Abstract
AtoC has a dual function as both an antizyme, the posttranslational inhibitor of polyamine biosynthetic enzymes, and the transcriptional regulator of genes involved in short-chain fatty acid catabolism (the atoDAEB operon). We have previously shown that AtoC is the response regulator of the AtoS-AtoC two-component signal transduction system that activates atoDAEB when Escherichia coli is exposed to acetoacetate. Here, we show that the same cis elements control both promoter inducibility and AtoC binding. Chromatin immunoprecipitation experiments confirmed the acetoacetate-inducible binding of AtoC to the predicted DNA region in vivo. DNase I protection footprinting analysis revealed that AtoC binds two 20-bp stretches, constituting an inverted palindrome, that are located at −146 to −107 relative to the transcription initiation site. Analyses of promoter mutants obtained by in vitro chemical mutagenesis of the atoDAEB promoter verified both the importance of AtoC binding for the inducibility of the promoter by acetoacetate and the σ54 dependence of atoDAEB expression. The integration host factor was also identified as a critical component of the AtoC-mediated induction of atoDAEB.
Escherichia coli utilizes a variety of carbon-containing compounds, including fatty acids, which are degraded by β-oxidation into acetyl coenzyme A (acetyl-CoA) (3). The utilization of long-chain (C12 to C18) and medium-chain (C7 to C11) fatty acids requires the actions of several proteins encoded by the fad regulon genes, whose products are involved in the transport, activation, and β-oxidation of fatty acids (reference 3 and references therein). The use of short-chain (C4 to C6) fatty acids also requires a membrane transporter (AtoE) and two catabolic enzymes, i.e., the acetyl-CoA-acetoacetate-CoA transferase (AtoD-AtoA) and the 3-ketoacyl-CoA thiolase II (AtoB) (12, 13). These proteins are encoded by the atoDAEB operon, which is highly inducible by acetoacetate but not by other saturated short-chain fatty acids, such as butyrate and valerate, whose catabolism also requires enzymes encoded by the atoDAEB operon genes (13, 25). Genetic analyses identified AtoC, a 48-kDa protein encoded by atoC, as the positive gene regulatory factor required for the inducible expression of the atoDAEB operon (13). Subsequent work identified the antizyme (Az) AtoC as the response regulator (RR) of a two-component system (TCS) that consists of AtoC and the sensor histidine kinase (HK) AtoS (15, 16).
Az was first described by Fong et al. and Heller et al. as a polyamine-inducible noncompetitive, proteinaceous inhibitor of ornithine decarboxylase, the key enzyme in polyamine biosynthesis (6, 10). In an attempt to identify the E. coli Az gene, we previously isolated and sequenced a 6.4-kb genomic DNA fragment encompassing part of the atoDAEB operon and the genes of a putative HK and an RR, which proved to encode AtoS and AtoC, respectively (2). The identified E. coli Az gene showed significant sequence homology to genes for RRs of the NtrC-NifA family of the σ54 RNA polymerase transcriptional activators (24, 35, 40), and its genetic locus coincided with atoC (2). A gene that showed significant sequence similarities to genes for sensor HKs (24, 35) and localized just upstream of atoC was assigned the name atoS since it was presumed to encode the sensor HK that modulates AtoC activity (2). Recently, we presented both biochemical evidence on the phosphorylation of AtoC by AtoS and functional data on the positive regulation of the atoDAEB operon by the AtoS-AtoC TCS upon acetoacetate induction (16). Using AtoS-enriched membrane preparations as a kinase source and wild-type and mutant forms of AtoC as substrates in the phosphorylation reactions, we showed that mutant AtoC that lacks the conserved aspartic acid-55 (D55) due to a D55G substitution can be phosphorylated. Histidine-73 (H73) appears to be a second AtoC phosphorylation site since it is located within a characteristic “H-box” and since its alteration in the D55G/H73L double AtoC mutant leads to almost complete inhibition of AtoC phosphorylation in vitro (16).
Transcriptome and phenotypic microarray analyses (21, 45), however, have indicated that the role of atoSC may not be limited to the regulation of atoDAEB expression. Indeed, E. coli mutants with the atoSC locus deleted display altered gene expression (11 up-regulated and 32 down-regulated genes) (21), as well as reduced motility, sodium chloride sensitivity, and increased susceptibility to membrane-acting agents and an aminoglycoside antibiotic (21, 45). We have also demonstrated previously that atoSC can positively modulate the biosynthesis of poly-(R)-3-hydroxybutyrate, a biopolymer with many physiological roles in E. coli (38).
The aim of this work was to further advance our understanding of the role of the AtoS-AtoC TCS in gene regulation by (i) characterizing the cis elements within the atoDAEB promoter regulatory region that are responsible for the induction of the operon by acetoacetate, (ii) mapping and identifying the AtoC binding site(s) within this region and, (iii) identifying additional factors that affect AtoC function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The genotypes of the E. coli strains used in this work are described in Table 1. E. coli cells were grown at 37°C either in modified M9 mineral medium supplemented with 0.1 mM CaCl2, 1 mM MgSO4, 0.4% (wt/vol) glucose, 1 μg of thiamine/ml, 80 μg of proline/ml, and 1.7 μΜ FeSO4 (16) or in Luria-Bertani broth (28).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers used in this work
| Strain, plasmid, or oligonucleotide | Characteristics or description | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔΜ15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG λ− | Invitrogen |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| N99 | Wild type; λ− F−sup0galK2 | M. Gottesman (9) |
| N7193 | N99 Δhip-3::CAT | M. Gottesman (9) |
| BW25113 | lacIqrrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | H. Aiba (21) |
| BW28878 | BW25113 Δ(atoSC)569 | H. Aiba (21) |
| Plasmids | ||
| pZErO 2.1 | Cloning vector | Invitrogen |
| pZero-patoD1 | atoD1p PCR product (−206 to +73 of atoDAEB) cloned into the EcoRV site of pZErO 2.1 | This work |
| pZero-patoD2 | atoD2p PCR product (−120 to +73 of atoDAEB) cloned into the EcoRV site of pZErO 2.1 | This work |
| pZero-patoD3 | atoD3p PCR product (−47 to +73 of atoDAEB) cloned into the EcoRV site of pZErO 2.1 | This work |
| pMLB1034 | Medium-copy-number vector containing a promoterless lacZ | 34 |
| pMLB-atoD1 | atoD1p (EcoRI-BamHI fragment from pZero-patoD1) cloned into pMLB1034 | This work |
| pMLB-atoD2 | atoD2p (EcoRI-BamHI fragment from pZero-patoD2) cloned into pMLB1034 | This work |
| pMLB-atoD3 | atoD3p (EcoRI-BamHI fragment from pZero-patoD3) cloned into pMLB1034 | This work |
| pCPG3 | rcsC regulatory sequences (−656 to +570) cloned into pMLB1034 | This work |
| pCPG4 | atoS regulatory sequences (−729 to +189) cloned into pMLB1034 | This work |
| pCPG5 | atoDAEB regulatory sequences (−419 to +1186) cloned into pMLB1034 | This work |
| pCPG6 | atoC regulatory sequences (−1150 to +187) cloned into pMLB1034 | This work |
| Oligonucleotides | ||
| U1patoD | 5′-TCGGAATTCATTGATGTATAAACTCCAGGAA-3′ | This work |
| U2patoD | 5′-AGGGAATTCAATTTTCTGCATAGCCGCTCAT-3′ | This work |
| U3patoD | 5′-TACGAATTCAACTAAATCCAATAATCTCATT-3′ | This work |
| LpatoD | 5′-AAGGGATCCGTGGCGTCTTGTAATGTCAT-3′ | This work |
Plasmid constructions.
The atoD1, atoD2, and atoD3 regions upstream of the atoDAEB operon, extending, respectively, from −206, −120, and −47 to +73 relative to the predicted transcriptional start site, were PCR amplified from plasmid pUC-Az (2) by using different forward primers (U1patoD, U2patoD, and U3patoD) and a common reverse primer (LpatoD) (Table 1). The PCR products were cloned into the EcoRV site of pZErO 2.1 (Invitrogen) to construct plasmids pZero-patoD1, pZero-patoD2, and pZero-patoD3, respectively, and then subcloned into the medium-copy-number plasmid vector pMLB1034 (EcoRI and BamHI sites) containing a promoterless lacZ gene (34) to yield plasmids pMLB-atoD1, pMLB-atoD2, and pMLB-atoD3 (Table 1).
The 930-bp EcoRI-HincII fragment from pCPC-J (2) containing the atoS promoter was introduced into EcoRI-SmaI-digested pMLB1034 to produce pCPG4 (atoS lacZ). The 1.2-kb EcoRI-HincII fragment from pCPC-Az (2) containing the atoDAEB promoter and sequences coding for AtoD and the N-terminal 21 amino acids of AtoA was cloned into EcoRI-SmaI-cut pMLB1034 to generate pCPG5 (atoA lacZ). The 1.3-kb EcoRV fragment from pUC-Az containing the atoC promoter was introduced into SmaI-cut pMLB1034 to yield pCPG6 (atoC lacZ). The 1.2-kb BamHI-EcoRI fragment from pUC-Az containing the rcsC (19) promoter was cloned into EcoRI-BamHI-cut pMLB1034 to generate pCPG3 (rcsC lacZ).
β-Galactosidase assays.
β-Galactosidase assays were performed using permeabilized E. coli cells carrying the appropriate plasmids, as described previously (16, 20). The cells had been grown in modified M9 mineral medium (28) supplemented with 0.1 mM CaCl2, 1 mM MgSO4, 0.4% (wt/vol) glucose, 1 μg of thiamine/ml, 80 μg of proline/ml, and 1.7 μΜ FeSO4 in the absence or presence of 10 mM acetoacetate.
AtoC purification and electrophoretic mobility shift assays (EMSAs) of DNA binding.
The recombinant AtoC (rAtoC) was purified (16) and dialyzed against a mixture of 10 mM HCl, 50% (vol/vol) glycerol, and 5 mM β-mercaptoethanol. Following the determination of the rAtoC concentration and an analysis of its purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the protein was stored in aliquots at −80°C.
The DNA binding activity of the purified rAtoC (16) was determined by nonradioactive EMSAs using biotin-labeled probes. The probes were prepared by PCR amplification using pZero-patoD1 DNA as the template and primer pairs consisting of a biotinylated M13 universal primer and either the U1patoD, U2patoD, or U3patoD primer (see above). The DNA binding reactions took place in a 20-μl final volume for 30 min at 4°C by incubating purified rAtoC (0.008 to 0.7 μM) in a mixture of 10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 5% (vol/vol) glycerol, 1 mM EDTA, 4 mM dithiothreitol, 0.2 mg of bovine serum albumin/ml, and 10 ng of the probe (approximately 3 nM) in the presence of 2 μg of a nonspecific DNA competitor (sonicated salmon sperm DNA). In competition assays, the mixtures were incubated with the appropriate concentration of a competitor prior to the addition of the probe to the reaction mixture. Supershift analyses were performed as previously described (23) using either a specific monoclonal antibody (MAb) such as HIS-1 (Sigma) that recognizes the rAtoC polyhistidine tag or the nonspecific 58S MAb that recognizes an unrelated protein, i.e., the herpes simplex virus type 1 transcriptional regulator ICP4 (31).
Following electrophoretic separation on 4% (wt/vol) polyacrylamide gels with 0.25× Tris-borate-EDTA buffer, the DNA-protein complexes were electroblotted onto 0.45-μm-pore-size Biodyne B membranes (PALL) for 1 h at 30 V in 0.5× Tris-borate-EDTA. The membranes were cross-linked with UV light (0.120 J) and blocked for 5 min with a solution of 5% (wt/vol) sodium dodecyl sulfate, 125 mM NaCl, 17 mM Na2HPO4, and 8 mM NaH2PO4, pH 7.2 (0.1 ml/cm2), and then the blots were developed using the Phototope-Star detection kit according to the instructions of the manufacturer (NEB).
DNase I footprinting.
PCR amplification was performed to prepare the DNA probes used in the DNase I footprinting analysis of the atoDAEB promoter, along with one nonradioactive and one 5′-end-32P-labeled primer (U1patoD and LpatoD) in each reaction. Labeled probes were purified using a Nucleospin extract kit (Macherey-Nagel) and mixed with various amounts of rAtoC (0.07 to 3.5 μM) before the DNase I protection footprinting analysis (22).
ChIP assays.
Chromatin immunoprecipitation (ChIP) from atoSC+ or ΔatoSC bacteria carrying either the pMLB-atoD1 or the pMLB-atoD2 plasmid was performed as previously described (8). Briefly, in vivo cross-linking of the bacterial nucleoproteins took place by the addition of formaldehyde (final concentration, 1% [vol/vol]) directly to the E. coli cultures growing in modified M9 mineral medium (28). After 20 min, glycine (300 mM) was added to quench the cross-linking reactions and the cells were collected by centrifugation at 4°C and washed twice with ice-cold Tris-buffered saline. Cell lyses, chromatin preparations, and ChIPs were performed as previously described (8) using an anti-AtoC rabbit polyclonal antibody (2). The presence of the plasmid-borne atoDAEB promoter sequences in the chromatin immunoprecipitates was detected by atoDAEB promoter-lacZ fusion-specific PCR amplification using a primer pair consisting of U2patoD and a universal M13 forward primer. Following 27 cycles of amplification, the PCR products were analyzed on 3% (wt/vol) low-melting-point agarose gels (Nusieve) and visualized by ethidium bromide staining.
Hydroxylamine mutagenesis.
Plasmid pZero-patoD1 (described above), carrying the atoDAEB promoter region from −206 to +73, was mutagenized in vitro with hydroxylamine (33). The mutagenized promoter sequences were excised by EcoRI-BamHI digestion and cloned into pMLB1034 (34). Competent E. coli Top10 cells were transformed with the ligation products, and the cells were plated onto M9 mineral medium-agar plates containing 10 mM acetoacetate and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Plasmid DNA was isolated from white or pale blue colonies, and the atoDAEB promoter region was sequenced using standard methodology.
RESULTS
Identification of the acetoacetate-responsive cis element within the atoDAEB promoter regulatory region.
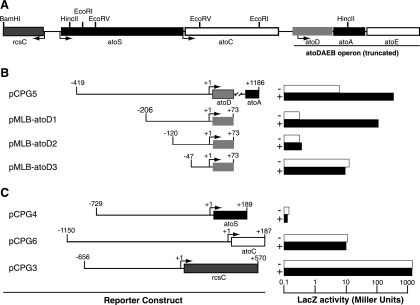
To define the acetoacetate-responsive cis element of atoDAEB, we constructed a series of galactosidase reporter translational fusions with atoDAEB subregions and monitored reporter expression in the absence or presence of the inducer. The reporter constructs pCPG5 and pMLB-atoD1 (Table 1), carrying the −419-to-+1186 and −206-to-+73 regions of atoDAEB, respectively (Table 1), were inducible by acetoacetate (Fig. 1B). However, further deletion of 86 or 159 bp from the 5′ end rendered the resulting constructs, pMLB-atoD2 and pMLB-atoD3 (Table 1), unresponsive to acetoacetate induction (Fig. 1B). Therefore, the acetoacetate-responsive element maps, at least partly, within the 86-bp region from −206 to −120 of the atoDAEB promoter (Fig. 1B).
FIG. 1.
Mapping of the acetoacetate-responsive cis element within the atoDAEB promoter. (A) Organization of the E. coli genomic sequences originally cloned into pUC-Az (2). The restriction enzyme sites used in the generation of reporter constructs are indicated. (B and C) E. coli Top10 cells carrying the indicated reporter constructs were grown in modified M9 mineral medium (28) in the absence (−; empty bars) or presence (+; filled bars) of 10 mM acetoacetate, and β-galactosidase (LacZ) activity in permeabilized cells was measured as described previously (20). (B) Susceptibility of reporter constructs carrying various regions of the atoDAEB promoter fused to the lacZ reporter gene to induction by acetoacetate. (C) Effects of acetoacetate on the activity of the atoS, atoC, and rcsC promoters.
To determine whether acetoacetate affects atoDAEB indirectly by altering the expression of the genes encoding its regulatory AtoS-AtoC TCS (15, 16), we also generated lacZ reporter fusion constructs with the −729-to-+189 region of atoS (pCPG4) or the −1150-to-+187 region of atoC (pCPG6). The expression of the lacZ reporter from these constructs was unaffected by the presence of acetoacetate (Fig. 1C). The expression of rcsC (36), a neighboring gene that is transcribed in the opposite direction (Fig. 1A) and that was also monitored as a specificity control, was not affected by acetoacetate (Fig. 1C).
In vitro analysis of AtoC binding to atoDAEB regulatory sequences.
To locate the AtoC binding sites within the atoDAEB promoter, we generated three overlapping DNA fragments that shared the same 3′ end (at +73 relative to the putative transcription start site) but differed at the 5′ end (Fig. 1) and tested them for the ability to bind AtoC. The affinity-purified rAtoC (16) used in these experiments was in its unphosphorylated state, since we were unable to phosphorylate it to a significant extent in vitro either with its cognate AtoS HK or with low-molecular-weight phosphodonors like carbamoyl phosphate, phosphoramidate, and acetyl phosphate (data not shown). Unphosphorylated RRs retain their ability to bind specifically their DNA target sequences, although phosphorylation enhances their affinity for DNA (27, 32, 43).
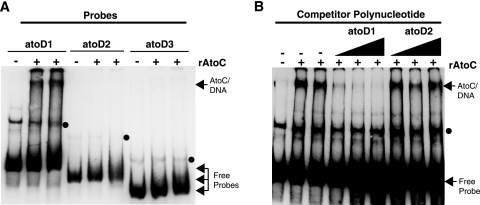
EMSA analysis revealed that while AtoC binds atoD1 (−206 to +73), it fails to bind atoD2 (−120 to +73) and atoD3 (−47 to +73) (Fig. 2A), suggesting that the sequences necessary for AtoC binding lie within the region from −206 to −120 relative to the predicted atoDAEB transcriptional start site. These data indicate that the same cis element, i.e., the region from −206 to −120, is involved in both AtoC binding and the acetoacetate responsiveness of atoDAEB and support the notion that AtoC is the transcription factor involved in the induction of atoDAEB by acetoacetate.
FIG. 2.
Mapping of the AtoC binding site(s) within the atoDAEB promoter. (A) The binding of AtoC (35 nM) to atoDAEB promoter regions atoD1 (−206 to +73), atoD2 (−120 to +73), and atoD3 (−47 to +73) was analyzed by nonradioactive EMSAs. (B) The DNA binding reactions were performed in the presence of 10 ng of the atoD1 probe (approximately 3 nM) and increasing amounts (5-, 10-, and 20-fold molar excesses) of either unlabeled specific (atoD1) or nonspecific (atoD2) competitors (see Materials and Methods) and analyzed by EMSAs. The AtoC-DNA complexes and the free probes are indicated by arrows, whereas the protein-independent bands are indicated by dots. +, present; −, absent.
The specificity of the above-described DNA-AtoC interactions was verified by competition experiments to evaluate the association of AtoC with biotinylated atoD1 in the presence of increasing concentrations of an unlabeled specific (atoD1) or nonspecific (atoD2) DNA competitor. The addition of increasing amounts of the specific atoD1 competitor greatly decreased the abundance of AtoC-DNA complexes, whereas similar concentrations of the nonspecific atoD2 competitor produced no visible effect (Fig. 2B). Finally, to further verify the presence of AtoC in the atoD1 EMSA complexes, we included in the reaction mixtures either a MAb that specifically recognizes polyhistidine stretches, such as the His10 tag of rAtoC, or a nonspecific MAb (58S) that recognizes an unrelated viral protein (31). While the nonspecific 58S MAb had no effect on the AtoC-DNA complexes, the specific anti-polyhistidine MAb clearly decreased the abundance of these complexes (data not shown). These data further demonstrate the presence of rAtoC in the EMSA complexes with the atoD1 probe.
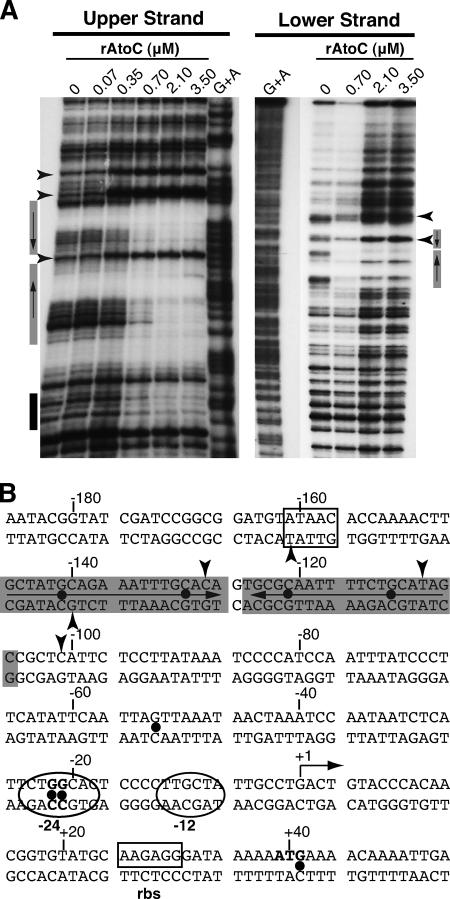
Characterization of the AtoC binding sites by DNase I footprinting.
Having shown that the region from −206 to −120 of atoDAEB is involved in both physical interactions with AtoC and promoter inducibility by acetoacetate, we performed DNase I protection footprinting analysis to identify the exact contacts of AtoC with promoter sequences. Probes labeled in either the upper or lower strand were subjected to DNase I nicking in the presence of increasing concentrations of AtoC (Fig. 3A). The analysis of the upper-strand-labeled promoter indicated that AtoC protects mainly the region from −146 to −110 from DNase I digestion. This region contains an inverted palindrome of the consensus sequence GCTATGCAGAAAWTTGCRCA in regions −146 to −127 and −126 to −107, between which the two halves are separated by 1 bp (Fig. 3B). The protection of these sites was verified by footprinting analysis of the lower strand (Fig. 3A). The locations of these AtoC binding sites are in good agreement with the relative positions of enhancer-like elements (ELEs) present in other σ54-dependent promoters (references 26 and 41 and references therein). AtoC binding induces significant DNase I hypersensitivity (Fig. 3), especially at positions −101, −109, and −128, indicating that significant structural DNA alterations take place upon AtoC binding. It is possible that such structural changes facilitate the interaction of AtoC with the σ54 RNA polymerase holoenzyme that is bound to the region from −24 to −12 (Fig. 3B).
FIG. 3.
DNase I footprinting analysis of the AtoC binding sites within the atoDAEB promoter regulatory region. (A) DNase I footprinting was performed with increasing concentrations of rAtoC and promoter fragments labeled in the coding (upper) or noncoding (lower) strand. Boxes indicate the protected areas, and arrowheads indicate the hypersensitivity positions. The gray boxes indicate the inverted palindrome protected by AtoC (see the text). (B) The atoDAEB sequence (−186 to +54) and its regulatory elements are shown. Note that the transcription initiation site shown in this figure is predicted and has not been validated experimentally. Shaded sequences represent the AtoC-protected areas, and arrowheads indicate AtoC-induced hypersensitivity. The positions of some predicted functional elements, such as the promoter region from −24 to −12 (in ovals), the putative ribosome binding site (boxed), and the initiator ATG (bold), are indicated. The positions of the hydroxylamine-induced mutations that render the promoter inactive and/or unresponsive to acetoacetate (Fig. 5 and 6) are marked by dots.
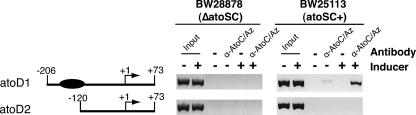
ChIP analysis of in vivo AtoC binding to atoDAEB regulatory sequences.
The above-described in vitro determination of the AtoC binding site within the atoDAEB regulatory sequences was also investigated in vivo by performing ChIP experiments (8) using atoSC+ (BW25113) or ΔatoSC (BW28878) bacteria carrying plasmids that either contained the AtoC binding site (pMLB-atoD1) or lacked it (pMLB-atoD2). Figure 4 shows that AtoC binds in vivo to the plasmid-borne atoDAEB sequence containing an AtoC binding site (atoD1, −206 to +73) whereas it fails to bind sequences lacking this site (atoD2, −120 to +73). Furthermore, the abundance of the AtoC-DNA complexes increased significantly in cells growing in the presence of acetoacetate. The specificity of these interactions was underscored by the lack of PCR products in the absence of antibody or in immunoprecipitates from the ΔatoSC E. coli strain BW28878 that lacks AtoC (Fig. 4). Thus, AtoC displayed the same sequence requirements for DNA binding both in vitro and in vivo, and the abundance of the AtoC-DNA complexes increased upon the activation of the AtoS-AtoC TCS by acetoacetate.
FIG. 4.
ChIP analysis of AtoC binding to atoDAEB promoter fragments in vivo. The binding of AtoC to plasmid-borne atoDAEB promoter fragments that either carry (atoD1) or lack (atoD2) the predicted binding site of AtoC (filled oval) in the presence (+) and absence (−) of acetoacetate (inducer) was determined. The atoSC+ strain BW25113 and its ΔatoSC isogenic counterpart BW28878, carrying either the plasmid pMLB-atoD1 or pMLB-atoD2 (Table 1), were used in this experiment. Immunoprecipitations were performed in the absence (−) or presence of anti-AtoC (α-AtoC/Az) antibodies, and they were followed by PCR amplification of the immunoprecipitated DNA fragments by using one atoDAEB-specific (U2patoD [Table 1]) and one plasmid-specific (universal M13 forward) primer. Chromatin input controls are indicated.
Characterization of the atoDAEB promoter regulatory elements by chemical mutagenesis.
To further characterize the functional cis elements within the atoDAEB promoter, we used in vitro hydroxylamine mutagenesis (33) and selected for severely reduced promoter activity in the presence of acetoacetate. This relatively unbiased approach was expected to yield two major types of mutations: (i) those that alter the core promoter elements, i.e., the sequence from −24 to −12 making contact with the σ54 holoenzyme of RNA polymerase, and (ii) those that render the promoter refractory to acetoacetate induction, e.g., by altering the AtoC binding site or the binding site of an AtoC cofactor protein. Indeed, sequence analysis of 24 mutants that displayed no, or greatly reduced, promoter activity in the presence of acetoacetate indicated that the majority of the mutants (16 out of 24) carried single mutations whereas the rest carried two or more mutations.
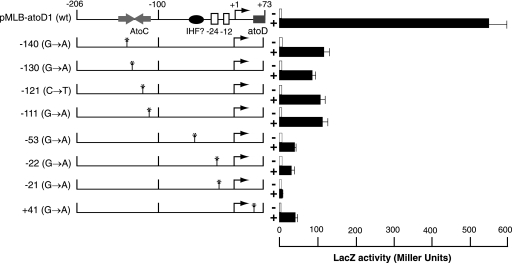
The majority of the single mutants (9 of 16) carried G-to-A transitions at positions −21 and −22 relative to the predicted transcription start site (Fig. 3B). Upon acetoacetate induction, the expression levels of the lacZ reporter in these mutants were severely reduced compared with that of the wild-type atoDAEB promoter regulatory sequence (Fig. 5). Four mutations that mapped within the AtoC binding site (−111, −121, −130, and −140) (Fig. 3B) resulted in significant decreases (≥80%) in the induced atoDAEB promoter activity (Fig. 5). Interestingly enough, one mutation that mapped at position −53, i.e., between the AtoC and the polymerase binding sequences (Fig. 3B), produced a more pronounced phenotype than any of the AtoC binding site mutations (Fig. 5). This result may be due either to a mutation-induced gross alteration in the DNA structure that also affects the AtoC binding or to the destruction of the binding site of another protein participating in the induction of the atoDAEB operon.
FIG. 5.
Acetoacetate inducibility of atoDAEB promoter mutants obtained by chemical mutagenesis. E. coli Top10 cells carrying wild-type or mutant atoDAEB promoters fused to the lacZ reporter gene were grown in modified M9 mineral medium (28) in the absence (−; empty bars) or presence (+; filled bars]) of 10 mM acetoacetate, and β-galactosidase (LacZ) activity in permeabilized cells was measured as described previously (20). The position of each mutation is indicated on the left. wt, wild type.
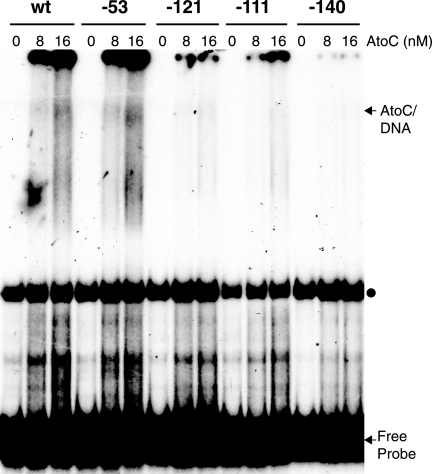
To determine whether the mutation at position −53 affects AtoC binding, as well as to confirm that the AtoC binding site mutations do affect the binding of AtoC, we tested the ability of probes carrying wild-type or mutant sequences to bind AtoC in vitro. The results of this experiment indicated that AtoC binding to probes carrying a single mutation within the AtoC binding site (at position −140, −121, or −111) was greatly reduced, whereas AtoC binding to a probe carrying the −53 mutation was indistinguishable from AtoC binding to the wild-type probe (Fig. 6). Therefore, the phenotype corresponding to the mutation at position −53 was not due to reduced AtoC binding.
FIG. 6.
In vitro binding of AtoC to wild-type and mutant atoDAEB promoter fragments. The binding of purified rAtoC (8 and 16 nM) to wild-type or mutant atoDAEB promoter (−206-to-+73) probes was analyzed by EMSAs. The mutations are identified at the top of each lane. AtoC-DNA complexes and free probes are indicated by arrows, whereas the positions of protein-independent bands are indicated by dots. wt, wild type.
The integration host factor (IHF) is required for the induction of the atoDAEB operon.
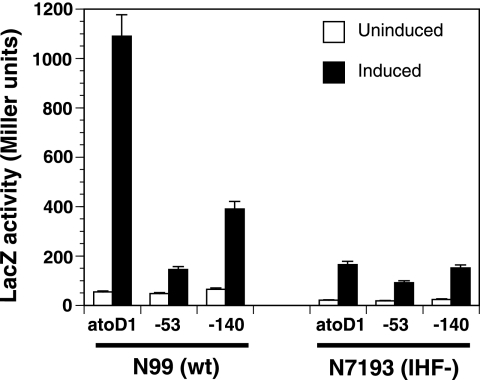
The atoDAEB promoter contains two sequence stretches (−62 to −50 and −37 to −25) that bear homology to IHF binding sites (data not shown). The presence of architectural factors, like IHF, is often required for the optimal contact of the ELE-bound NtrC-type transcription factor with the promoter-bound σ54 RNA polymerase (11, 26, 29, 41). Since the −53 mutation lies within one putative IHF binding site, we tested whether IHF is involved in the acetoacetate-induced expression of atoDAEB. Plasmids carrying the lacZ reporter under the control of either the wild-type promoter (atoD1) or a promoter with one mutation (−53 or −140) were introduced into both the wild-type E. coli strain N99 and its isogenic counterpart, N7193 (Table 1), which has an IHF null phenotype due to an insertion in the hip (himD) gene (9). LacZ activities in cells growing in the absence or presence of acetoacetate were monitored. The results of this experiment indicated that (i) the induction of atoDAEB in the IHF null N7193 strain was greatly reduced compared to that in wild-type N99 cells and (ii) in the IHF null cells, the wild-type atoDAEB promoter could not be differentiated from a mutant carrying an AtoC binding site mutation on the basis of acetoacetate inducibility (Fig. 7). This finding is in contrast with results for induced wild-type N99 cells, in which the LacZ reporter activity corresponding to the promoter with the −140 AtoC binding site mutation was more than 70% lower than that corresponding to the wild-type promoter (Fig. 7). These data indicated that the presence of IHF is required for the AtoC-mediated induction of the atoDAEB operon.
FIG. 7.
Effect of the IHF on the acetoacetate inducibility of wild-type and mutant atoDAEB promoters. A pair of isogenic wild-type (N99 [wt]) or IHF-negative (N7193 [IHF−]) E. coli strains (Table 1) were used to monitor the activities of the wild-type atoDAEB promoter (atoD1) and mutant atoDAEB promoters (with a mutation at position −53 or −140) in the absence (empty bars) or presence (filled bars) of 10 mM acetoacetate. The promoters had been fused to the lacZ reporter gene, and the activity of β-galactosidase (LacZ) in permeabilized cells was measured as described previously (20).
DISCUSSION
AtoC possesses diverse biological roles both as a posttranslational inhibitor of polyamine biosynthetic enzymes and as a transcriptional regulator of atoDAEB expression. The aim of the experiments reported here was to identify regulatory elements and factors that are involved in the transcriptional regulator function of AtoC.
The expression of the atoDAEB operon, whose gene products participate in the catabolism of short-chain fatty acids, is highly inducible by acetoacetate (13, 25). This induction requires AtoC (13), a transcription factor homologous to NtrC-NifA-type bacterial transcriptional regulators of the σ54 RNA polymerase holoenzyme (15, 26). These transactivators are usually part of two-component signal transduction systems that typically consist of a sensor HK and an RR, the latter of which is often a transcription factor (24, 35, 40). Upon sensing environmental or other signals, these HKs dimerize and autophosphorylate by trans-phosphorylation on conserved histidine residues. The phosphorylated HKs then transfer their phosphoryl groups to cognate RRs, usually to a conserved aspartate residue, leading to the multimerization and activation of the RRs. The activated RRs subsequently trigger the expression of the target operon(s), thus mounting a cell response to the received stimuli (24, 35, 40). We have previously shown that AtoC phosphorylation by the HK AtoS is required for the induction of atoDAEB expression upon the exposure of E. coli to acetoacetate (15, 16). By performing a deletion analysis of the atoDAEB promoter regulatory region, it was found that acetoacetate-mediated transcriptional activation requires cis elements mapping within the region from −206 to −120. Moreover, we showed that this induction is not due to altered levels of the AtoS-AtoC TCS proteins, since acetoacetate does not augment the expression of their genes, i.e., atoS and atoC.
We and others have reported previously that the in vitro AtoC phosphorylation by its cognate AtoS HK is rather inefficient (references 16 and 42 and unpublished data). Additionally, low-molecular-weight phosphodonors (5, 17, 18) such as carbamoyl phosphate, phosphoramidate, and acetyl phosphate that have been shown to phosphorylate a number of RRs in vitro failed to phosphorylate AtoC as well as they phosphorylate the other RRs (data not shown). It is not clear yet whether this failure to obtain significant amounts of phosphorylated AtoC represents a lack of phosphorylation or rapid AtoC-P dephosphorylation due to either chemical instability or phosphatase activity. By necessity, therefore, we used unphosphorylated AtoC in the in vitro protein-DNA binding experiments performed in the course of this work. Although RRs normally require prior phosphorylation by an HK to bind their target sequences in vivo (reference 35 and references therein), in vitro experiments have shown that unphosphorylated RRs retain their sequence-specific DNA binding activities, albeit with lower affinity (27, 32, 43). The observation that active forms of RRs exist in unphosphorylated preparations of these proteins has led to the suggestion of the two-state equilibrium hypothesis, which postulates that phosphorylation alters the equilibrium between the active and inactive conformations of an RR (7, 39). Indeed, our in vivo data have indicated that overexpressed AtoC can activate its target atoDAEB promoter even when it is unable to phosphorylate due either to a lack of its cognate AtoS HK or to phosphorylation site mutations (16).
The in vitro DNA binding data presented here demonstrated that purified rAtoC binds specifically within the same sequences that are involved in the acetoacetate responsiveness of the atoDAEB operon, i.e., the region from −206 to −120. Note that the binding of AtoC to its target sequences is not enhanced when acetoacetate is included in the in vitro binding reactions (data not shown), which is consistent with the notion that acetoacetate acts by triggering the AtoS-AtoC TCS. The in vitro findings were verified by ChIP experiments, which demonstrated that the exposure of E. coli to acetoacetate induces the binding of AtoC to the atoDAEB regulatory sequences from −206 to −120 in vivo. Thus, AtoC appears to display the same DNA binding specificity both in vivo and in vitro.
The exact location of AtoC binding was examined further using DNase I footprinting analysis, which revealed that AtoC binds two 20-bp regions constituting an inverted palindrome that are located at −146 to −127 and −126 to −107 relative to the predicted atoDAEB transcription start site (Fig. 3). The sequence of each 20-bp AtoC binding site conforms to the consensus GCTATGCAGAAAWTTGCRCA, and their locations are in good agreement with the relative positioning of other ELEs present in σ54-dependent promoters (references 26 and 41 and references therein). The importance of the presence of both copies of the AtoC binding site for the acetoacetate-dependent inducibility of atoDAEB was established from the results of chemical mutagenesis experiments. These results indicated that even single-base mutations within either of the two AtoC binding sites significantly reduced both the in vitro binding of AtoC to the mutant sites and the inducibility of the mutant promoters by acetoacetate. Thus, the integrity of both AtoC binding sites within the atoDAEB regulatory region is critical both for AtoC binding to DNA and for its effect on gene expression, possibly because this integrity is required for the cooperative DNA binding of this transcription factor (43).
We found another interesting mutation at −53 that reduced the inducibility of atoDAEB without affecting the binding of AtoC. This mutation lay within one of the two putative IHF binding sites that are located at −62 to −50 and −37 to −25, between the AtoC and RNA polymerase binding sites. IHF has been shown to bind several promoters that are regulated by NtrC-type transcription factors, and its binding is believed to facilitate the optimal contact of the ELE-bound transcription factor with the promoter-bound σ54 RNA polymerase (11, 26, 29, 41). Indeed, IHF is required for atoDAEB induction, so that E. coli cells lacking functional IHF due to an insertion in the hip (himD) gene (9) are severely impaired with regard to the levels of atoDAEB promoter inducibility by acetoacetate.
The chemical mutagenesis data also provide experimental evidence of the transcription of the atoDAEB promoter by the σ54 RNA polymerase, a relationship that was suspected based on sequence similarities but lacked experimental verification thus far (26). This confirmation stems primarily from the finding that the majority of the analyzed mutations that impair the expression of atoDAEB are located in a conserved GG doublet (at positions −21 and −22) whose significance in σ54 holoenzyme complex formation was recognized quite early (reference 14 and references therein) and was verified recently following the compilation and analysis of a large number of σ54-dependent bacterial promoter sequences (1).
Recent data posit the involvement of the AtoS-AtoC TCS in the expression of a number of E. coli genes other than those of the atoDAEB operon. Indeed, the deletion of the E. coli atoSC locus results in the up-regulation of 11 genes and the down-regulation of 32 genes (21), suggesting both positive and negative roles for the AtoS-AtoC TCS. While some of these effects may be indirect, it has been shown previously that RRs can be involved in both positive and negative gene regulation. For example, NtrC activates the σ54-dependent glnAp2 promoter but represses the σ54-dependent glnAp1 promoter in E. coli (30), while BvgA functions as both a transcriptional activator of multiple virulence factors and a repressor of genes required for motility and/or survival in several Bordetella species (4). This differential regulation may be due to a strategic placement of the binding sites and/or the concentration and the phosphorylation state of the RR. While BLAST analysis indicated that there are no palindromes consisting of two inverted AtoC binding sites other than the one at atoDAEB in the E. coli genome, it did show several regions with significant homology to the 20-bp AtoC binding site (data not shown). It is not clear whether any of these sites are recognized in vivo by either AtoC or an AtoC-containing complex or if they represent targets of regulation. However, it is clear that AtoC is more than a regulator of the genes that participate in short-chain fatty acid catabolism since it affects a number of other physiological processes, such as flagellum biosynthesis and motility, the biosynthesis of the biopolymer poly-(R)-3-hydroxybutyrate, and processes that influence the sodium sensitivity but not the potassium sensitivity of E. coli cells or their susceptibility to membrane-acting agents and aminoglycoside antibiotics (21, 38, 45).
It remains to be elucidated why AtoC evolved both as a posttranslational regulator of polyamine biosynthetic enzymes and as a transcriptional factor for genes that encode proteins involved in other aspects of the E. coli physiology and metabolism. It is tempting to speculate that the reason relates to the diverse effects exerted by the polyamines on the expression of the E. coli genes sometimes described as the polyamine modulon (44), which includes genes involved in flagellar synthesis (44), and the expression of promoters transcribed by the σ54 RNA polymerase (37).
Acknowledgments
We thank Saul Silverstein, Department of Microbiology, Columbia University, New York, NY, for the gift of the 58S MAb and his continuous support. We are also grateful to Hirofumi Aiba (Nagoya University, Japan) for the E. coli strains BW25113 and BW28878 and Max Gottesman (Columbia University, New York, NY) for strains N99 and N7193. We also thank Howard Shuman (Columbia University, New York, NY) for his constructive comments.
This work was supported in part by an IRAKLITOS doctoral research grant from the Hellenic Ministry of Education (M.K.M. and C.A.P.).
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canellakis, E. S., A. A. Paterakis, S. C. Huang, C. A. Panagiotidis, and D. A. Kyriakidis. 1993. Identification, cloning, and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 90:7129-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, D. P., and J. E. Cronan, Jr. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 4.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 5.Da Re, S. S., D. Deville-Bonne, T. Tolstykh, M. Veron, and J. B. Stock. 1999. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 457:323-326. [DOI] [PubMed] [Google Scholar]
- 6.Fong, W. F., J. S. Heller, and E. S. Canellakis. 1976. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim. Biophys. Acta 428:456-465. [DOI] [PubMed] [Google Scholar]
- 7.Gardino, A. K., B. F. Volkman, H. S. Cho, S. Y. Lee, D. E. Wemmer, and D. Kern. 2003. The NMR solution structure of BeF3−-activated Spo0F reveals the conformational switch in a phosphorelay system. J. Mol. Biol. 331:245-254. [DOI] [PubMed] [Google Scholar]
- 8.Grainger, D. C., T. W. Overton, N. Reppas, J. T. Wade, E. Tamai, J. L. Hobman, C. Constantinidou, K. Struhl, G. Church, and S. J. Busby. 2004. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 186:6938-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffo, G., A. B. Oppenheim, and M. E. Gottesman. 1989. Repression of the lambda pcin promoter by integrative host factor. J. Mol. Biol. 209:55-64. [DOI] [PubMed] [Google Scholar]
- 10.Heller, J. S., W. F. Fong, and E. S. Canellakis. 1976. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc. Natl. Acad. Sci. USA 73:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoover, T. R., E. Santero, S. Porter, and S. Kustu. 1990. The integration host factor stimulates interaction of RNA polymerase with NifA, the transcriptional activator for nitrogen fixation operons. Cell 63:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins, L. S., and W. D. Nunn. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins, L. S., and W. D. Nunn. 1987. Regulation of the ato operon by the atoC gene in Escherichia coli. J. Bacteriol. 169:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lioliou, E. E., and D. A. Kyriakidis. 2004. The role of bacterial antizyme: from an inhibitory protein to AtoC transcriptional regulator. Microb. Cell Fact. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lioliou, E. E., E. P. Mimitou, A. I. Grigoroudis, C. H. Panagiotidis, C. A. Panagiotidis, and D. A. Kyriakidis. 2005. Phosphorylation activity of the response regulator of the two-component signal transduction system AtoS-AtoC in E. coli. Biochim. Biophys. Acta 1725:257-268. [DOI] [PubMed] [Google Scholar]
- 17.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch, A. S., and E. C. Lin. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 22.Panagiotidis, C. A., S. Artandi, K. Calame, and S. J. Silverstein. 1995. Polyamines alter sequence-specific DNA-protein interactions. Nucleic Acids Res. 23:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panagiotidis, C. A., E. K. Lium, and S. J. Silverstein. 1997. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J. Virol. 71:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 25.Pauli, G., and P. Overath. 1972. ato operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur. J. Biochem. 29:553-562. [DOI] [PubMed] [Google Scholar]
- 26.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rombel, I., P. Peters-Wendisch, A. Mesecar, T. Thorgeirsson, Y. K. Shin, and S. Kustu. 1999. MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J. Bacteriol. 181:4628-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Santero, E., T. R. Hoover, A. K. North, D. K. Berger, S. C. Porter, and S. Kustu. 1992. Role of integration host factor in stimulating transcription from the sigma 54-dependent nifH promoter. J. Mol. Biol. 227:602-620. [DOI] [PubMed] [Google Scholar]
- 30.Shiau, S. P., P. Chen, and L. J. Reitzer. 1993. Effects of insertions and deletions in glnG (ntrC) of Escherichia coli on nitrogen regulator I-dependent DNA binding and transcriptional activation. J. Bacteriol. 175:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 34.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 36.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terui, Y., K. Higashi, S. Taniguchi, A. Shigemasa, K. Nishimura, K. Yamamoto, K. Kashiwagi, A. Ishihama, and K. Igarashi. 2007. Enhancement of the synthesis of RpoN, Cra, and H-NS by polyamines at the level of translation in Escherichia coli cultured with glucose and glutamate. J. Bacteriol. 189:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theodorou, M. C., C. A. Panagiotidis, C. H. Panagiotidis, A. A. Pantazaki, and D. A. Kyriakidis. 2006. Involvement of the AtoS-AtoC signal transduction system in poly-(R)-3-hydroxybutyrate biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1760:896-906. [DOI] [PubMed] [Google Scholar]
- 39.Volkman, B. F., D. Lipson, D. E. Wemmer, and D. Kern. 2001. Two-state allosteric behavior in a single-domain signaling protein. Science 291:2429-2433. [DOI] [PubMed] [Google Scholar]
- 40.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 41.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448-1456. [DOI] [PubMed] [Google Scholar]
- 43.Yang, X. F., Y. Ji, B. L. Schneider, and L. Reitzer. 2004. Phosphorylation-independent dimer-dimer interactions by the enhancer-binding activator NtrC of Escherichia coli: a third function for the C-terminal domain. J. Biol. Chem. 279:36708-36714. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, M., K. Kashiwagi, A. Shigemasa, S. Taniguchi, K. Yamamoto, H. Makinoshima, A. Ishihama, and K. Igarashi. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 279:46008-46013. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]