Abstract
The genome of the gastric pathogen Helicobacter pylori contains a homologue of the gene luxS, which has been shown to be responsible for production of the quorum-sensing signal autoinducer 2 (AI-2). We report here that deletion of the luxS gene in strain G27 resulted in decreased motility on soft agar plates, a defect that was complemented by a wild-type copy of the luxS gene and by the addition of cell-free supernatant containing AI-2. The flagella of the luxS mutant appeared normal; however, in genetic backgrounds lacking any of three flagellar regulators—the two-component sensor kinase flgS, the sigma factor σ28 (also called fliA), and the anti-sigma factor flgM—loss of luxS altered flagellar morphology. In all cases, the double mutant phenotypes were restored to the luxS+ phenotype by the addition of synthetic 4,5-dihydroxy-2,3-pentanedione (DPD), which cyclizes to form AI-2. Furthermore, in all mutant backgrounds loss of luxS caused a decrease in transcript levels of the flagellar regulator flhA. Addition of DPD to luxS cells induced flhA transcription in a dose-dependent manner. Deletion of flhA in a wild-type or luxS mutant background resulted in identical loss of motility, flagella, and flagellar gene expression. These data demonstrate that AI-2 functions as a secreted signaling molecule upstream of FlhA and plays a critical role in global regulation of flagellar gene transcription in H. pylori.
Many bacterial species regulate aspects of their behavior through an intercellular signaling system called quorum sensing (4). Bacteria produce small molecules, termed autoinducers (AIs), that travel across the bacterial membrane and accumulate in the environment. Increases in cell density result in corresponding increases in AI concentration in the extracellular environment and cell cytoplasm. At threshold AI concentrations, the interaction between the AI and its transduction machinery results in the coordinated transcription of target genes required for specific group behaviors (52).
A quorum-sensing system that is widespread across the bacterial kingdom and has been proposed to function as a “bacterial Esperanto,” uses a collection of molecules collectively called AI-2 (4). AI-2-dependent processes include biofilm formation, motility, and type III secretion (18). AI-2 is synthesized by the LuxS protein (47), and luxS genes have been identified in at least 250 bacterial genomes to date (50). LuxS functions to cleave S-ribosylhomocysteine, an intermediate in the S-adenosylmethionine (SAM) pathway, into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) (40). Through processes of dehydration and cyclization, DPD exists in equilibrium with several molecules, some or all of which function as the molecular signal AI-2 (40). AI-2 has been extensively studied in Vibrio harveyi, where it has been shown to regulate bioluminescence and other traits (5, 20, 21, 28), and a V. harveyi reporter strain that expresses bioluminescence in response to the addition of AI-2 has become the standard method for detecting AI-2 production by other bacterial species (3).
Although AI-2 production is common among bacterial species, it remains controversial as to how many of these bacteria utilize AI-2 as a signaling molecule, as opposed to producing it as a metabolic by-product of the SAM pathway (15, 50). One such controversial case is Helicobacter pylori, a gram-negative pathogen of the human stomach. Two independent groups demonstrated that H. pylori secretes AI-2 into its extracellular environment by a luxS-dependent mechanism (19, 25). A subsequent study reported that an H. pylori luxS mutant exhibited reduced production of the flagellin protein FlaA and reduced expression of an flaA transcriptional reporter construct (29). In addition, an H. pylori luxS mutant is reported to form biofilms more efficiently than wild-type cells (10). Recently, two groups reported that mutation of the luxS genes in H. pylori strains SS1 and TK1402 exhibited motility defects on soft agar plates and colonization defects in mouse and gerbil models of infection, respectively (27, 37). However, Lee et al. found that a luxS mutant in a third H. pylori strain, X47, did not exhibit these defects. Based on the phenotypic variability they observed between strains, these authors concluded that luxS mutant phenotypes were the result of fitness defects due to the disruption of the SAM metabolic pathway, and they argued that AI-2 was unlikely to function as a quorum-sensing molecule in H. pylori. Evidence for a signaling function for AI-2 would require a demonstration that purified extracellular AI-2 is sufficient to reverse a luxS-associated phenotype. To date, besides the demonstration that AI-2-containing cell-free supernatant could modestly increase flaA reporter gene expression (29), no such evidence has been reported.
The possibility that H. pylori flagellar gene expression is regulated by an extracellular signaling molecule is intriguing in light of a long-standing proposal that H. pylori may regulate the composition of its flagellar filament in response to environmental cues (44). Unlike the regulation of flagellar gene expression in the model organisms Salmonella and Caulobacter crescentus, in which a single master regulator controls transcription of an entire hierarchy of genes in response to signals such as growth phase, in H. pylori the top-tier regulators of flagellar gene transcription, as well as chemotaxis- and motor protein-encoding genes, are constitutively expressed under the control of the housekeeping sigma factor, σ80, and are referred to as class 1 genes (36). Class 2 genes are controlled by σ54 (RpoN) and contain “middle” structural flagellar genes that encode components of the rod, hook, and sheath and the minor flagellin FlaB. Class 3 genes are controlled by σ28 (FliA) and the anti-sigma factor FlgM (11, 24), and this group contains “late” flagellar structural genes such as the major flagellin FlaA. A final intermediate class, including both structural and regulatory genes, is controlled by both σ54 and σ28 (36). The closest candidate for a master regulator in this hierarchy is FlhA, a component of the basal body export apparatus, that is required for flagellum formation and for expression of most class 2 and class 3 genes and the intermediate class of flagellar genes (36, 41). A possible point through which environmental signals could be channeled into the flagellar transcriptional hierarchy is the unconventional cytoplasmic sensor kinase FlgS (HP0244), which coregulates σ54-dependent genes of class 2 and the intermediate classes (6, 36).
Motility is an important prerequisite for H. pylori colonization of the stomach, as has been demonstrated for mutants lacking flagellar genes in multiple animal models (reviewed in reference 38). Thus, dissecting regulation of motility in H. pylori is important for understanding this pathogen's virulence strategies. By characterizing flagellar morphology and flagellar gene transcription in wild-type, luxS mutant, and AI-2-supplemented cells lacking key flagellar regulators, we demonstrate that AI-2 signaling influences the flagellar regulon through modulation of the transcription of the top-tier regulator gene flhA. Our data provide strong support for the model that H. pylori uses AI-2 as an intercellular signaling molecule.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori strain G27 (14) was used in this study as the wild-type strain from which all mutants were derived, as listed in Table 1. All strains were maintained on blood agar plates consisting of Columbia agar (Difco) and 5% defibrillated horse blood (Hemostat) supplemented with 0.02 mg ml−1 β-cyclodextrin (Sigma), 8 mg ml−1 amphotericin B (Sigma), and 20 μg ml−1 vancomycin (Sigma) and incubated at 37°C and 10% CO2. Liquid medium (hereafter, BB medium) for H. pylori growth consisted of filter-sterilized Brucella broth (Difco) supplemented with 10% fetal bovine serum (Gibco) and 20 μg ml−1 vancomycin (Sigma). Liquid cultures were grown in 50-ml conical tubes with loosened lids (BD Falcon) and shaking at 37°C in anaerobic jars (Oxoid) with CampyGen microaerobic sachets (Oxoid). Selective plates were made with 10 μg ml−1 kanamycin (Fisher), 0.08 M sucrose, or 18 μg ml−1 metronidazole (Sigma).
TABLE 1.
Strains used in this study
| H. pylori strain | Description | Reference or source |
|---|---|---|
| Wild type | G27 | 14 |
| luxS strain | luxS::kan sacB | This study |
| ΔluxS strain | Deletion of HP0105 | This study |
| luxS* strain | rdx::luxS and deletion of luxS | This study |
| fliA strain | fliA::kan | This study |
| fliA ΔluxS strain | fliA::kan and deletion of luxS | This study |
| flgS strain | flgS::kan | This study |
| flgS ΔluxS strain | flgS::kan and deletion of luxS | This study |
| flgM strain | flgM::kan | This study |
| flgM ΔluxS strain | flgM::kan and deletion of luxS | This study |
| flhA strain | flhA::kan | This study |
| flhA ΔluxS strain | flhA::kan and deletion of luxS | This study |
Construction of the ΔluxS, luxS*, and flagellar mutant strains.
All primers used for strain construction are given in Table 2. The luxS gene (HP0105) was PCR amplified using the primers LuxSSpeR and LuxSEcoF and ligated into the TOPO vector using a TOPO TA cloning kit (Invitrogen Life Technologies, Carlsbad, CA) to create the vector pBR003. The primers TopoXbaF and TopoBlgR were used to amplify outward from HP0105 and around pBR003. A kan-sacB construct (12) encoding kanamycin resistance (Kanr) and sucrose sensitivity (Sucs) was PCR amplified using the primers FBglIIpKan+ and PFA3 (12). The pBR003 and Kan-SacB PCR products were digested and ligated together to make the vector pBR007. pBR007 was purified and used to disrupt the HP0105 locus in strain G27 through natural transformation and selection of Kanr colonies. The resulting luxS::kan-sacB strain was verified by PCR and sequencing of the genomic locus. To create the ΔluxS deletion strain, a deletion construct consisting of 265 bp immediately 5′ of the HP0105 open reading frame fused to 255 bp 3′ of HP0105 was created by PCR amplifying these sequences from G27 genomic DNA and then stitching them together in a subsequent PCR using primers listed in Table 2. This PCR product was used to transform the luxS::kan-sacB mutant strain, and Sucs colonies were isolated and validated as above. The luxS gene is not located in an apparent operon in either of the two H. pylori strains for which the genome has been sequenced (1, 49), and thus the ΔluxS deletion is unlikely to have polar effects.
TABLE 2.
Primers used for cloning and RT-PCR
| Primer | Sequence (5′-3′) |
|---|---|
| LuxSSpeR | AAAATCATTCCCCAAACCAATGATCAGGG |
| LuxSEcoF | CCGGAATTCAGAGCAAGCGTTCGCTAAAA |
| TopoXbaF | GCTCTAGACTGAGATCATCCGCAACCAT |
| TopoBlgR | GAAGATCTTTGGCATGTCCATGTGATCT |
| FBglIIpKan+ | GAAGATCTTCCCGGGCGAACCATTTGAGGTGA |
| PFA3 | GCTCTAGACCCGGGTATAAGCCCATTTTCATGC |
| FLS2Bam | CGGGATCCGAGGCGGTTGCTTTTGTAGA |
| LuxS3AB | CGACATAACTGGCATGGTTGGCGAACGCTTGCTCTAAA |
| RLS2Xho2 | TTTATCTTCTCGAGCGCCCATGAATGTCTGAAGT |
| RdxA1F | TAAACGAGCGCCATTCTTG |
| RdxA1LR | GATTAAAAGCGCTTCAGCGTGAGGCGGTTGCTTTTGTAGA |
| RdxA2LF | CTTCAGACATTCATGGGCGATTCAACCACAGCATGCAAA |
| RdxA2R | CGATCAAGCATGCGATTTTA |
| FliaAF | TGCTTCGTTTCAACCATGAG |
| FliaAK | AAGCAACAACACCACCATCACAGGAAACAGCTATGACA |
| FliaBK | CTGGCCGTCGTTTTACTCGCGCATTTCTCAAATC |
| FliaBR | ACTCACACGATTAGGGCGTTT |
| HP0244AF | CGATGAGATTTATGCGTCCA |
| HP0244AK | CCTTATTTGAAGCGTTCCCACAGGAAACAGCTATGAC |
| HP0244BK | ACTGGCCGTCGTTTTACGAGAGCGAACAGGGTCAA |
| HP0244BR | GCTAACCCTAGGCCGTTACC |
| FlgMAF | GTTGTGGTTGCTCATGTTCG |
| FlgMAK | AATGCCGTTTCTTCTCTTGCAGGCAACAGCTATGA |
| FlgMBK | ACTGGCCGTCGTTTTACTCTCACAAAATGGCAAAGGA |
| FlgMBR | TCCTAACTTCATGATTGA |
| FlhAIntF | CCTAGTGCGGTGAGCGATATTATC |
| FlhAIntR | GCTCACGCTAAATTCCCCAA |
| UreAIntF | AAAGACTGCGGCTGAATTG |
| UreAIntR | CCATCACATCATCCGGTTT |
| FlhABgl2 | GAAGATCTACCTCACAAGAACCCGTATCGT |
| FlhAXho2 | ATTCCGCTCGAGCAATGGAAACTCTCAGCCACTTC |
| M13Nhe1 | CTAGCTAGCGTAAAACGACGGCCAGT |
| M13Acs1 | TTGGCGCGCCCAGGAAACAGTTATGAC |
| 1320f | CCATGAAGTCGGAATCGCTAG |
| 1431r | ACTCCCATGGTGTGACGG |
The complemented luxS* strain was constructed by inserting a wild-type copy of the luxS gene in the rdx locus of the ΔluxS chromosome, as described previously (43). PCR products corresponding to the first 56 bp of the rdxA gene, 988 bp of the full-length luxS gene (including 265 bp of upstream sequence), and the final 94 bp of the rdxA gene were amplified in three separate PCRs and then stitched together in two subsequent PCRs using the primers listed in Table 2. The final PCR product was used to disrupt the rdx locus of ΔluxS by selecting for metronidazole-resistant colonies, which were validated as above.
The flhA (HP1041) gene was amplified using the FlhAXho2 and FlhABgl2 primers and ligated into the TOPO vector using a TOPO TA cloning kit (Invitrogen Life Technologies, Carlsbad, CA) to create the vector KHR1. A Kanr nonpolar cassette with two internal ribosomal binding sites from pUC18 K-2 to ensure expression of downstream genes (33) was used to disrupt the flhA locus. The Kanr nonpolar cassette was amplified by the primers M13Asc1 and M13Nhe1, digested along with KHR1, and ligated into the vector. The flhA::Kanr construct was PCR amplified and introduced into the flhA locus through natural transformation. The flgS (HP0244), flgM (HP1122), and fliA (HP1032) mutants were all constructed using the Kanr nonpolar cassette as described previously (35). In each case the 5′ and 3′ ends of the flagellar gene and Kanr nonpolar cassette were PCR amplified in separate reactions and then stitched together in two subsequent PCRs using primers listed in Table 2.
Motility assays.
Motility assays were performed on 0.4% agar plates consisting of Brucella broth, 0.02 mg ml−1 β-cyclodextrin (Sigma), 8 mg ml−1 amphotericin B (Sigma), and 20 μg ml−1 vancomycin (Sigma). Plates were seeded with 1 μl of liquid culture grown to an optical density at 600 nm (OD600) of 1.0, corresponding to early stationary phase growth (24 h as shown in Fig. 1), and incubated for 5 to 8 days under microaerophilic conditions. The area of outward migration was recorded with a digital camera and calculated relative to a standard size reference area on the plate using Metamorph imaging software.
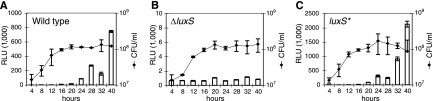
FIG. 1.
AI-2 production in H. pylori is luxS dependent. AI-2 production in the wild-type strain G27 (A), the ΔluxS mutant (B), and the complemented luxS* strain (C) was measured as the ability of CFS harvested at the indicated times after inoculation of liquid H. pylori cultures to induce bioluminescence in the V. harveyi reporter strain BB170 (relative light units [RLU]; bars). The cell densities of the cultures across the growth curves were determined by viable plate counts (CFU/ml; diamonds). Error bars indicate standard deviations of triplicate or duplicate samples for the bioluminescence and cell density measurements, respectively.
Bacterial growth curve and bioluminescence bioassay.
Bacterial cultures were started from an overnight liquid culture, diluted to an OD600 of 0.05 in BB medium, and at various time points samples were taken and plated to determine the number of CFU per milliliter. At those same time points cell suspensions were harvested and filtered through a 0.2-μm-pore-size filter in SpinX columns and centrifuged at 6,000 × g (Nalgene Nunc International). Cell-free supernatant (CFS) was collected from all strains and assayed for AI-2 activity with the V. harveyi BB170 bioassay as previously described (45, 46). Briefly, an overnight V. harveyi culture was diluted 1:10,000 in fresh AB medium (22a). CFS preparations were added to the diluted V. harveyi to a 10% (vol/vol) final concentration, and these cultures were shaken at 30°C for 3 h, after which 3-ml aliquots were assayed in triplicate for light production using an Optocomp I luminometer (MGM Instruments, Inc.).
AI-2 complementation experiments.
To test for complementation of the ΔluxS motility phenotype, soft agar plates consisting of 0.4% agar were made using 70% BB medium and 30% CFS collected from wild-type or ΔluxS cultures at an OD600 of 1.0. Synthetic DPD was prepared as described previously (42). DPD activity was quantified with the V. harveyi BB170 bioassay and compared to CFS from wild-type H. pylori cultures grown to an OD600 of 1.0, the point in the growth curve at which we measured maximal AI-2 activity. DPD was added to H. pylori liquid cultures of various genotypes to a final concentration of 0.01 mM, 0.1 mM, or 1.0 mM. Cultures were incubated for 4 h, after which RNA was extracted or bacterial cells were visualized by transmission electron microscopy (TEM).
RNA collection and extraction.
RNA was collected from H. pylori strains grown in BB medium to an OD600 of 1.0. Cells were passed onto 0.45-μm-pore-size cellulose nitrate membrane filters (Whatman, Maidstone, England) by vacuum. Filters were placed in 50-ml conical tubes and stored at −80°C. To extract RNA, filters were thawed on ice, appropriate amounts of Trizol were added, and RNA was extracted following the manufacturer's instructions (Gibco). Contaminating genomic DNA was removed using a Turbo DNA Free kit (Ambion).
RT-PCR.
Primers used for reverse transcription-PCRs (RT-PCRs) are listed in Table 2. All RT reactions were performed using 1 μg of RNA, Superscript II (Invitrogen), and gene-specific primers, and mixtures were incubated for 50 min at 42°C. Quantitative RT-PCR (qRT-PCR) reaction mixtures contained 5 μl of cDNA template, gene-specific primers, 12.5 μl of SYBR Green Master Mix (Applied Biosystems), and distilled H20 to a 25-μl total volume. qRT-PCR assays were performed in triplicate with an ABI Prism 7900HT Sequence Detector (Applied Biosystems). Transcript levels were normalized to ureA or the 16S rRNA gene in each sample (using ΔΔCT analysis as described in ABI User Bulletin 2, available at http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). The ureA transcript was chosen for normalization because it amplified consistently across different genotypes at the same growth phase. The flhA mutants were an exception in that they expressed reduced levels of ureA transcript, consistent with a previous report implicating flhA in regulation of ureA transcription (32); for these mutants the 16S rRNA gene was chosen for normalization, using primer sequences 1320f and 1431r as described previously (13). Other primers used for qRT-PCR were designed using Primer Express software (Applied Biosystems); all primer sequences are listed in Table 2.
TEM.
All samples for TEM were grown overnight in BB medium from a starting OD600 of 0.02. A 10-μl sample of each strain was applied to copper grids (400 mesh) with Butvar support film coated with carbon, rinsed twice, and negatively stained for 1 min with 1% phosphotungstate (pH 6.97). Grids were visualized and photographed in a Philips CM12 TEM at an acceleration voltage of 80 kV. Bacterial counts for each genotype and treatment group were conducted on three separate grids, counting a minimum of 100 cells, for at least two independent experiments.
RESULTS
luxS is responsible for AI-2 production in H. pylori strain G27.
To investigate whether AI-2 functions as a signaling molecule in H. pylori, we generated a clean deletion of the luxS gene in the laboratory strain G27 (hereafter referred to as the ΔluxS strain). To verify that any phenotypes we observed with the ΔluxS strain were due to the loss of the luxS gene product, we constructed a genetically complemented strain (hereafter referred to as the luxS* strain) by placing the full-length luxS gene and upstream sequences in the ΔluxS mutant chromosome at the rdx locus, which encodes a nonessential gene that, when mutated, results in metronidazole resistance without causing a growth disadvantage (2, 43). We isolated CFS from wild-type, ΔluxS, and luxS* cultures at different time points during growth in liquid medium and assayed their abilities to induce bioluminescence when incubated with the V. harveyi reporter strain BB170. The wild-type strain produced AI-2 in a growth-stage-dependent manner, with significant AI-2 activity accumulating during stationary phase (Fig. 1A). The ΔluxS mutant, unlike the wild-type parent strain, was not able to activate bioluminescence (Fig. 1B, note difference in y-axis scale). The luxS* strain activated bioluminescence in the reporter at levels higher than the wild-type strain (Fig. 1C), suggesting that the insertion of the luxS gene and its promoter at a heterologous position in the chromosome resulted in elevated expression of the gene product. The complementation experiment confirmed that LuxS is responsible for AI-2 production in H. pylori. Neither the interruption of the luxS gene nor the insertion of luxS at the rdx locus caused any growth defect under the conditions examined (Fig. 1).
The ΔluxS mutant has reduced motility.
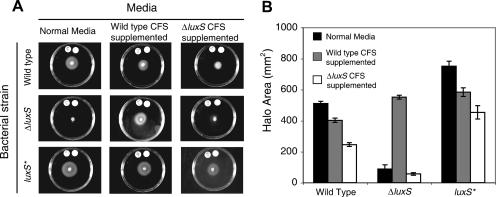
Based on the report that luxS may regulate flaA expression (29), we measured motility of the H. pylori wild-type, ΔluxS, and luxS* strains. Liquid bacterial cultures growing at early stationary phase were spotted onto the center of soft agar plates and incubated for 6 days, and the resulting swarm halo areas were quantified. The ΔluxS mutant consistently produced a reduced swarm halo compared to the wild-type strain in repeated trials (one representative trial is shown in Fig. 2). Interestingly, the luxS* strain, which produces higher levels of AI-2 than the wild type (Fig. 1), also produced a larger swarm halo than the wild-type strain. To test whether the ΔluxS motility defect could be complemented by a secreted factor produced by wild-type but not ΔluxS mutant cells, we prepared soft agar plates composed of 70% fresh BB medium and 30% CFS from each strain. Both the wild-type and luxS* strains formed halos approximately 25% smaller when grown on wild-type CFS-supplemented plates than on normal medium (Fig. 2), which could be due to AI-2, depletion of an essential nutrient, or to the presence of a noxious by-product of bacterial metabolism in the spent medium, or a combination of all three. Both strains formed halos approximately 50% smaller when grown on ΔluxS CFS relative to normal medium, which could be due to similar noxious effects of the wild-type CFS without the positive effect of AI-2 or to differences in the chemical composition of the CFS from the two strains, as illustrated by the global metabolic difference between wild-type and ΔluxS E. coli (51). Notably, the ΔluxS mutant formed halos over 600% larger on wild-type CFS-supplemented plates than on normal medium or ΔluxS CFS-supplemented plates (Fig. 2). These data indicate that a substance in the wild-type but not ΔluxS CFS, likely AI-2, can complement the motility defect of the ΔluxS mutant.
FIG. 2.
The ΔluxS mutant has a motility defect that is rescued by wild-type CFS. Wild-type, ΔluxS, and luxS* bacteria were seeded onto soft agar plates composed of normal medium or supplemented with CFS from wild-type or ΔluxS cultures. After 6 days of incubation, the halos of migration were visualized (A) and quantified (B). Error bars indicate the standard deviation of halo sizes of triplicate cultures in one experiment.
AI-2 influences flagellum morphogenesis.
We used TEM to examine the flagellar morphology of the wild-type, ΔluxS mutant, and luxS* strains. In cell populations of all three genotypes, the flagella appeared normal, as scored for at least 100 cells per genotype (Fig. 3A to D). We reasoned, however, that the flagellum might be especially sensitive to the presence of AI-2 in genetic backgrounds in which its integrity was already compromised. To test this idea, we constructed mutations in four key flagellar regulators in both the wild-type and the ΔluxS mutant genetic backgrounds. Using a nonpolar cassette designed to allow translation of downstream genes in the operon (33), we created insertion mutations in flgS (HP0244), encoding the sensor kinase that regulates class 2 and the intermediate class gene transcription (6, 36); in fliA, the σ28 responsible for class 3 and the intermediate class gene expression; in flgM, the anti-sigma factor for fliA (11, 24); and in flhA, the flagellar master regulator (36, 41). All flagellar single and ΔluxS double mutants were tested for motility on soft agar plates. In all cases, the mutants were nonmotile, as expected based on previous reports, with no differences between the single and double mutants (data not shown).
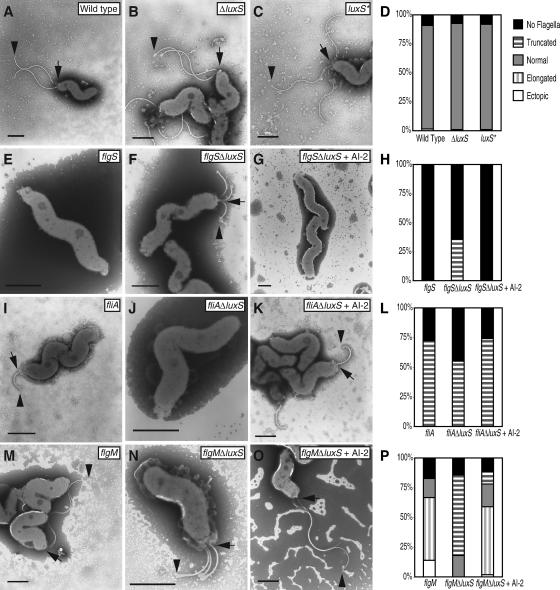
FIG. 3.
AI-2 modulates flagellar morphogenesis. Representative flagella, as visualized by TEM, are shown for each of the genotypes indicated grown in normal medium (A to C, E, F, I, J, M, and N) or for 4 h in the presence of 0.1 mM DPD (G, K, and O). In all cells with flagella, the base is marked by an arrow, and the tip is marked by an arrowhead. Scale bars, 1 μm. The flagellar morphologies were scored for at least 100 cells for each treatment group (D, H, L, and P).
We assessed the flagellar morphologies of the single and double mutants by TEM (at least 100 cells of each genotype in at least two independent experiments). In the cases of fliA, flgM, and flgS, we found that the flagellar ΔluxS double mutants had phenotypes that differed from the single mutant phenotypes as described below. The results for one representative experiment are shown in Fig. 3. The flgS single mutant did not produce any flagella (Fig. 3E), as reported previously (36). In contrast, 36% of the flgS ΔluxS double mutant population produced truncated flagella (Fig. 3F and H). The fliA single mutant exhibited truncated flagella (Fig. 3I), as has been previously described (11, 24). In the fliA ΔluxS double mutant population, the proportion of cells completely lacking flagella increased, with a corresponding decrease in the fraction of cells carrying truncated flagella (Fig. 3J and L). As previously reported, the flgM single mutant produces abnormally long flagella (11), and a small proportion displays ectopic flagella at both cell poles (24) (Fig. 3 M). In contrast, the majority of the flgM ΔluxS double mutant cells exhibited truncated flagella (Fig. 3N and P). As previously reported, the flhA mutant did not produce flagella (41). The flhA ΔluxS double mutant cells were also completely aflagellated (data not shown).
To test whether the flgS ΔluxS, fliA ΔluxS, and flgM ΔluxS double mutant flagellar phenotypes were due to the loss of AI-2 signaling, we treated each of the double mutant cultures with synthetic DPD, which spontaneously converts to AI-2. For these experiments we used a final concentration of 0.1 mM DPD that corresponded to the maximal concentration of AI-2 produced by H. pylori cultures (see Fig. 4C). Cultures were incubated with DPD for 4 h and visualized by TEM. In all cases, the addition of DPD completely reversed the ΔluxS double mutant flagellar phenotypes (Fig. 3G, K, and O). These results demonstrate that AI-2, acting as an extracellular signal, modulates flagellar morphogenesis.
FIG. 4.
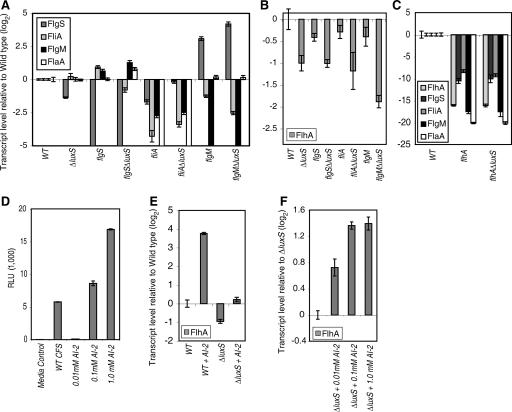
AI-2 modulates flagellar gene transcription. Transcript levels of flgS, fliA, flgM, and flaA (A) and of flhA (B) were determined by qRT-PCR normalized to the levels of ureA in each of the genotypes indicated and are represented as the log2 difference relative to the levels in the wild-type strain. Levels of the same transcripts were determined in the indicated genotypes and normalized to the levels of the 16S rRNA gene in each sample (C). AI-2 activity in CFS from wild-type early-stationary-phase H. pylori was measured relative to different concentrations of DPD (D). flhA transcript levels were measured as in panel B in wild-type and ΔluxS cultures untreated or treated for 4 h with 0.1 mM DPD (E) and in ΔluxS cultures treated with a range of DPD concentrations (F). Error bars indicate the standard deviations for three triplicate samples.
AI-2 regulates flagellar gene transcription by modulating flhA expression.
To determine the molecular mechanism underpinning the changes in flagellar morphology in the ΔluxS mutants, we used semiquantitative RT-PCR to screen for alterations in gene expression of representative genes of the four flagellar classes in the 10 genotypes described above. The genes that showed marked differences in transcript levels between the genotypes were reanalyzed by qRT-PCR in an independent experiment. Transcript abundances were normalized to ureA, which displayed consistent levels across the different genotypes at the same growth phase. The flhA and flhA ΔluxS mutants were exceptions and exhibited marked reduction in ureA transcription, consistent with a previous report of reduced urease activity in flhA mutants (32); for these mutants 16S rRNA transcript was used for normalization. The class 1 genes involved in chemotaxis and motor function (cheW, cheA, motA, motB, tlpA, and hpaA, encoding a flagellar sheath-associated protein) did not exhibit differences in expression levels between the genotypes. In contrast, dynamic changes were observed across the genotypes in multiple genes involved in flagellar biosynthesis, including genes regulated by all three sigma factors (Fig. 4A). The transcriptional differences between the various mutants demonstrate the complexity of the flagellar gene transcription hierarchy and its sensitivity to perturbations. For example, the flgS ΔluxS double mutant, which formed some truncated flagella in contrast to the aflagellated flgS mutant, exhibited elevated levels of flaA transcript encoding the major flagellin. Whereas elimination of flgS in a ΔluxS background derepressed flagellar production, flgS levels were elevated in both the fliA ΔluxS and flgM ΔluxS double mutants relative to the luxS+ single mutants. We note that elevation of flgS transcription was associated with loss or truncation of flagella in the double mutants, suggesting a role for FlgS as a negative regulator of flagellar biosynthesis.
Notably, the top tier regulator flhA showed a consistent pattern of reduced transcription in the ΔluxS genotypes (Fig. 4B). We demonstrated that flhA was required for expression of the flagellar genes flgS, fliA, flgM, and flaA (Fig. 4C), confirming its role as a key activator of the flagellar regulon. The flhA ΔluxS double mutant exhibited a pattern of flagellar gene transcription identical to that of the flhA single mutant (Fig. 4C). To test whether loss of AI-2 signaling could account for the reduction in flhA transcript in the flgS ΔluxS, fliA ΔluxS, and flgM ΔluxS double mutants, we added back an equivalent amount of synthetic DPD to ΔluxS cultures to restore the normal level of AI-2 in the medium. To do this, we compared the level of AI-2 activity in CFS derived from early stationary phase wild-type H. pylori, which we had shown to produce maximal AI-2 (Fig. 1A), to various concentrations of synthetically prepared DPD. We found that a 0.1 mM concentration of DPD and wild-type CFS caused roughly equivalent levels of expression of bioluminescence in the V. harveyi BB170 reporter strain (Fig. 4D). Indeed, incubation of the ΔluxS mutant with this concentration of DPD was sufficient to restore flhA transcript levels to that of wild-type cells (Fig. 4E). Furthermore, exposure of wild-type cells to 0.1 mM DPD resulted in elevated flhA transcript levels (Fig. 4E). To further characterize the dose responsiveness of flhA transcription to AI-2, we exposed ΔluxS mutant cultures, which are devoid of endogenous AI-2, to a range of DPD concentrations. flhA transcript levels increased in a dose-dependent manner between 0.01 and 0.1 mM DPD (Fig. 4F). No additional increase in flhA transcript levels was observed at 1 mM DPD, suggesting that the effects of DPD on flhA transcription were saturable or possibly even inhibitory. These data show that AI-2 alone is sufficient to modulate flagellar gene expression in a dose-dependent manner.
DISCUSSION
In this study we show that AI-2 signaling is necessary for normal levels of motility in H. pylori strain G27. The basis of the ΔluxS mutant phenotype could be due to a defect in the morphogenesis or function of the motility apparatus. We observed no abnormalities in the morphology of the ΔluxS mutant flagella by TEM or of the luxS* strain that produces elevated levels of AI-2, and the mutant cells were motile when observed in wet mount, although we did observe some differences in their swimming behavior, which is the focus of a separate study (B. A. Rader and K. Guillemin, unpublished data). However, when we deleted the luxS gene in mutants lacking any of three key flagellar regulatory genes—the sensor kinase flgS, the flagellar sigma factor fliA, and its anti-sigma factor, flgM—we observed strikingly different flagellar phenotypes in the double versus the single mutants. All of the double mutant phenotypes were reversed to that of the luxS+ single mutants by the addition of purified AI-2, indicating that the regulation of flagellar morphology is mediated by this extracellular signal. We confirmed flhA's role as a top-tier regulator of flagellar gene transcription and demonstrated that an flhA mutant and an flhA ΔluxS double mutant had identical phenotypes, consistent with the idea that the AI-2 signal acts upstream of FlhA. We further demonstrated that purified AI-2 is sufficient to induce expression of flhA.
Our finding that AI-2 regulates flagellar gene expression in H. pylori argues that this molecule functions as an intercellular signaling molecule in this species. In addition, our data suggest that AI-2 signaling regulates motility, as indicated by our finding that wild-type but not ΔluxS CFS can restore spreading of ΔluxS mutants on soft agar plates. It is unlikely, however, that AI-2 regulation of flagellar gene expression explains the motility defect of the ΔluxS mutants, given the similarity in the wild-type and mutant flagellar morphologies. Our data do not rule out additional metabolic requirements for luxS in this species that could explain the colonization defects observed with luxS mutant H. pylori strains (27, 37). The finding that a luxS mutant in the X47 strain did not exhibit motility or colonization defects (27) indicates that AI-2 signaling or LuxS metabolic activities are not required in all H. pylori strains, reflecting a phenotypic heterogeneity in this species that we have also observed (2).
Integration of environmental signals into the flagellar gene transcription hierarchy.
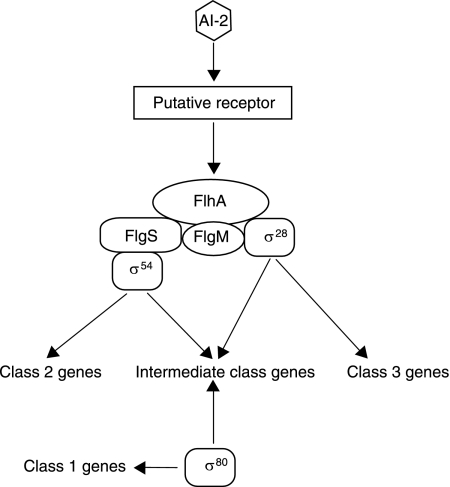
Our genetic and molecular data suggest a model, shown in Fig. 5, in which AI-2 regulates flagellar morphology by influencing initial steps in the flagellar gene transcription hierarchy. Consistent with the model that the AI-2 signal functions exclusively upstream of FlhA, we observed identical phenotypes in the flhA single and flhA ΔluxS double mutants in their lack of motility and flagella and reduced expression of flagellar genes. In contrast, the flagellar morphology of the ΔluxS double mutants in combination with flagellar regulators in parallel pathways that branch downstream of FlhA differed from those of the corresponding single mutants, consistent with the model that the AI-2 signal functions both upstream and in parallel to FlgS, FliA, and FlgM. In addition, our finding that AI-2 is sufficient to activate expression of flhA places it upstream of this top-tier flagellar regulator.
FIG. 5.
Model of AI-2 modulation of the H. pylori flagellar regulon. The role of AI-2 in transcriptional regulation of components of the flagellar gene hierarchy of H. pylori is shown, based on Neihus et al. (36). AI-2 signals upstream of FlhA, which, in turn, regulates branching pathways of gene transcription regulated by FlgS, FlgM, and FliA/σ28.
The comparisons between the wild-type strain and mutants in flgS, fliA, and flgM revealed interdependence between these regulators: in every case in which one of the regulators was removed, the transcript levels of the other two were altered. In the absence of luxS, loss of flgS resulted in increased transcription of the major flagellin, flaA, and increased flagellum formation, whereas elevated flgS levels in the fliA ΔluxS and flgM ΔluxS mutants correlated with shorter flagella than with the luxS+ genotypes. All three regulators were also required for wild-type levels of flhA expression, and their effects were at least additive with that of luxS. Since flhA is required for expression of most of the flagellar gene regulon (36), AI-2 regulation of flhA transcription is likely to contribute to the differences in flagellar gene expression and flagellar morphology observed in the different ΔluxS mutants, although the complexity of this regulatory network and the importance of posttranscriptional regulatory events make it difficult to formulate specific predictions about the consequence of reducing flhA levels in each mutant background analyzed. Our finding that AI-2 itself is sufficient to increase flhA transcript levels in a dose-dependent manner represents a new signal that can influence H. pylori flagellar biosynthesis.
Multiple complex and diverse hierarchical regulatory pathways of flagellar gene expression have been described in bacteria (31), many of which are regulated by master regulators whose expression is, in turn, influenced by environmental conditions such as nutrient availability and cell cycle (7, 30). In several systems, AI-2 has been implicated in regulating flagellar biosynthesis. An luxS mutant of Campylobacter jejuni, a close relative of H. pylori, also exhibits decreased motility on soft agar plates and regulates flaA expression in an luxS-dependent manner (17, 23). Treatment of Escherichia coli strain K-12 with purified AI-2 has been shown to increase this bacterium's motility on soft agar plates through a mechanism that involves a novel motility quorum-sensing regulator, MqsR, regulating qseBC and flhDC expression (22).
Environmental regulation of H. pylori flagellar morphogenesis.
Motility is an absolute requirement for H. pylori colonization in several animal models (16, 38, 39) and is thus considered an important virulence trait of this pathogen. Little is known, however, about the regulation of H. pylori flagellar biosynthesis within the gastric environment. Unlike the flagellar gene hierarchies of the enteric Gammaproteobacteria or Caulobacter, which possess the master regulators FlhCD and CtrA, respectively, responsible for all flagellar gene expression, many of the H. pylori flagellar and chemotaxis genes (the class 1 genes) are under the control of the housekeeping sigma factor, σ80, and are constitutively expressed (36). Environmental factors, however, clearly modulate expression of flagellar genes. Growth of H. pylori in acidic medium induces expression of many of the class 2 and intermediate genes and is associated with increased motility (34), whereas growth on cultured gastric epithelial cells represses expression of the class 3 gene flaA (26). flaA expression increases in the switch from exponential growth to stationary phase (48), during the period when we observe significant AI-2 accumulation, and AI-2-containing CFS induces transcription of an flaA reporter (29). We report here that AI-2 regulates expression of the class 1 gene flhA and influences flagellar morphogenesis. In addition, the transcript levels of another class 1 gene, flgS, fluctuated dramatically between the different genotypes we analyzed, indicating that expression of these top-tier regulators is sensitive to concentrations of an environmental signal.
H. pylori may use AI-2, which we have shown it produces in a cell density-dependent manner, in conjunction with other environmental cues to regulate flagellar structures as a survival strategy within the human stomach. At high cell densities and high AI-2 concentrations, it may be beneficial for H. pylori to increase flagellum production as a strategy to promote motility and dispersal throughout the stomach. Also, despite the textbook picture of the stomach as devoid of bacteria, a recent 16S rRNA enumeration study demonstrated a diverse gastric microbiota (8), which may contribute to AI-2 levels in the stomach and influence H. pylori motility in this habitat. Patterns of H. pylori dispersal within the gastric environment are correlated with disease outcome: ulcers are associated with concentrated bacterial populations in the distal stomach and gastric cancer arises from infections in which the bacteria are distributed throughout the stomach (9). An understanding of the environmental cues, both bacterial and host derived, that regulate H. pylori flagellar morphogenesis in the human stomach will contribute to our ability to predict and control H. pylori-associated disease outcomes.
Acknowledgments
We thank Jeannie Selker and Kurt Langworthy for assistance with electron microscopy; Kevin Hicks for assistance with the construction of the flhA and flhA ΔluxS mutants; David Baltrus, Kevin Bourzac, and Kyle Mouery for help with genetic manipulations of H. pylori; and the entire Guillemin laboratory for helpful discussions.
K.G. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and was supported by Research Scholar Grant RSG-03-101-01-MBC from the American Cancer Society and NIH grant R56 DK075667-01. B.L.B. is funded by the NIH, NSF, and HHMI.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baltrus, D. A., and K. Guillemin. 2006. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255:148-155. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125:237-246. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 6.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergara, F., C. Ibarra, J. Iwamasa, J. C. Patarroyo, R. Aguilera, and L. M. Marquez-Magana. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J. Bacteriol. 185:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colland, F., J. C. Rain, P. Gounon, A. Labigne, P. Legrain, and H. De Reuse. 2001. Identification of the Helicobacter pylori anti-sigma28 factor. Mol. Microbiol. 41:477-487. [DOI] [PubMed] [Google Scholar]
- 12.Copass, M., G. Grandi, and R. Rappuoli. 1997. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect. Immun. 65:1949-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Keersmaecker, S. C., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14:114-119. [DOI] [PubMed] [Google Scholar]
- 16.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 18.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsyth, M. H., and T. L. Cover. 2000. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect. Immun. 68:3193-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 21.Freeman, J. A., B. N. Lilley, and B. L. Bassler. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 35:139-149. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 23.Jeon, B., K. Itoh, N. Misawa, and S. Ryu. 2003. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol. Immunol. 47:833-839. [DOI] [PubMed] [Google Scholar]
- 24.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 25.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, N., E. A. Marcus, Y. Wen, D. L. Weeks, D. R. Scott, H. C. Jung, I. S. Song, and G. Sachs. 2004. Genes of Helicobacter pylori regulated by attachment to AGS cells. Infect. Immun. 72:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, W. K., K. Ogura, J. T. Loh, T. L. Cover, and D. E. Berg. 2006. Quantitative effect of luxS gene inactivation on the fitness of Helicobacter pylori. Appl. Environ Microbiol. 72:6615-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 29.Loh, J. T., M. H. Forsyth, and T. L. Cover. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 31.McCarter, L. L. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 32.McGee, D. J., C. Coker, T. L. Testerman, J. M. Harro, S. V. Gibson, and H. L. Mobley. 2002. The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J. Med. Microbiol. 51:958-970. [DOI] [PubMed] [Google Scholar]
- 33.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouery, K., B. A. Rader, E. C. Gaynor, and K. Guillemin. 2006. The stringent response is required for Helicobacter pylori survival of stationary phase, exposure to acid, and aerobic shock. J. Bacteriol. 188:5494-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 37.Osaki, T., T. Hanawa, T. Manzoku, M. Fukuda, H. Kawakami, H. Suzuki, H. Yamaguchi, X. Yan, H. Taguchi, S. Kurata, and S. Kamiya. 2006. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J. Med. Microbiol. 55:1477-1485. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, P. W., M. C. Lane, and S. Porwollik. 2000. Helicobacter pylori motility. Microbes Infect. 2:1207-1214. [DOI] [PubMed] [Google Scholar]
- 39.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz, A., C. Josenhans, and S. Suerbaum. 1997. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J. Bacteriol. 179:987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semmelhack, M. F., S. R. Campagna, M. J. Federle, and B. L. Bassler. 2005. An expeditious synthesis of DPD and boron binding studies. Org. Lett. 7:569-572. [DOI] [PubMed] [Google Scholar]
- 43.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suerbaum, S. 1995. The complex flagella of gastric Helicobacter species. Trends Microbiol. 3:168-171. [DOI] [PubMed] [Google Scholar]
- 45.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 47.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 50.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 51.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]