactions, they could be involved in host-pathogen interactions, as has been suggested for raxST in Xanthomonas oryzae pv. oryzae (14).
Quantitative analysis of GUS expression of selected IVX strains.
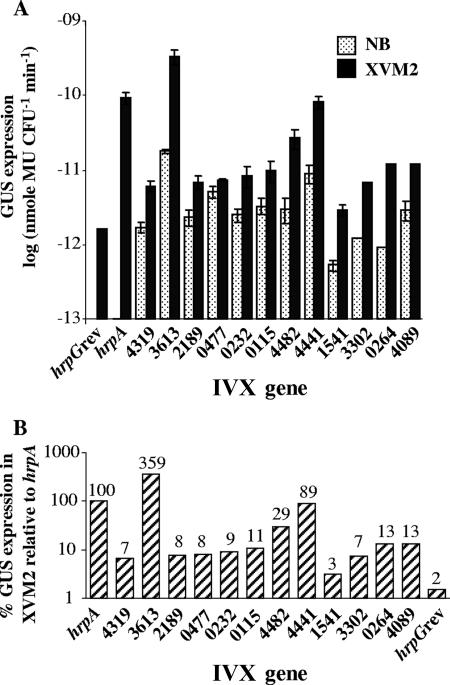
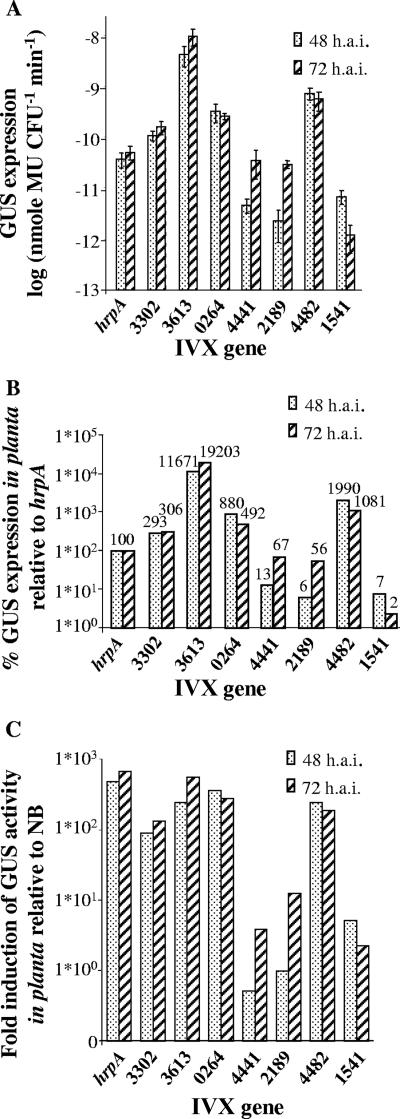
A selected group of genes from different functional categories was further analyzed. The originally isolated strains containing the tnpR-uidA transcriptional fusions were used to reconfirm and to better determine their induction profiles. For this purpose, GUS expression was first quantified in rich (NB) and in minimal (XVM2) media. As shown in Fig. 4A, all strains showed GUS expression profiles of stronger expression in XVM2 than in NB medium. Further, a comparison with GUS expression resulting from induction driven by the hrpA promoter (strain ResPhrpA) showed GUS expression as high as 356% and as low as only 3% of that of ResPhrpA in minimal medium (Fig. 4B), the latter indicating either that the method is very sensitive to low levels of induction or that these promoters could be induced by some plant-derived signal absent in the minimal medium.
FIG. 4.
GUS expression analysis of selected X. campestris pv. vesicatoria IVX genes. Genes are denoted based on their corresponding gene numbers in the sequence of X. campestris pv. vesicatoria strain 85-10 (only the number designations are shown on the x axis, without the X. campestris pv. vesicatoria prefix, Xcv). (A) GUS expression following growth in rich (NB) or minimal (XVM2) medium. Data represent the average and standard errors of at least two independent experiments. (B) Percentage of GUS expression relative to the hrpA gene, as measured in the ResPhrpA control strain.
A set of IVX strains was selected for in vivo analysis. Bacterial suspensions at 105 CFU/ml were syringe infiltrated into tomato leaves, and samples were taken 24, 48, and 72 hpi. At this inoculum concentration, samples collected after 24 h gave results that were below the sensitivity level of detection. For most tested strains, the induction after 72 h was stronger than after 48 h (Fig. 5A). However, Xcv1541 showed a stronger induction after 48 h than after 72 h. It is possible that this transient induction would not have been noticed without the irreversible capture by TnpR. In planta GUS expression of the IVX genes was compared to that of the hrpA gene as measured in the ResPhrpA strain (Fig. 5B). The induction of the hrpA promoter, as well as of some of the tested IVX genes stayed on the same order of magnitude as observed before for expression in minimal medium. However, some of the genes, such as Xcv4482, Xcv3302, Xcv0264, and Xcv3613, were induced at higher levels than hrpA in planta but not in minimal medium, suggesting that a plant-derived signal, absent in the medium, may be enhancing their expression. Finally, all tested IVX genes showed a greater induction in planta 48 or 72 hpi than in rich medium, ranging from less than 1 to 2 orders of magnitude, thus validating the RIVET approach and confirming its sensitivity to low levels of induction (Fig. 5C).
FIG. 5.
(A) GUS expression following growth in planta for 48 and 72 hpi. Data shown are means and standard errors of three independent assays. (B) Percentage of GUS expression in planta relative to the hrpA gene, as measured in the ResPhrpA control strain. (C) Increase in induction (n-fold) of GUS activity in planta relative to rich medium. The in planta GUS activity at 48 or 72 hpi. was divided by the activity in rich medium (NB) to give relative induction values. IVX genes are denoted based on their corresponding gene numbers in the sequence of X. campestris pv. vesicatoria strain 85-10 (without the Xcv prefix). hai, hours after inoculation.
Mutational analysis.
Some of the genes were chosen for mutational analysis to determine their contribution to the virulence of X. campestris pv. vesicatoria. Genes were selected based on the involvement of genes with similar functions in virulence of other pathogenic bacteria or for other reasons specified below. In total, 13 genes were mutated (Table 3). Of the genes falling within the category of transport, the following were knocked out: Xcv4319, a tatC homologue; Xcv4089, encoding an ABC transporter of antimicrobial peptides; Xcv3613, a citH homologue encoding a citrate-H+ symporter; and Xcv1541, Xcv1540, and Xcv1543, components of a RND-type multidrug efflux transporter, similar to mexF, mexE, and oprN, respectively. The mexE and oprN homologues flank the identified mexF homologous gene and were interrupted for further investigation of the role of this RND efflux pump.
The tatC homolog encodes a translocase, which is the most conserved component of the Tat (twin arginine translocation) protein translocation pathway. This pathway enables Sec-independent translocation of fully folded proteins across the cytoplasmic membrane or their insertion into the membrane (7). In Pseudomonas aeruginosa the Tat system was shown to be important for the virulence of this pathogen on mouse (32). Interestingly, in a recent IVET study aimed at identifying genes induced in Ralstonia solanacearum during growth in tomato, four putative transmembrane proteins that are predicted substrates for the Tat secretion pathway were identified (10). Genes related to multidrug efflux were interrupted based on findings, mentioned previously in this report, on the role of these multidrug efflux pumps in virulence. The citH homologue (Xcv3613) is predicted to have a C4-dicarboxylate transport activity and has a 61% identity to the characterized citrate transporter of Bacillus subtilis, CitN (accession number P42308) (9). To the best of our knowledge, there are no studies that examined the role of citrate transporters in virulence. However, this gene was interrupted for several reasons: (i) its promoter showed the strongest induction in minimal medium of all tested IVX strains and a higher level of induction than the hrpA promoter (Fig. 4); (ii) organic acids play numerous roles in rhizosphere-microbial interactions (26); (iii) in different Pseudomonas species, tricarboxylic acid cycle (TCA) intermediates were shown to be the preferred carbon source over carbohydrates (35); (iv) different IVET studies, looking into bacterial interaction with diverse niches—R. solanacearum in the xylem, E. amylovora in fruit tissue, Pseudomonas syringae pv. syringae in the phylosphere, and Pseudomonas fluorescens in the rhizoshere—identified genes involved in C4-dicarboxylate transport (10, 29, 36, 54). In contrast, these genes were not detected during bacterial interaction with animal hosts, suggesting that possibly these genes are specific to plant-microbial interactions (10).
Of the genes falling under metabolism, Xcv0264, Xcv4441, and Xcv4482, homologous to malate synthase, recD, and phoN, respectively, were mutated. Malate synthase, an enzyme in the glyoxylate cycle, was shown to be important for virulence and persistence in several animal and plant pathogens (47). The recD homologue is found within close proximity to several other genes identified in this screen and to xopQ, a gene encoding a type III secretion effector (39). The phoN homolog encodes an acidic phosphatase. In addition, Xcv3302, a cypX homologue that encodes an enzyme that synthesizes a secondary metabolite, was interrupted. This gene product is 35% identical to cytochrome P450 107A1 from Saccharopolyspora erythraea, involved in the synthesis of erythromycin. Finally, three of the regulatory genes identified, Xcv2189, Xcv0232, and Xcv0115, were also knocked out.
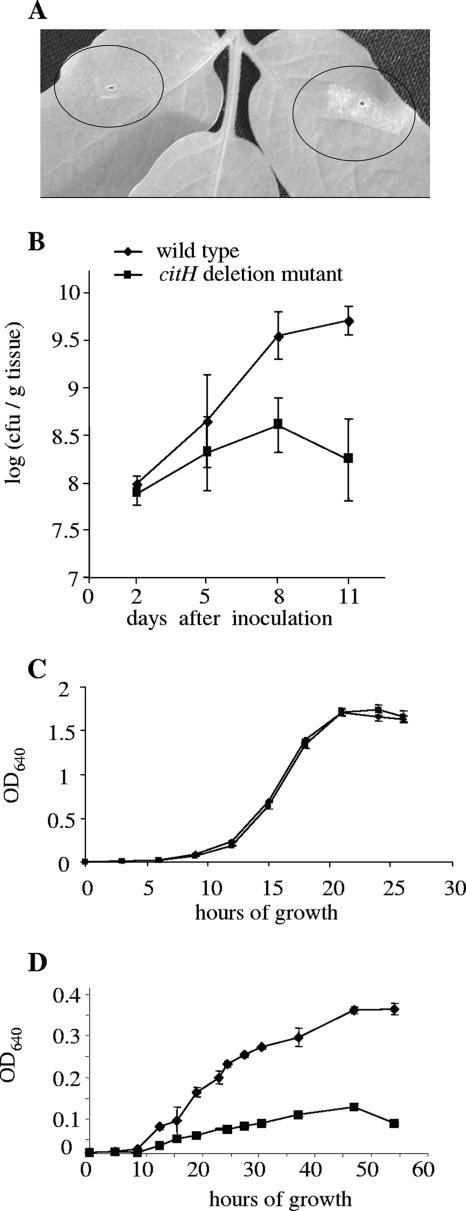
Knockout strains were analyzed for their ability to grow in planta and for their ability to elicit disease symptoms. Of the tested mutants, the citH deletion mutant was shown to be significantly impaired in its ability to grow in planta and cause disease symptoms compared to the wild-type strain. We observed a ∼0.5- to 1.5-log difference between the mutant and the wild-type strain, 5 to 11 dpi (Fig. 6B) and a significant reduction in its ability to cause disease symptoms (Fig. 6A). The citH mutant did not differ from the wild-type strain in its ability to grow in rich medium (Fig. 6C) or in minimal medium supplemented with l-malic acid, succinic acid, fructose, or sucrose as the sole carbon sources (not shown); however, this mutant was unable to grow in minimal medium with citrate as the sole carbon source (Fig. 6D). The rest of the mutants were not attenuated in their ability to grow in the plant following dip inoculation, a phenomenon that was also reported in other genomewide in vivo screening methods. We did observe, however, a trend by which the cypX (Xcv3302) deletion mutant, the malate synthase mutant (Xcv0264), and the mutant defective in the transcriptional regulator encoded by Xcv2189 induced fewer and smaller lesions than the wild-type strain following syringe infiltration at 105 CFU/ml. The former induced reduced symptoms in 80 to 90% of the inoculation sites, the malate synthase mutant displayed reduced symptoms in 70 to 75% of the inoculation sites, and the transcriptional regulator mutant displayed reduced symptoms in 35 to 55% of the inoculation sites in two independent experiments (with approximately 50 inoculation sites per strain per experiment).
FIG. 6.
Symptom development and growth of the citH mutant strain compared to the wild-type strain on tomato leaflets. (A) Symptom development 9 days after syringe infiltration at 105 CFU/ml. Circled areas show the citH mutant strain (left) and wild-type strain (right). (B) In planta growth of the citH mutant strain (▪) compared to the wild-type strain (⧫) following dip inoculation at 105 CFU/ml. Data (means and standard errors) from one experiment of two with similar results are shown. (C) Growth of the citH mutant strain (▪) compared to the wild-type strain (⧫) in rich medium (NB). (D) Growth of the citH mutant strain (▪) compared to the wild-type strain (⧫) in modified M9 minimal medium supplemented with 10 mM sodium citrate as the sole carbon source. OD640, optical density at 640 nm.
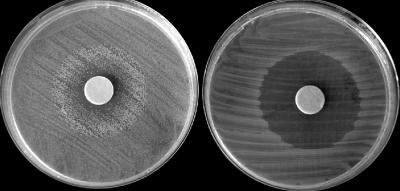
The mexF mutant was also tested for its ability to grow in the presence of Cm and nalidixic acid. Compared to the wild-type strain, this mutant displayed a similar inhibition zone, of a radius of about 1.5 cm after 24 h of growth on NA with 300 μg/μl nalidixic acid or 100 μg/μl Cm. However, after 48 h, the inhibition zone of the mexF deletion mutant remained unchanged, while colonies of the wild-type strain developed toward the paper discs, resulting in reduced inhibition zones of 0.5 and 0.9 cm around nalidixic acid and Cm discs, respectively (not shown). This trend continued over the course of 96 h for both antibiotics (shown for nalidixic acid in Fig. 7).
FIG. 7.
Inhibition zone around a disc containing 3 mg of nalidixic acid 96 h after growth on NA medium at 28°C. The disks were added immediately after cells were streaked on the medium. Left, wild-type strain; right, the mexF mutant strain. Note that, in contrast to the mutant, the wild-type strain was able to partially overcome the inhibition by nalidixic acid.
DISCUSSION
We have tailored a RIVET approach to study the interaction between X. campestris pv. vesicatoria and tomato. This is the first RIVET method set up and applied to a phytopathogenic bacterium. Like other variations of IVET, it has the advantage of identifying genes that are induced in vivo. The main advantage of RIVET over other IVET strategies resides in its sensitivity, since it enables the detection of genes that are only slightly or transiently induced. This allowed us to broaden our knowledge of the gene machinery in action during this plant-microbe interaction. It sheds light on the various aspects of the infectious process from the pathogen's perspective: initial surviving and adapting to this hostile environment, followed by propagating and overcoming host defenses, and finally damaging the plant to gain access to its nutritional reservoirs. Understanding the broadest gene machinery is essential for understanding the entire disease process, since most of it is not governed by single essential genes but, rather, by a number of coordinated gene arrays, consisting of genes not associated with virulence per se.
The vast majority of genes found in this study delineate the different processes that allow the pathogen to survive, propagate to large cell densities, and cause disease. For instance, among the genes identified, different genes were found to encode proteins that may function to better sense and perceive the environment and to respond to it. Several genes have been identified that may play a role in sensing the extracellular milieu. Two genes, Xcv0232 and Xcv0115, encoding proteins of two-component regulatory systems have been identified in this study. These systems are ubiquitous among gram-negative and gram-positive bacteria and play a key role in sensing environmental signals and transducing them within the cell. Two-component regulatory systems regulate many biological processes, including those related to pathogenicity (11).
X. campestris pv. vesicatoria propagates in the host plant apoplast. To reach the cell densities required to conquer the host, it has to be able to adapt to the intercellular environment and to utilize the available nutritional sources. The avrBs2 gene, identified as being upregulated in this screen, encodes AvrBs2, which is a known type III-secreted virulence effector that was suggested to function in the modification of glucans to promote Xanthomonas adaptation to the apoplast in a compatible interaction (44). Other genes identified in this screen may point to the different strategies X. campestris pv. vesicatoria uses to gain access to nutrients. One strategy is by activating alternative metabolic pathways. A gene encoding malate synthase, Xcv0264, was identified in this work as being upregulated in vivo. This enzyme is part of the glyoxylate cycle, a conserved metabolic pathway that functions as a TCA shunt. This shunt allows the conversion of C2 compounds into glucose through the gluconeogenic pathway. It has been shown to be important for virulence and persistence in several animal and plant pathogens by allowing dietary adjustment in the host (47). Interestingly, the second enzyme of the glyoxylate pathway, isocitrate lyase (aceA), was found to be upregulated in Pseudomonas putida in the maize rhizosphere (37).
Another means of obtaining nutrients is through the degradation of macromolecules. A bga2 homologue (Xcv4356), encoding an enzyme with polysaccharide-degrading activity, was identified. Also, genes encoding the exonucleases recD and recC (Xcv4441 and Xcv4443, respectively) and the endonuclease nucH (Xcv0477) were identified and could be involved in metabolic adaptation to the host environment. Finally, the pathogens can use carbon sources in their available form. In this study, the gene citH encoding a citrate transporter was identified. We are unaware of studies dealing with concentrations of organic acids in the apoplast of tomato leaves. It is known, however, that citrate is transported from the roots with the transpiration stream, and when transpiration stops, citrate accumulates in the xylem, suggesting a diurnal exchange between leaf apoplast and symplast. It is also known that citrate complexed with metal ions allows a more efficient transport of these micronutrients (26). It is unclear what role apoplastic organic acids may play in plant-microbe interactions, but in different Pseudomonas species, TCA intermediates have been shown to be the preferred carbon sources over carbohydrates (35). Genes for C4-dicarboxylate transporters have also been identified in several other IVET studies looking into bacterial interactions with diverse plant niches, such as R. solanacearum in the xylem, E. amylovora in fruit tissue, P. syringae pv. syringae in the phylosphere, and P. fluorescens in the rhizoshere (10, 29, 36, 54). It is possible that phytopathogenic bacteria have evolved to utilize the available carbon sources in their host.
Here, we show that an X. campestris pv. vesicatoria citH knockout strain was attenuated in its ability to grow in planta and cause disease symptoms compared to the wild-type strain. In contrast to the wild type, the citH mutant was unable to grow in minimal medium with citrate as the sole carbon source (Fig. 5C). Furthermore, we observed that in minimal medium with citrate as the sole carbon source, growth of the wild-type strain resulted in an increase of the medium pH. In contrast, growth of the citH mutant did not affect the pH of the medium (unpublished data). These phenotypic changes were observed despite the fact that, according to the X. campestris pv. vesicatoria genome annotation, this bacterium contains another putative citH gene in its genome (Xcv3602, encoding a protein that is 72% homologous to a citrate-H+ sympoter from Klebsiella pneumoniae). We intend to further study this mutant to determine why this mutation affects X. campestris pv. vesicatoria virulence and to understand the role of citH in the X. campestris pv. vesicatoria-tomato interaction. One possibility is that citrate serves as an important carbon source in the apoplast. However, we cannot discard the possibility that citrate uptake and utilization by the pathogen contribute to the generation of favorable conditions for the pathogen by changing the pH. It is well known that alkalinization of the host apoplast is required for successful establishment of many phytopathogenic bacteria (2, 3).
Until studies are available that show how the identified genes really function to promote disease, clues as to the function of genes will come from sequence or structural homologies and, often, from their genomic location. It is interesting that four promoters of genes within the same genomic region were identified: Xcv4441 and Xcv4443, recD and recC homologues, respectively; Xcv4437, coding for a putative aminopeptidase precursor; and Xcv4439, coding for a putative DNA binding protein. Interestingly, these genes are flanking the xopQ gene, which encodes a previously identified type III-secreted effector. xopQ is conserved among many plant pathogens and is structurally homologous to an inosine-uridine nucleoside N-ribohydrolase enzyme, a protein implicated in facilitating rescue of nucleotides from their environment. Roden et al. (39) suggested that xopQ possibly functions as a scavenging hydrolase in planta or interferes with plant cell processes by binding and sequestering nucleosides important for plant signaling and/or metabolism. The genes recC and recD encode an exodeoyribonuclease involved in DNA metabolism and repair. The putative aminopeptidase is presumably a membrane-associated protease and may be involved in signaling. The putative DNA binding protein encodes a protein with a DNA binding domain and the VapI (virulence associated protein) conserved domain predicted to function as a plasmid maintenance system antidote. A BLASTX search of this protein showed that it has 40% (32/80) identity to VapA′ of Dichelobacter nodosus, present in all virulent isolates of this animal pathogen but absent from most of the benign isolates (23). Knockout of recD in our study did not result in reduced virulence under the conditions tested. However, it would be interesting to test whether any of these xopQ flanking genes is also translocated via the type III secretion system and whether they interact with host factors to alter host response.
In response to the perceived extracellular milieu, some of the genes found in this study may function in defense against toxic plant compounds and/or endophytic microbial populations that might be competing for nutrients. For instance, Xcv4089 encodes an ABC transporter of antimicrobial peptides. In the fungal phytopathogens Gibberella pulicaris and Botrytis cinerea, multidrug efflux ABC transporters were shown to contribute to virulence by enhancing fungal tolerance to phytoalexins (15, 41). Llama-Palacios et al. (25) showed that ybiT in E. chrysanthemi encodes a putative ABC transporter that contributes to the pathogen's ability to compete against endophytic and saprophytic bacteria. Production and secretion of antimicrobial compounds is another means of reducing competition. Xcv3302, annotated as cypX, is 56% identical to a putative cytochrome P450 133B1 from Xylella fastidiosa and 35% identical to cytochrome P450 107A1 from S. erythraea, which is involved in the synthesis of erythromycin. In this study, an X. campestris pv. vesicatoria cypX deletion mutant was not impaired in its growth in planta compared to the wild-type strain but showed a reduced ability to induce symptoms following leaf infiltrations. A similar result was obtained with a gene encoding a transcriptional regulator (Xcv2189) and the gene encoding a malate synthase (Xcv0264). As stated above, these mutants did not differ in in planta growth relative to the wild-type strain. It is possible that the assays used in our study for determining in planta growth are not sensitive enough to detect subtle phenotypes although the assays were carried out with relatively low inoculum concentrations and without vacuum application. Therefore, further experiments should be performed to explore the role of these genes in the interaction between X. campestris pv. vesicatoria and its hosts.
As indicated above, Xcv1541 encodes an MexF homologue, one of the components of a tripartite RND-type multidrug efflux transporter. This type of efflux pump is required for tolerance of E. chrysanthemi and E. amylovora to plant-derived toxins (6, 12, 27). In our study, the X. campestris pv. vesicatoria mexF deletion mutant did not display a reduced virulence in comparison with the wild-type strain. As stated above, we cannot discard the possibility that the available pathogenicity assays are not sensitive enough for the detection of subtle phenotypes. Indeed, most knockout mutants generated in this study did not show reduced abilities to grow in planta and to elicit disease symptoms compared to the wild type. These results were expected and were also reported in other genomewide in vivo screening methods. One possibility is that these genes do not actually contribute significantly to virulence. Alternatively, the lack of defective phenotypes could have other explanations. Due to the multifactorial nature of the infection process of plant pathogenic bacteria (2), many virulence-associated genes contribute to a given functionality only slightly and quantitatively. Functional redundancy in the genome could be another reason. It is also possible that an apparent phenotype would have been detected with a different experimental setup or on a more suitable host or in different host developmental stages and environmental conditions. Nevertheless, preliminary studies with the X. campestris pv. vesicatoria mexF mutant suggest that this gene facilitates the efflux of antimicrobial compounds. This was evident by the ability of the wild-type strain, but not of the mexF mutant, to partially overcome the growth inhibition of the antibiotics Cm and nalidixic acid (Fig. 6).
Last, we should refer to the relatively high number of identified fusions, 80, in which the sequence trapped had considerable overlap with predicted genes on the opposite DNA strand, according to the X. campestris pv. vesicatoria genome annotation. This is not the first IVET study to detect a significant number of such sequences. However, with few exceptions, these sequences have been generally discarded as they were thought to represent false positives of the approach. Osorio and Camilli (33) were the first to refer to these kind of sequences and suggested that these “opposite” IVX fusions might represent a kind of antisense regulation, controlled by so-called cryptic promoters. Later, in an IVET study with the soil bacterium P. fluorescens, Silby and Levy (42) revealed 10 opposite IVX fusions among a total of 22 with elevated expression in the soil compared to growth in culture. To the best of our knowledge, no attempts have been made to date to investigate the biological significance of these findings.
In conclusion, we have shown the applicability of the RIVET approach to the study of the X. campestris pv. vesicatoria-tomato interaction. The genes identified in this study reflect the complexity of the infectious process. As expected, because of the multifactorial nature of virulence determinants in phytopathogenic bacteria and sensitivity limitations of available pathogenicity assays, among other factors, most of the generated knockout strains did not differ from the wild-type strain in their ability to induce symptoms or to grow in planta under the specific conditions used in this study. Nevertheless, some of the identified genes were found to contribute to the virulence of the pathogen in this interaction. In the future, RIVET can be used to expand even further the list of IVX genes of this and of other phytopathogenic bacteria. This will enrich our knowledge of the gene machinery required by phytopathogenic bacteria to grow in the plant and to promote disease and potentially provide new tools for disease management.
Acknowledgments
We thank Andrew Camilli and Barbara Kunkel for providing plasmids for this study. We also thank Guido Sessa for valuable technical advice and Marganit Levy for valuable comments during preparation of the manuscript.
This work was supported by Start-Up Grant 2003138 from the United States-Israel Binational Science Foundation.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Abu Kwaik, Y., and L. L. Pederson. 1996. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol. Microbiol. 21:543-556. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astua-Monge, G., G. V. Minsavage, R. E. Stall, M. J. Davis, U. Bonas, and J. B. Jones. 2000. Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol. Plant-Microbe Interact. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 6.Barabote, R. D., O. L. Johnson, E. Zetina, S. K. San Francisco, J. A. Fralick, and M. J. San Francisco. 2003. Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J. Bacteriol. 185:5772-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 8.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44:73-88. [DOI] [PubMed] [Google Scholar]
- 9.Boorsma, A., M. E. van der Rest, J. S. Lolkema, and W. N. Konings. 1996. Secondary transporters for citrate and the Mg2+-citrate complex in Bacillus subtilis are homologous proteins. J. Bacteriol. 178:6216-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. G., and C. Allen. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641-1660. [DOI] [PubMed] [Google Scholar]
- 11.Burdman, S., Y. Shen, S. W. Lee, Q. Xue, and P. Ronald. 2004. RaxH/RaxR: a two-component regulatory system in Xanthomonas oryzae pv. oryzae required for AvrXa21 activity. Mol. Plant-Microbe Interact. 17:602-612. [DOI] [PubMed] [Google Scholar]
- 12.Burse, A., H. Weingart, and M. S. Ullrich. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant-Microbe Interact. 17:43-54. [DOI] [PubMed] [Google Scholar]
- 13.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.da Silva, F. G., Y. Shen, C. Dardick, S. Burdman, R. C. Yadav, A. L. de Leon, and P. C. Ronald. 2004. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant-Microbe Interact. 17:593-601. [DOI] [PubMed] [Google Scholar]
- 15.Fleissner, A., C. Sopalla, and K. M. Weltring. 2002. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol. Plant-Microbe Interact. 15:102-108. [DOI] [PubMed] [Google Scholar]
- 16.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 18.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 19.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 20.Jones, J. B., H. Bouzar, R. E. Stall, E. C. Almira, P. D. Roberts, B. W. Bowen, J. Sudberry, P. M. Strickler, and J. Chun. 2000. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 50:1211-1219. [DOI] [PubMed] [Google Scholar]
- 21.Jones, J. B., G. H. Lacy, H. Bouzar, R. E. Stall, and N. W. Schaad. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27:755-762. [DOI] [PubMed] [Google Scholar]
- 22.Jones, J. B., R. E. Stall, and H. Bouzar. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36:41-58. [DOI] [PubMed] [Google Scholar]
- 23.Katz, M. E., C. L. Wright, T. S. Gartside, B. F. Cheetham, C. V. Doidge, E. K. Moses, and J. I. Rood. 1994. Genetic organization of the duplicated vap region of the Dichelobacter nodosus genome. J. Bacteriol. 176:2663-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel, B. N., A. F. Bent, D. Dahlbeck, R. W. Innes, and B. J. Staskawicz. 1993. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5:865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llama-Palacios, A., E. Lopez-Solanilla, and P. Rodriguez-Palenzuela. 2002. The ybiT gene of Erwinia chrysanthemi codes for a putative ABC transporter and is involved in competitiveness against endophytic bacteria during infection. Appl. Environ. Microbiol. 68:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Bucio, J., M. F. Nieto-Jacobo, V. V. Ramirez-Rodriguez, and L. Herrera-Estrella. 2000. Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 160:1-13. [DOI] [PubMed] [Google Scholar]
- 27.Maggiorani Valecillos, A., P. Rodriguez Palenzuela, and E. Lopez-Solanilla. 2006. The role of several multidrug resistance systems in Erwinia chrysanthemi pathogenesis. Mol. Plant-Microbe Interact. 19:607-613. [DOI] [PubMed] [Google Scholar]
- 28.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 29.Marco, M. L., J. Legac, and S. E. Lindow. 2003. Conditional survival as a selection strategy to identify plant-inducible genes of Pseudomonas syringae. Appl. Environ. Microbiol. 69:5793-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrell, D. S., and A. Camilli. 2000. Detection and analysis of gene expression during infection by in vivo expression technology. Philos. Trans. R Soc. Lond. B 355:587-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osorio, G., and A. Camilli. 2003. Hidden dimensions of Vibrio cholerae pathogenesis. ASM News 69:396-401. [Google Scholar]
- 34.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 35.Rahme, L. G., M. N. Mindrinos, and N. J. Panopoulos. 1992. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 174:3499-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 37.Ramos-Gonzalez, M. I., M. J. Campos, and J. L. Ramos. 2005. Analysis of Pseudomonas putida KT2440 gene expression in the maize rhizosphere: in vivo expression technology capture and identification of root-activated promoters. J. Bacteriol. 187:4033-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rediers, H., P. B. Rainey, J. Vanderleyden, and R. De Mot. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden, J. A., B. Belt, J. B. Ross, T. Tachibana, J. Vargas, and M. B. Mudgett. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA 101:16624-16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schoonbeek, H., G. Del Sorbo, and M. A. De Waard. 2001. The ABC transporter BcatrB affects the sensitivity of Botrytis cinerea to the phytoalexin resveratrol and the fungicide fenpiclonil. Mol. Plant-Microbe Interact. 14:562-571. [DOI] [PubMed] [Google Scholar]
- 42.Silby, M. W., and S. B. Levy. 2004. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J. Bacteriol. 186:7411-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 44.Swords, K. M., D. Dahlbeck, B. Kearney, M. Roy, and B. J. Staskawicz. 1996. Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 178:4661-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Buttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klosgen, T. Patschkowski, C. Ruckert, O. Rupp, S. Schneiker, S. C. Schuster, F. J. Vorholter, E. Weber, A. Puhler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 47.Vereecke, D., K. Cornelis, W. Temmerman, M. Holsters, and K. Goethals. 2002. Versatile persistence pathways for pathogens of animals and plants. Trends Microbiol. 10:485-488. [DOI] [PubMed] [Google Scholar]
- 48.Wengelnik, K., C. Marie, M. Russel, and U. Bonas. 1996. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9:704-712. [DOI] [PubMed] [Google Scholar]
- 50.Whalen, M. C., R. W. Innes, A. F. Bent, and B. J. Staskawicz. 1991. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, S., N. T. Perna, D. A. Cooksey, Y. Okinaka, S. E. Lindow, A. M. Ibekwe, N. T. Keen, and C. H. Yang. 2004. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol. Plant-Microbe Interact. 17:999-1008. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 53.Yu, Z. H., J. F. Wang, R. E. Stall, and C. E. Vallejos. 1995. Genomic localization of tomato genes that control a hypersensitive reaction to Xanthomonas campestris pv. vesicatoria (Doidge) dye. Genetics 141:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, Y., S. E. Blumer, and G. W. Sundin. 2005. Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187:8088-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]