Abstract
During genetic transformation of Streptococcus pneumoniae, single strands from native donor DNA enter competent cells, where they associate with an unidentified protein with a molecular mass of 15 to 20 kDa to form the eclipse complex. Using Western blotting, we identify the principal protein cofractionating with donor DNA in this complex as SsbB.
During genetic transformation of Streptococcus pneumoniae (pneumococcus), single-stranded (ss) fragments extracted from double-stranded donor DNA (dsDNA) are transported into competent cells (10). Though its genetic information is intact, this ssDNA intermediate has much less transforming activity than the parental dsDNA (13) and is therefore described as being “in eclipse” (6); it emerges from eclipse during integration into the resident dsDNA. Little is known about the mechanism of this efficient recombination reaction or even about the precise role(s) of the four competence-induced proteins that are likely to participate in it, CoiA (4), DprA (1), RecA (11, 19), and SsbB (1, 2; unpublished data from the University of Illinois at Chicago and unpublished data from the Laboratoire de Microbiologie et Génétique Moléculaires [LMGM]).
While in eclipse, donor DNA strands are recoverable from cell lysates as a nuclease-resistant nucleoprotein complex (14, 15, 18). This eclipse complex (EC), which arises intracellularly, not in lysates (14), contains a single [35S]methionine-labeled protein with an apparent molecular mass (MM) of 15.7 or 19.5 kDa whose identity has not been established (16, 17). The ability to interact with ssDNA has been documented for only three of the four above-mentioned competence-specific proteins, RecA, SsbB, and DprA. RecA is an ATP-dependent DNA recombinase whose sequence is 60% identical to that of the RecA protein from Escherichia coli (22). SsbB is, with SsbA, one of two paralogous ssDNA-binding proteins identified in S. pneumoniae (8); in contrast to SsbA, it is not essential. DprA was recently characterized as another ssDNA-binding protein able to protect ssDNA from nucleases in vitro (20). Recently, evidence that DprA is a presynaptic protein that conveys incoming ssDNA to RecA was obtained (21). On the other hand, CoiA appears to act later, as the EC accumulates to normal levels in a coiA mutant but progresses 100-fold less efficiently to integration (4, 5; unpublished data from LMGM).
The labeled component of the EC seems more likely to be SsbB (MM, 14,925 Da) than DprA (MM, 31,063 Da) or RecA (MM, 41,950 Da), but the basis for this argument is indirect, resting on the observation of a single prominent competence-specific protein in the 12- to 20-kDa region with exactly the mobility of the 35S-labeled EC component (16). To identify more precisely the component(s) of the EC, we scaled up the EC purification described previously and detected protein components both by Western blotting with specific antisera and by nonspecific protein-staining methods.
Initial fractionation of cell extract.
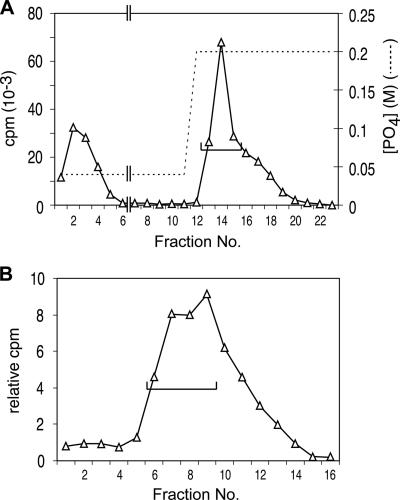
The purification procedure described previously (15) was modified slightly to allow an increase of scale by 2 orders of magnitude. Specifically, sucrose gradient fractionation was replaced as the first step by an initial adsorption to hydroxylapatite (HAP). This capture step eliminated from the lysate both material not binding to HAP in 0.04 M sodium phosphate buffer (PB) and material not released by 0.2 M PB (Fig. 1A). Remaining components were separated by gel exclusion chromatography as described previously, but using Sephacryl S400HR (Fig. 1B). As the exclusion limit for this material is 300 bp for DNA but 107 kDa for globular proteins, this method is expected to separate nucleoprotein filaments from most nonadhering proteins.
FIG. 1.
Initial fractionation of cell extract. (A) An 800-ml culture of R1501 (3) in C medium supplemented with bovine serum albumin and CaCl2 at pH 7.9 (9) was treated with 80 μg of CSP-1 at an optical density of 0.11 at 37°C for 12 min. The competent culture was cooled to 25°C by agitation in ice-water for 64 s in two 500-ml Schott bottles, exposed to 160 μg [3H]DNA (20,000 cpm μg−1; uniformly labeled chromosomal DNA prepared as described previously [5]) for 6 min and then to 3 mg DNase I for 30 s, and cooled further for centrifugation at 5,000 × g for 15 min. The pellet was washed with 40 ml cold SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and resuspended in 8 ml lysis buffer containing 0.1 M NaCl, 0.05 M Tris-HCl (pH 7.5), 0.01 M EDTA, 0.5% Sarkosyl, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 50 μg ml−1 RNase A, and 10% glycerol. The clear lysate obtained after 5 min at 37°C was passed 20 times through a 5-cm 21G hypodermic needle to reduce viscosity. The sheared lysate was brought to 0.04 M PB (pH 6.8) and loaded on a 9-ml HAP column. The column was washed at room temperature with 3.3 column volumes of 0.05% Sarkosyl-0.04 M PB, followed by 23 ml of the same buffer adjusted to 0.20 M PB. The flowthrough and subsequent eluates were collected in 4-ml (lanes 1 to 8) or 2-ml (lanes 9 to 23) fractions. Total numbers of cpm per fraction are displayed. Peak fractions (lanes 13 to 15) containing ∼40% of donor DNA label (bracket) were pooled. (B) S400 gel exclusion fractionation. Pooled fractions from panel A (6 ml) were applied to a 48-ml (1.5-cm diameter) column of Sephacryl S400HR and eluted with 0.15 M NaCl-0.05% Sarkosyl at 0.25 ml min−1. The first 16 2-ml fractions collected are shown. Fractions 6 to 9 were combined for analysis by gradient elution from HAP.
Identification of the major EC protein.
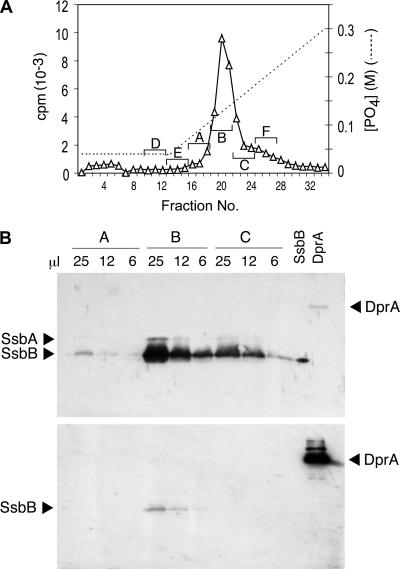
Finally, the excluded-volume peak was fractionated on HAP with a sodium phosphate gradient, as described previously (14). As shown in Fig. 2A, a single peak of donor DNA label emerged near 0.12 M PB, earlier than ssDNA, which elutes at 0.19 M PB. Previous studies explained the lower affinity for HAP as arising from the protein component of the EC. To identify proteins associated with donor DNA in the EC, both the peak fraction pool and two side fraction pools from the HAP gradient elution were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for Western blotting analysis, using 1% of each pool per lane. As shown in Fig. 2B (top) (as well as in Fig. S2B in the supplemental material), an anti-ssDNA-binding-protein (SSB)-positive protein comigrating with authentic recombinant pneumococcal SsbB was present; its amount, as judged by this enhanced chemiluminescence stain, was greatest in the peak fraction pool and much less in the pools of the side fractions, while antisera of similar sensitivity yielded no indication of any DprA (Fig. 2B, bottom) or RecA (not shown). As 0.5 ng DprA would have been detected in the Western blots, we conclude that there was less than 2 ng DprA in each pooled fraction (A to C).
FIG. 2.
Identification of the major EC protein. (A) HAP chromatography of EC. The S400HR excluded-volume peak was brought to 0.04 M PB and loaded on a 2-ml HAP column; the column was washed with 9 ml 0.04 M PB and eluted with a 40-ml linear gradient of PB from 0.04 to 0.3 M PB. The nominal PB gradient was estimated for each collected fraction. Three-fraction pools were made as indicated by letters and brackets and concentrated 40-fold by centrifugal ultrafiltration (Amicon Centricon; 10,000-Da MM cutoff) with 0.05% SDS and 100 ng bovine serum albumin. (B) Western blot analyses were classically performed as described in reference 11, using samples of the indicated volumes from 100-μl pools A, B, and C (from panel A); 10 ng of purified SsbB protein and 20 ng of purified DprA protein (20) were included as controls. Rabbit polyclonal antibodies raised against purified Bacillus subtilis SSB were used at a 1/2,000 serum dilution to detect SsbB and SsbA proteins (upper). Antibodies were then stripped off, and the membrane was incubated with rabbit polyclonal anti-DprA antibodies raised against two synthetic peptides of DprA (residues 3 to 18 and residues 118 to 131) at a 1/5,000 serum dilution to detect DprA protein (lower) (note that the band observed at the position of SsbB might correspond to incomplete removal of anti-SSB antibodies).
In a typical experiment (see Fig. S2 in the supplemental material), comparison with the intensity of the band of authentic recombinant SsbB protein in the same gel suggests that the amount of SsbB was about 2,800 ng in the peak pool (fraction B) but only 410 and 235 ng in side fraction pools A and C, respectively. This peak amount value corresponds to about 800 molecules of SsbB per cell engaged in the EC. In parallel, the total number of 3H[DNA] counts per minute (cpm) recovered in the peak pool corresponds to about 8,000 nucleotides internalized per cell. Within the limits of our calculation, the ratio of one SsbB tetramer to 40 nucleotides in the EC is consistent with the reported SSB65-like mode of binding of SsbB to ssDNA (7).
Search for an additional EC protein(s).
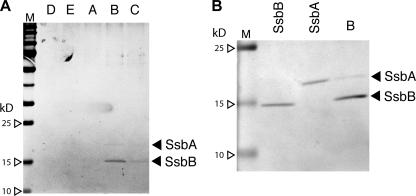
To identify an additional protein(s) that would not have been detected by our antisera, SDS-PAGE gels prepared with larger samples of the same HAP pools were examined for protein content by silver staining (Fig. 3) and Coomassie blue staining (not shown). They revealed a major band that comigrated in PAGE with SsbB and was at the highest level in the peak pool. We conclude that the principal protein cofractionating with donor DNA in the EC is SsbB. There was a second band, of lower intensity, that also centered on the peak pool. Curiously, this protein comigrated in SDS-PAGE with recombinant pneumococcal SsbA (Fig. 3).
FIG. 3.
Silver-stained SDS-PAGE gels of 10-μl samples of HAP pools. Panels A and B correspond to HAP pools from independent experiments (whose results are shown in Fig. 2A and Fig. S2B in the supplemental material, respectively). The amount of SsbA and SsbB in reference lanes was 200 ng. For silver staining, gels were fixed in 40% ethanol-0.05% formaldehyde solution and incubated in 0.02% Na2S2O3, followed by incubation in 0.1% AgNO3 and visualization in 3% Na2CO3-0.02% Na2S2O3-0.05% formaldehyde solution, as described previously (12). M, MM standards.
Among the proteins synthesized in pneumococci developing competence, only a single protein, with an MM estimated variously at 15.7 and 19.5 kDa, copurifies with the EC (16, 17). The present results identify that protein as SsbB. Does the complex contain other proteins as well? Neither stained gel revealed any protein present in a greater quantity than SsbB, and only a single band of lower intensity was visible under silver staining. Thus, it appears that there may be a single additional component, possibly SsbA, that is present at less than 1/10 the amount of SsbB (from silver staining) (Fig. 3B). Of course, the degree to which additional proteins can be excluded depends on the mass of the EC available and on the sensitivities of the staining methods used. The possible presence of SsbA should be interpreted with caution, as its comigration with EC material is by no means proof of the binding of SsbA to incoming ssDNA. This material could instead result from the capture of replication intermediates (recipient chromosomal ssDNA) to which SsbA is presumably bound. Such material is very likely to exist (e.g., the estimate for Escherichia coli is about 3 kb ssDNA per cell) and may not behave differently from SsbB-ssDNA complexes during HAP chromatography.
Significance of the SsbB-ssDNA EC?
Although the processing of donor DNA between its release from the transport pore as a single strand and its incorporation within duplex resident target DNA is entirely unknown, the participating competence-specific proteins can now be associated with several distinct stages or transitions on the basis of physical or genetic evidence. The significance of our present finding that internalized strands of donor DNA are complexed with SsbB is unclear. As EC ssDNA has been shown to be insensitive to nucleases (18), it is tempting to attribute to SsbB a protective role. However, the reduction in the yield of recombinants in ssbB mutant cells, which was initially reported to be 10- to 30-fold (2), was subsequently found to be only 3- to 5-fold (1; unpublished data from the University of Illinois at Chicago and unpublished data from LMGM). It is therefore difficult to conclude that SsbB plays an essential role in the protection of incoming ssDNA from nucleases.
In contrast, uptake of DNA by competent cells of dprA or recA mutants appears to be entirely destructive; neither ssDNA, an EC, nor any other macromolecular product of uptake has been found (1). Together with the observation that DprA binds ssDNA and protects it from nucleases (20), this would be consistent with a prominent role for DprA in protection of internalized donor DNA. It is therefore puzzling that neither DprA nor RecA is associated with EC material. These competence pheromone-induced proteins could have an indirect role in stabilizing donor strands during or immediately after uptake, perhaps by favoring the loading of SsbB on the incoming DNA strand. Although there is no direct proof of any role for DprA or RecA during the integration process, indirect evidence suggests that both proteins play important roles in the process, in addition to being required for nondestructive uptake. Thus, DprA is presumably involved in the loading of RecA onto ssDNA (21), and RecA, which is also known to be required for homologous recombination outside competence (23), has the potential to form heteroduplex joints in vitro (22). Future work should unravel further the role(s) of each dancer in the transformation ballet.
Supplementary Material
Acknowledgments
We thank Patrice Polard and his group (Laboratoire de Génétique Microbienne, INRA, Jouy en Josas, France) for the generous gift of rabbit polyclonal antibodies raised against purified Bacillus subtilis SSB and of purified S. pneumoniae SsbA and SsbB.
This work was supported in part by a grant (RB/CD/2003/09/001) from the Ministère Délégué à la Recherche et aux Nouvelles Technologies (Programme Microbiologie 2003-2004), by a grant from the Agence Nationale de la Recherche (projet no. BLAN06-3_141806), and by a grant from the NSF Microbial Genetics Program (MCB 0543187).
Footnotes
Published ahead of print on 29 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bergé, M., I. Mortier-Barrière, B. Martin, and J. P. Claverys. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming single strands. Mol. Microbiol. 50:527-536. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 3.Dagkessamanskaia, A., M. Moscoso, V. Hénard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 4.Desai, B. V., and D. A. Morrison. 2006. An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 188:5177-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, B. V., and D. A. Morrison. 2007. Transformation in Streptococcus pneumoniae: formation of eclipse complex in a coiA mutant implicates CoiA in genetic recombination. Mol. Microbiol. 63:1107-1117. [DOI] [PubMed] [Google Scholar]
- 6.Ephrussi-Taylor, H. 1960. L'état du DNA transformant au cours des premières phases de la transformation bactérienne. C. R. Soc. Biol. 154:1951-1955. [PubMed] [Google Scholar]
- 7.Grove, D. E., and F. R. Bryant. 2006. Effect of Mg2+ on the DNA binding modes of the Streptococcus pneumoniae SsbA and SsbB proteins. J. Biol Chem. 281:2087-2094. [DOI] [PubMed] [Google Scholar]
- 8.Grove, D. E., S. Willcox, J. D. Griffith, and F. R. Bryant. 2005. Differential single-stranded DNA binding properties of the paralogous SsbA and SsbB proteins from Streptococcus pneumoniae. J. Biol. Chem. 280:11067-11073. [DOI] [PubMed] [Google Scholar]
- 9.Guiral, S., V. Hénard, C. Granadel, B. Martin, and J. P. Claverys. 2006. Inhibition of competence development in Streptococcus pneumoniae by increased basal-level expression of the ComDE two-component regulatory system. Microbiology 152:323-331. [DOI] [PubMed] [Google Scholar]
- 10.Lacks, S., B. Greenberg, and K. Carlson. 1967. Fate of donor DNA in pneumococcal transformation. J. Mol. Biol. 29:327-347. [DOI] [PubMed] [Google Scholar]
- 11.Martin, B., P. García, M. P. Castanié, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367-379. [DOI] [PubMed] [Google Scholar]
- 12.Merril, C. R., D. Goldman, S. A. Sedman, and M. H. Ebert. 1981. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science 211:1437-1438. [DOI] [PubMed] [Google Scholar]
- 13.Miao, R., and W. R. Guild. 1970. Competent Diplococcus pneumoniae accept both single- and double-stranded deoxyribonucleic acid. J. Bacteriol. 101:361-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison, D. A. 1977. Transformation in pneumococcus: existence and properties of a complex involving donor deoxyribonucleate single strands in eclipse. J. Bacteriol. 132:576-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison, D. A. 1978. Transformation in pneumococcus: protein content of eclipse complex. J. Bacteriol. 136:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison, D. A., and M. Baker. 1979. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature 282:215-217. [DOI] [PubMed] [Google Scholar]
- 17.Morrison, D. A., M. Baker, and B. Mannarelli. 1979. A protein component of the pneumococcal eclipse complex, p. 43-52. In S. W. Glover and L. O. Butler (ed.), Transformation—1978. Cotswold Press Ltd., Oxford, United Kingdom.
- 18.Morrison, D. A., and B. Mannarelli. 1979. Transformation in pneumococcus: nuclease resistance of deoxyribonucleic acid in eclipse complex. J. Bacteriol. 140:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortier-Barrière, I., A. de Saizieu, J. P. Claverys, and B. Martin. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159-170. [DOI] [PubMed] [Google Scholar]
- 20.Mortier-Barrière, I., M. Velten, P. Dupaigne, S. McGovern, B. Martin, E. Le Cam, P. Polard, and J. P. Claverys. 2006. Exploration of the DNA binding properties of DprA, a competence-induced protein essential for the production of transformants in Streptococcus pneumoniae, abstr. A56, p. 67-68. 7th ASM Conference on Streptococcal Genetics, St. Malo, France, 18 to 21 June 2006.
- 21.Mortier-Barrière, I., M. Velten, P. Dupaigne, N. Mirouze, O. Piétrement, S. McGovern, G. Fichant, B. Martin, P. Noirot, E. Le Cam, P. Polard, and J. P. Claverys. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell, in press. [DOI] [PubMed]
- 22.Steffen, S. E., and F. R. Bryant. 2000. Purification and characterization of the RecA protein from Streptococcus pneumoniae. Arch. Biochem. Biophys. 382:303-309. [DOI] [PubMed] [Google Scholar]
- 23.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.