Abstract
The opportunistic pathogen Pseudomonas aeruginosa produces colorful, redox-active antibiotics called phenazines. Excretion of pyocyanin, the best-studied natural phenazine, is responsible for the bluish tint of sputum and pus associated with P. aeruginosa infections in humans. Although the toxicity of pyocyanin for other bacteria, as well as its role in eukaryotic infection, has been studied extensively, the physiological relevance of pyocyanin metabolism for the producing organism is not well understood. Pyocyanin reduction by P. aeruginosa PA14 is readily observed in standing liquid cultures that have consumed all of the oxygen in the medium. We investigated the physiological consequences of pyocyanin reduction by assaying intracellular concentrations of NADH and NAD+ in the wild-type strain and a mutant defective in phenazine production. We found that the mutant accumulated more NADH in stationary phase than the wild type. This increased accumulation correlated with a decrease in oxygen availability and was relieved by the addition of nitrate. Pyocyanin addition to a phenazine-null mutant also decreased intracellular NADH levels, suggesting that pyocyanin reduction facilitates redox balancing in the absence of other electron acceptors. Analysis of extracellular organic acids revealed that pyocyanin stimulated stationary-phase pyruvate excretion in P. aeruginosa PA14, indicating that pyocyanin may also influence the intracellular redox state by decreasing carbon flux through central metabolic pathways.
Redox transformations are a defining feature of the creation of biomass. To form precursors for incorporation into cellular material, heterotrophic organisms catalyze the oxidation of organic carbon sources, generating reducing power. This reducing power can be released in fermentation products or transferred to an externally supplied oxidant via the respiratory chain. The fluid exchange of electrons between intra- and extracellular environments permits organisms to maintain a buffered intracellular redox state, a condition required for the stability and function of biological macromolecules (7, 47). Under traditional laboratory culture conditions, electron donors and acceptors are often provided in excess, allowing microorganisms to control intracellular redox conditions. However, it is becoming clear that energy starvation, i.e., the limitation of substrates for oxidative or substrate-level phosphorylation, more closely mirrors conditions encountered by many bacteria in their natural habitats (37). How do bacteria maintain redox homeostasis under these conditions?
Bacteria of the genus Pseudomonas, like most heterotrophic bacteria (12, 24), oxidize organic carbon sources via the activity of the Entner-Doudoroff pathway and the citric acid cycle. Several of the oxidative steps in these pathways are coupled to the reduction of NAD+ to NADH, and NADH must be reoxidized so that these pathways can proceed and generate anabolic precursors. In pseudomonads, the primary mechanism whereby this is accomplished is through reduction of one of the NADH dehydrogenases at the start of the respiratory chain, which ultimately transfers the electrons to oxygen or nitrate (71). It has therefore been assumed that pseudomonads, organisms that rely on respiration for growth under most conditions, accumulate NADH when terminal electron acceptors become limiting.
Given that the NADH/NAD+ redox couple plays a major role in central metabolism, the ratio of the reduced form to the oxidized form is thought to be representative of the intracellular redox state (16). In previous studies, measurements of the NADH/NAD+ ratio in a variety of bacterial species have distinguished the opportunistic pathogen Pseudomonas aeruginosa from organisms such as Clostridium welchii, Klebsiella aerogenes, Escherichia coli, and Staphylococcus albus as the only species with a steady-state NADH/NAD+ ratio greater than 1 (73). However, another characteristic feature of some pseudomonad strains, which distinguishes them from all of the other genera mentioned above, is the ability to produce redox-active antibiotics known as phenazines (44). Some of these compounds, including pyocyanin, the best-studied phenazine due to its role in the pathology of P. aeruginosa infections (38), have been shown to react with NADH in vitro (15, 36). This has led to the hypothesis that electron transfer to phenazines may represent an adaptation that allows bacteria to modulate their intracellular redox state (23, 52, 66, 67). This physiological role would be consistent with the fact that phenazine biosynthesis is regulated such that phenazines are produced at high cell densities (9, 33, 51, 69), a condition that typically correlates with electron acceptor limitation (63).
Much research has focused on the toxic effects of pyocyanin as a virulence factor in the eukaryotic host (38, 42, 48, 56, 62) as well as in microorganisms (2, 3, 28, 35, 55). These effects have been attributed to the production of reactive oxygen species such as superoxide in the presence of pyocyanin (26, 27), and physiological studies have shown that P. aeruginosa resists the toxicity of this compound with increased superoxide dismutase and catalase activities under pyocyanin-producing conditions (29, 30). Additionally, recent gene expression studies have uncovered a role for pyocyanin in intercellular signaling (18). However, little is known about the role of this compound in pseudomonad metabolism or whether P. aeruginosa derives a benefit from the utilization of pyocyanin as an electron acceptor.
The facile reversibility of phenazine redox reactions allows these compounds to oxidize major intracellular reductants and subsequently reduce extracellular oxidants, thereby acting as redox mediators (25, 31, 39, 46, 53, 61). The redox potentials of pyocyanin and phenazine-1-carboxylic acid, the two major phenazines produced by P. aeruginosa PA14, are −34 mV (22) and −116 mV (Y. Wang and D. K. Newman, unpublished data), respectively, versus the standard hydrogen electrode at pH 7. These potentials are high enough to allow reduction by NADH (E0′ = −320 mV) and glutathione (E0′ = −240 mV) (1, 65) but low enough to allow electron transfer to environmentally relevant oxidants, including oxygen, nitrate, and ferric iron. Therefore, electron shuttling via phenazines may be a mechanism whereby pseudomonads can utilize electron acceptors that, due to low concentrations or solubility, might otherwise be inaccessible via conventional biochemical and enzymatic routes (13, 32, 52). The role of this electron transfer in P. aeruginosa energy metabolism has yet to be elucidated, but work carried out with another gammaproteobacterium, Shewanella oneidensis MR-1, indicated that phenazines, as well as similarly structured small molecules, facilitates reduction of insoluble ferric oxyhydroxides and stimulates growth (31, 41). Furthermore, a critical electron carrier function has been demonstrated for a membrane-bound phenazine derivative present in the electron transport chains of methanogenic archaea (4). In an effort to better understand the physiological significance of phenazine reduction in pseudomonads, we characterized the effects of pyocyanin on redox homeostasis and central metabolism in P. aeruginosa PA14.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For this study, we used P. aeruginosa strain UCBPP-PA14 (54), which produces approximately 10 times more pyocyanin in LB batch cultures than strain PAO1 (18). The P. aeruginosa PA14 mutant containing the MAR2xT7 transposon inserted in the ldhA gene in a ΔexoU background was obtained from a publicly available mutant library (40) and has mutant identification no. 5174. Generation of the P. aeruginosa PA14 ΔphzA1-1G1 ΔphzA2-2G2 deletion mutant (hereafter referred to as the Δphz mutant) was described previously (18). P. aeruginosa PA14 wild-type and mutant strains were grown aerobically at 37°C in Luria-Bertani broth (Fisher Scientific), or modified MOPS (morpholinepropanesulfonic acid) synthetic medium (50). Our modified MOPS synthetic medium contained 50 mM MOPS (Sigma) at pH 7.2, 93 mM NH4Cl, 43 mM NaCl, 2.2 mM KH2PO4, 1 mM MgSO4·7H2O, and 3.6 μM FeSO4·7H2O. Unless otherwise noted, 50 mM d-glucose was added to this medium as the sole carbon and energy source. Aerobic conditions were generated either through incubation with vigorous shaking at 250 rpm or in a BioFlo 110 fermentor (New Brunswick Scientific) set to agitate at 250 rpm and bubble with 100% air at a rate of 2 liters/minute. Aerobic culture volumes relative to vessel size are described below for specific experiments. Culture densities were followed at 500 or 600 nm in a Thermo Spectronic 20D+ or Beckman Coulter DU 800 spectrophotometer. Cultures with optical densities of greater than 0.8 were diluted 1:10 in fresh medium to allow accurate measurements. The method used to test strains for the ability to survive via pyruvate fermentation is described in the supplemental material.
Preparation of pyocyanin for reduction assays and NADH/NAD+ studies.
To maximize pyocyanin yields from P. aeruginosa cultures, we utilized a mutant, strain DKN370, which contains two copies of the gene phzM. PhzM converts phenazine-1-carboxylic acid to the precursor for pyocyanin, 5-methylphenazinium carboxylate (45). Purification of pyocyanin by organic extractions was carried out as described previously (18). High-pressure liquid chromatography (HPLC) analysis verified the purity of pyocyanin after the extraction step, so the HPLC purification step described in reference 18 was omitted. Purified pyocyanin was dissolved in MOPS buffer (MOPS synthetic medium without FeSO4, MgSO4, or glucose), and filtered (0.2-μm filter).
Whole-cell suspension assay for pyocyanin reduction.
Cell culture samples were concentrated or diluted in filtrates of supernatants from the same culture to normalize the optical density at 600 nm to 0.8. In an anaerobic chamber, the samples were transferred to cuvettes, and an anoxic solution of oxidized pyocyanin (in MOPS buffer) was added for a final pyocyanin concentration of about 0.1 mM. The cuvettes were stoppered to minimize oxygen exposure. Pyocyanin reduction was then followed as a decrease in absorbance at 690 nm over time in a DU 800 Beckman Coulter spectrophotometer. The rate of reduction could be calculated by converting the change in absorbance to μmol pyocyanin using the extinction coefficient for pyocyanin at this wavelength (ɛ = 4,310 M−1 cm−1 at pH 7) (49) and the volume of sample in the cuvette (1 ml).
Quantification of pyocyanin.
Pyocyanin concentrations in filtrates (0.2-μm pore) from LB and MOPS synthetic medium cultures were quantified as described previously (18). Briefly, absorbance in LB culture filtrates was measured spectrophotometrically at 690 nm, and pyocyanin concentrations were calculated using the extinction coefficient for pyocyanin (see above). Pyocyanin concentrations in 200-μl sample filtrates from MOPS synthetic medium cultures were determined by HPLC analysis on a Waters Symmetry C18 reverse-phase column by a gradient method (water versus acetonitrile containing 0.1% trifluoroacetic acid) and calculated based on absorbance values for purified standards diluted into MOPS buffer.
Extraction and quantification of intracellular NADH and NAD+.
Extraction of NADH and NAD+ was carried out according to the method described by San et al. (58). Two 1-ml samples of culture were placed in two separate microcentrifuge tubes and centrifuged at 16,000 × g for 1 min. Supernatant was removed, and pellets were resuspended in 300 μl of 0.2 M NaOH (for NADH extraction) or 0.2 M HCl (for NAD+ extraction). These extracts were incubated for 10 min at 50°C and then for 10 min on ice. While vortexing, 300 μl of 0.1 M HCl (for NADH) or 0.1 M NaOH (for NAD+) was added dropwise to neutralize the solutions. The samples were then centrifuged for 5 min at 16,000 × g. Supernatants were transferred to fresh tubes and stored at −80°C until quantification.
Relative or absolute NADH and NAD+ were quantified using a modification (58) of the enzyme cycling assay developed by Bernofsky and Swan (6), adapted for measurement in a microtiter plate. A master reagent mix was prepared with 1× Bicine buffer (2.0 M, pH 8.0), 8× water, 1× 80 mM EDTA, 2× 100% ethanol, 2× 4.2 mM thiazolyl blue, and 4× 16.6 mM phenazine ethosulfate. The reagent mix was warmed to 30°C, and then 90-μl aliquots were dispensed into individual wells of a 96-well microtiter plate. Five microliters of standard or sample was added to each well, and then the cycling reaction was started by the addition of 5 μl of alcohol dehydrogenase (Sigma no. A-3263) prepared at 347 units/ml in 0.1 M Bicine (pH 8.0). The microtiter plate was incubated at 30°C, the contents were mixed by brief shaking, and the plate was read every 30 to 60 seconds for absorbance at 570 nm, which is the spectral peak of thiazolyl blue that increases upon reduction. Slopes arising from plots of absorbance at 570 nm over time were generated for NADH and NAD+ standards as well as for all samples. Standard curves were used to calculate the absolute concentrations in μM, and values were normalized to the optical density of the original cell culture sample.
Relative quantification of dissolved oxygen in batch cultures.
Oxygen was measured in batch fermentor cultures using a Clark electrode (11). The electrode was calibrated such that the reading obtained by the computer without the probe attached was equal to zero, while the initial reading for the uninoculated medium (after aeration and agitation for 12 h) was set to 100 percent.
Analysis of small organic acids in culture filtrates.
Two hundred microliters was sampled from MOPS-glucose cultures (10 ml in an 18- by 150-mm test tube) at regular intervals and filtered (0.2-μm pore). In cases where repeated sampling from the same culture would alter the total culture volume by more than 10%, multiple identical cultures were inoculated from the same preculture and sampled sequentially. Twenty microliters of each filtrate was loaded onto a Bio-Rad Aminex HPX-87H column (300 by 7.8 mm) and subjected to an isocratic method in 5 mM H2SO4 at 35°C, using a Waters HPLC system. Compounds were detected by UV absorbance at 210 nm. Absolute concentrations of pyruvate were calculated using a standard curve for pyruvate diluted in MOPS buffer. The identity of the pyruvate peak was verified by coelution of an internal standard.
RESULTS
P. aeruginosa PA14 catalyzes pyocyanin reduction.
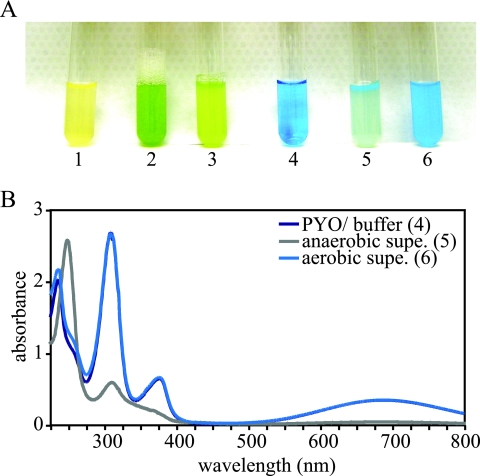
Stationary-phase LB cultures of P. aeruginosa PA14 turn bright blue-green due to production of the blue pigment pyocyanin specifically during this growth phase. P. aeruginosa PA14 also catalyzes the reduction of pyocyanin, a process that is readily observed when a stationary-phase culture is left standing without mixing or aeration by bubbling. Pyocyanin is converted from its blue (oxidized) form to a colorless (reduced) form (13). At the air-liquid interface, pyocyanin remains oxidized or becomes reoxidized by an abiotic reaction with oxygen, but respiration by the bacteria creates a steep oxygen gradient just below this interface such that pyocyanin below a few millimeters remains colorless. A demonstration of this process is depicted in Fig. 1A (tube 3). We centrifuged a stationary-phase culture, resuspended the cell pellet in a 100 μM solution of pyocyanin in MOPS buffer, and then allowed the culture to sit without shaking for 5 min at room temperature. A gradient formed that resembled those observed for cultures in growth media. After vortexing, the entire suspension regained its original blue color (Fig. 1A, tubes 5 and 6). A supernatant from this suspension had the absorbance spectrum characteristic of pyocyanin in the oxidation state most stable under atmospheric conditions. When we moved the culture into an anaerobic chamber and used a stoppered anaerobic cuvette to measure the absorbance spectrum of anaerobic culture supernatant, the sample showed decreased absorbance, indicating that pyocyanin had been reduced (Fig. 1B).
FIG. 1.
Stationary-phase P. aeruginosa PA14 cultures produce pyocyanin and directly catalyze its reduction. (A) Tube 1, exponential-phase LB culture; tube 2, stationary-phase LB culture, immediately after removal from a shaking incubator; tube 3, stationary-phase LB culture, left standing at room temperature for ∼5 min; tube 4, 100 μM pyocyanin in MOPS buffer, left standing at room temperature for ∼5 min; tube 5, same culture as in tubes 2 and 3, resuspended in buffer shown in tube 4 and left standing at room temperature for ∼5 min; tube 6, same suspension as in tube 5, after vortexing. (B) Absorbance spectra of buffer and supernatants from panel A, tubes 4 to 6. The suspension from tube 5 was centrifuged and placed in a stoppered cuvette under anaerobic conditions. The pyocyanin/buffer spectrum overlaps almost completely with that of the supernatant from the aerated culture. PYO, pyocyanin. Supe., supernatant.
Pyocyanin reduction rates increase in stationary phase.
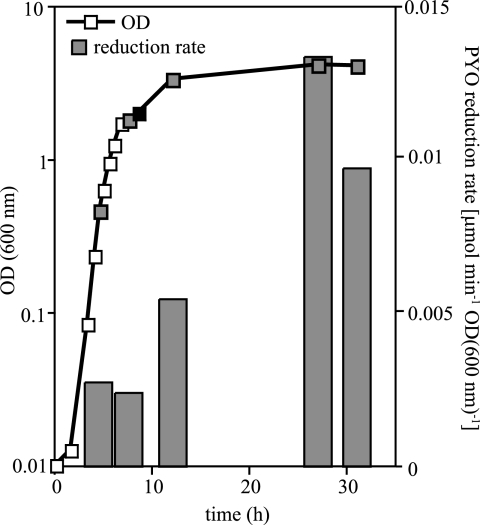
To quantify the rate of pyocyanin reduction by whole cells and test whether this process, like the biosynthesis of phenazines, was growth phase dependent, we sampled an LB culture at different stages of growth. Samples were diluted into their own supernatant, amended with pyocyanin, and transferred to an anaerobic cuvette. We followed the decrease in oxidized pyocyanin absorbance over time for each sample and observed a marked increase in the rate of pyocyanin reduction after the appearance of pyocyanin in stationary phase. This result indicates that the rate of pyocyanin reduction by whole cells is growth phase dependent (Fig. 2).
FIG. 2.
The rate of pyocyanin reduction increases in stationary phase in P. aeruginosa PA14. A 100-ml P. aeruginosa LB culture was grown in a 500-ml Erlenmeyer flask and sampled at various points in the growth curve. Cells were concentrated or diluted in culture supernatant to normalize their optical density (600 nm) to 0.8, amended with pyocyanin, and then transferred to anaerobic cuvettes and stoppered. Absorbance at 690 nm was measured over time and was converted to the amount of oxidized pyocyanin remaining in the cuvette. Gray squares indicate time points at which samples were taken for cell suspension assays. The black square indicates the first appearance of pyocyanin in the culture. Data shown are representative of three separate experiments. OD, optical density; PYO, pyocyanin.
Pyocyanin exposure balances the intracellular redox state.
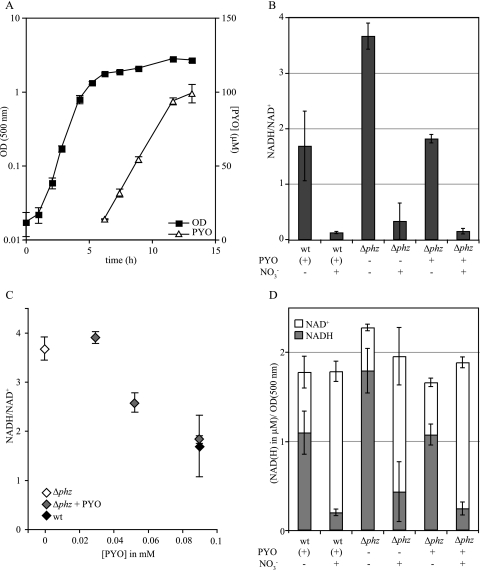
Strains of P. aeruginosa have been shown to vary in the timing and extent of phenazine production relative to the growth phase (9, 10, 18). We have observed that the appearance of pyocyanin in wild-type P. aeruginosa PA14 LB cultures correlates with entry into stationary phase and that pyocyanin production plateaus in late stationary phase, reaching concentrations ranging from ∼100 to 300 μM, depending on the growth conditions (Fig. 3A; see Fig. 4C).
FIG. 3.
Pyocyanin exposure effects redox balancing in stationary phase in a manner analogous to that of a known physiological electron acceptor. (A) Growth and pyocyanin production for wild-type P. aeruginosa PA14 grown aerobically in 10 ml LB in 18- by 150-mm tubes. (B) NADH/NAD+ ratios for cultures grown under the same conditions as those described for panel A. At 7 h, pyocyanin production in the wild-type cultures was visible by eye; 45 μM (half the expected final concentration) was added to the Δphz cultures to be tested for complementation, and 15 mM KNO3 was added to cultures to be tested for the effect of an additional electron acceptor. At 9 h, pyocyanin in the wild-type cultures had increased to near-maximum concentrations, so a second dose of pyocyanin or KNO3 was added to the appropriate cultures, for final concentrations of 90 μM and 30 mM, respectively. Water was added to negative controls. At 11 hours after inoculation and 2 h after the addition of the final dose of pyocyanin, NAD(H) was extracted and assayed for each culture. (C) NADH/NAD+ ratios for cultures treated as for panel B but with various concentrations of pyocyanin added. (D) NADH and NAD+ concentrations for cultures described for panel B, normalized to optical density (500 nm). Error bars represent the standard deviations from triplicate samples. OD, optical density; wt, wild type; PYO, pyocyanin.
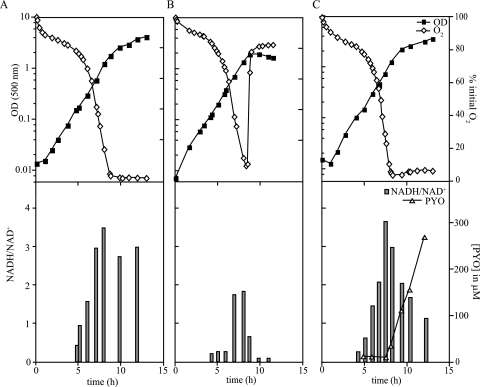
FIG. 4.
NADH accumulates in stationary phase in cultures limited for oxygen and defective in pyocyanin production. P. aeruginosa wild-type and Δphz cultures were grown in 1 liter of MOPS synthetic medium supplemented with either 50 or 10 mM glucose in a 3-liter fermentor with constant aeration and agitation. Cultures were sampled at various points in the growth curve to allow measurement of the optical density at 500 nm and extraction of NAD(H). Relative dissolved oxygen concentrations were measured throughout growth using a polarographic oxygen electrode. Optical density (500 nm), dissolved oxygen concentration, and NADH/NAD+ are shown for the Δphz mutant grown in medium containing 50 mM glucose (A) and the Δphz mutant grown in medium containing 10 mM glucose (B). For panel C, showing wild-type P. aeruginosa PA14 grown in medium containing 50 mM glucose, these parameters plus the concentration of pyocyanin produced by the culture are shown. OD, optical density; PYO, pyocyanin; O2, dissolved oxygen.
Given that NADH reacts with pyocyanin in vitro (36), one potential consequence of pyocyanin production and/or exposure would be a decrease in intracellular NADH levels. We tested this by growing cultures of wild-type P. aeruginosa and a Δphz mutant (with in-frame deletions of both phenazine biosynthetic loci [18]) and measuring intracellular NAD(H) at approximately 4 hours after the onset of stationary phase. The intracellular NADH/NAD+ ratio in the wild type was less than half that observed in the Δphz mutant. The growth curves for these cultures were virtually identical under the incubation conditions for this experiment (data not shown). Addition of 90 μM oxidized pyocyanin (the approximate concentration of pyocyanin produced by wild-type cultures under these conditions) to Δphz mutant cultures reduced the NADH/NAD+ ratio to the wild-type level (Fig. 3B). As a negative control, supernatant from the Δphz mutant was extracted according to the protocol for pyocyanin preparation and tested for an effect on intracellular NAD(H) concentrations; no difference was observed between cultures treated with “pyocyanin” preparations from the Δphz mutant and those treated with water (data not shown). In titration experiments, an inverse relationship was found to exist between the concentration of pyocyanin added to a Δphz mutant culture and the NADH/NAD+ ratio (Fig. 3C).
To test whether the effect of pyocyanin is similar to that of a physiologically relevant terminal electron acceptor, we added 30 mM nitrate (a concentration sufficient to support growth of P. aeruginosa via anaerobic nitrate respiration [70]) to a wild-type culture and added nitrate with or without pyocyanin to Δphz mutant cultures in stationary phase (Fig. 3B). Nitrate and pyocyanin both effected decreases in intracellular NADH/NAD+ ratios, apparently by catalyzing NADH oxidation, since decreases in absolute NADH concentrations correlated with increases in absolute NAD+ concentrations (Fig. 3D). Whereas pyocyanin effected a decrease when added in the micromolar range, nitrate did so only when added at millimolar concentrations (data not shown). Together, these results suggested that NADH can act as a source of electrons for pyocyanin reduction.
The intracellular NADH/NAD+ ratio is influenced by the relative availability of electron donor and acceptor.
Our observation that other electron acceptors, i.e., pyocyanin and nitrate, decreased the NADH/NAD+ ratio suggested that oxygen was limiting during stationary phase in our cultures. This could explain the accumulation of NADH 4 hours after the onset of stationary phase in the Δphz mutant (Fig. 3B). To confirm this, we grew a batch culture of the Δphz mutant in a fermentor, which allowed us to control temperature and aeration while simultaneously measuring dissolved oxygen in the culture. We sampled at regular intervals to measure optical density and extract NAD(H). As predicted, oxygen levels decreased slowly until the culture reached mid- to late exponential phase, at which time it plummeted to zero. This drop in oxygen correlated with an increase in the intracellular NADH/NAD+ ratio (Fig. 4A). To test whether the drop in oxygen depended on the availability of electron donors for oxygen reduction, we repeated the experiment and added 20% of the glucose concentration added to the medium in the initial experiment (10 mM versus 50 mM). When less electron donor was available, the oxygen concentration decreased in mid-exponential phase, but never reached zero, and rapidly increased again upon entry into stationary phase (Fig. 4B). This culture never reached the same growth yield achieved by the culture containing 50 mM glucose, implying that the carbon source was the limiting factor that led it to enter stationary phase. The culture experienced oxygen limitation only transiently, if at all, due to the lower ratio of electron donor to electron acceptor in the experiment depicted in Fig. 4B compared to that in Fig. 4A. As a result, the NADH/NAD+ ratio never reached the high level observed for the culture containing excess glucose.
Finally, we tested the wild-type strain in the presence of 50 mM glucose, and sampled for pyocyanin concentrations in addition to NAD(H) and cell density. The wild-type strain also exhibited increased NADH/NAD+ ratios upon entry into stationary phase, and these ratios correlated with oxygen limitation. However, unlike the Δphz mutant, the wild type showed a decrease in intracellular NADH/NAD+ that correlated with the appearance of pyocyanin in the culture. These results further support the hypothesis that pyocyanin can act as an alternate oxidant under conditions where the terminal electron acceptor for respiration has become limiting. This interpretation derives from the large difference in NADH/NAD+ ratios observed between the wild-type strain and Δphz after about 12 h of incubation and the correlation between decreasing NADH/NAD+ ratios and increasing pyocyanin concentrations in culture filtrates observed upon entry into stationary phase (Fig. 4C).
P. aeruginosa PA14 excretes, and then consumes, pyruvate in late stationary phase.
For fermentative organisms such as E. coli and Propionibacterium freudenreichii, the addition of the synthetic redox-cycling compound ferricyanide has been shown to alter carbon flux through central metabolic pathways. Particularly when the reoxidation of this compound is coupled to electron transfer to an electrode, ferricyanide shifted the fermentation balance away from ethanol and propionate, products that require NADH for their formation, toward acetate, a more oxidized product (19, 20). This implies that the ferricyanide acts as an electron shuttle from major pools of reductant inside the cell, such as NADH, to the electrode, thereby lessening the need for formation of more reduced fermentation products to dissipate cellular reductant.
To determine whether pyocyanin could play a similar role in P. aeruginosa, we analyzed filtered culture supernatants for small organic acids that are known fermentation products of P. aeruginosa metabolism. P. aeruginosa has been shown to ferment pyruvate under energy-starved conditions, converting it to lactate, acetate, and/or succinate. The production of lactate or succinate from pyruvate requires NADH as a substrate, while the conversion of pyruvate to acetate requires NAD+ (21). Therefore, the NADH/NAD+ ratio in the wild type would be more favorable for acetate production, whereas the NADH/NAD+ ratio in the Δphz mutant would favor production of lactate and succinate.
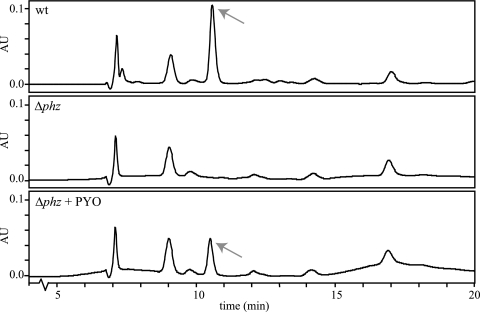
Surprisingly, we observed a marked difference between the wild type and the Δphz mutant with respect to the production of pyruvate itself. In late stationary phase (about 30 h after inoculation) after growth in a defined medium with 50 mM glucose, we observed pyruvate concentrations as high as 6 mM in wild-type culture filtrates (as indicated by a peak eluting at about 10.5 min) but were unable to detect any pyruvate in filtrates from Δphz mutant cultures. Adding pyocyanin to the Δphz mutant upon entry into stationary phase complemented the pyruvate excretion phenotype (Fig. 5), although incompletely because we added only about half the final concentration of pyocyanin produced by the wild type under these conditions (50 versus 100 μM). We also detected citrate, lactate, and acetate in both wild-type and Δphz mutant culture filtrates at similar concentrations, eluting at ∼9.1, 14.3, and 17.0 min, respectively. The peak eluting at 7.1 min was the MOPS buffer from the medium. We were unable to identify the compounds represented by the peaks eluting at approximately 7.3 min (wild-type filtrate only) and at 9.9 and 12.2 min (both wild-type and Δphz mutant filtrates). Standards containing 2-oxoglutarate and malate were run with the same method but did not coelute with any of these peaks.
FIG. 5.
Wild-type P. aeruginosa PA14 excretes pyruvate in stationary phase, and addition of pyocyanin to Δphz mutant cultures restores the pyruvate excretion phenotype. Cultures were inoculated into MOPS synthetic medium amended with 50 mM glucose (initial optical density at 500 nm, 0.03). To complement pyruvate excretion, 50 μM pyocyanin was added to the Δphz culture at the time when pyocyanin reached its maximum concentration in the wild-type cultures (approximately 12 h after inoculation). Twenty microliters of culture filtrates at the 24-hour time point was loaded onto an anion-exchange column and subjected to an isocratic gradient in 5 mM H2SO4. Pyruvate peaks are indicated by arrows. The elution time of pyruvate drifts slightly but averages around 10.5 min. Results shown are representative of three separate experiments. Other peak identities are described in the text. wt, wild type; PYO, pyocyanin.
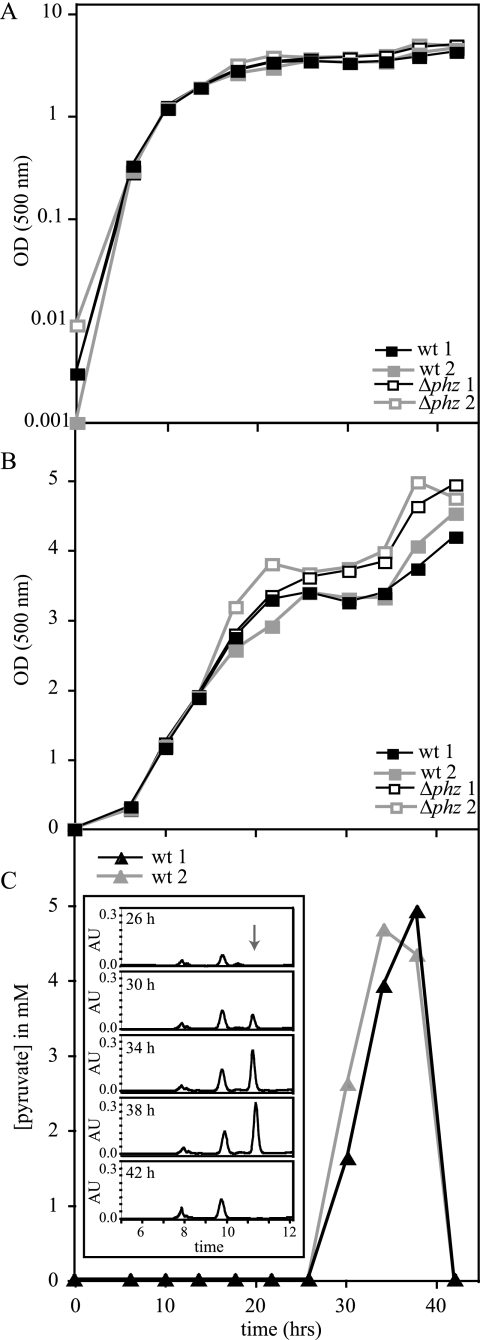
To better constrain the timing of metabolite excretion in the wild type and the Δphz mutant, we sampled every 4 h from duplicate cultures over the course of approximately 30 h in stationary phase (Fig. 6). Pyruvate appeared at detectable levels in wild-type cultures at between 22 and 26 h after inoculation and had increased to ∼5 mM after 38 h. However, by the 42-hour time point, the pyruvate in both replicates had decreased to levels below the detection limit (∼0.05 mM) (Fig. 6C). Abiotic degradation of pyruvate generates a peak eluting at approximately 8 min, which does not coelute with any of the peaks observed in traces from our culture filtrates (data not shown). Therefore, the disappearance of the pyruvate peak at the 42-hour time point implied that it had been metabolized by the bacteria.
FIG. 6.
P. aeruginosa PA14 cultures consume excreted pyruvate in very late stationary phase. Duplicate cultures were inoculated at an optical density (500 nm) of ∼0.001 in MOPS synthetic medium amended with 50 mM glucose. Approximately every 4 h, 100 to 200 μl of culture was sampled and filtered for HPLC analysis as described for Fig. 5. (A) Optical density at 500 nm for wild-type and Δphz cultures plotted on a logarithmic scale. (B) same data as in panel A plotted on a linear scale to show the lower growth yields consistently observed for wild-type PA14 under this condition. (C) Quantification of pyruvate production for the “wt 1” and “wt 2” cultures. Inset, chromatograms demonstrating the disappearance of pyruvate at 42 h for the “wt 1” culture. The arrow indicates the elution time of the pyruvate peak. OD, optical density; wt, wild type.
Another phenotype that became apparent under these growth conditions was the reproducible difference in cell yields between wild-type and Δphz mutant cultures. The optical densities of wild-type cultures were typically lower than those of the Δphz mutant cultures in stationary phase, a phenotype that becomes more apparent when the optical density is plotted on a linear scale (Fig. 6A and B).
Pyruvate fermentation facilitates survival in energy-starved P. aeruginosa PA14 cultures.
Recently, Schobert and colleagues have characterized genes implicated in a pyruvate fermentation pathway in P. aeruginosa strain PAO1 (21, 59). In this pathway, pyruvate is converted by multiple enzymes to succinate, acetate, and/or lactate. We do not suspect that these reactions were responsible for the consumption of pyruvate in late stationary phase in our cultures, because these compounds are detectable by our analytical HPLC method, and we did not see their concentrations increase as pyruvate disappeared (data not shown). We therefore hypothesize that pyruvate was completely oxidized through the utilization of the small amount of oxygen available to the cells. However, in environments with steep gradients of electron acceptor availability, such as those encountered in surface-attached or aggregated bacterial communities, excreted pyruvate may be utilized for substrate-level phosphorylation when respiratory electron acceptors become limiting. To verify that P. aeruginosa strain PA14 can utilize pyruvate for survival under strict anaerobic conditions, we incubated the wild type and an ldhA mutant, defective in the ability to reduce pyruvate to lactate, in stoppered serum bottles containing buffered LB amended with 20 mM pyruvate. As a control, we set up a wild-type culture with no pyruvate. We followed CFU in samples from these cultures over more than 3 weeks and found that, as had been previously reported for P. aeruginosa PAO1 (21, 59), a mutant with a disruption in the gene ldhA was defective in survival on pyruvate (see Fig. S1 in the supplemental material). The decline of this mutant was similar to that of the wild-type culture containing no added pyruvate. P. aeruginosa PA14 is therefore also able to survive under conditions of energy starvation through utilization of a lactate dehydrogenase-dependent pathway for pyruvate fermentation.
DISCUSSION
In this study, we have characterized the effects of a stationary-phase-specific metabolite on the carbon and energy metabolism of P. aeruginosa cultures. Metabolites formed in stationary phase historically have been categorized as products of “secondary” forms of metabolism that bear little relevance to energy generation. However, we have shown that the redox activity of pyocyanin, a phenazine produced after the exponential phase of growth in batch cultures, affects the metabolic status of its producer. Bacteria such as the pseudomonads, with limited capacities for fermentation, are generally thought to depend on terminal electron acceptors, and on the function of their membrane-bound respiratory chains, for the ability to maintain a balanced intracellular redox state. The redox-balancing effect of pyocyanin may be particularly important in bacterial communities limited for oxygen, an electron acceptor whose uptake rate outpaces its diffusion rate through dense cultures of respiring bacteria (63, 75).
As part of our characterization of the physiological effects of pyocyanin reduction in P. aeruginosa, we found that pyocyanin reductive activity in whole cells increases after entry into stationary phase and the appearance of phenazines in batch cultures (Fig. 2). While we cannot rule out that this increase is due merely to an increase in the concentration of the electron donor for this reaction, we know that pyocyanin induces expression of genes encoding multidrug efflux pumps and oxidoreductases that could be involved in the redox cycling of this compound (18). Such gene products may contribute to the observed increase in the pyocyanin reduction rate. We also found that pyocyanin exposure in stationary-phase cultures decreases the intracellular NADH/NAD+ ratio (Fig. 3). The increase in the NADH/NAD+ ratio that we observe for a mutant defective in phenazine production correlates with oxygen limitation (Fig. 4) and is relieved by the presence of pyocyanin (Fig. 3), suggesting that pyocyanin plays a role in redox balancing.
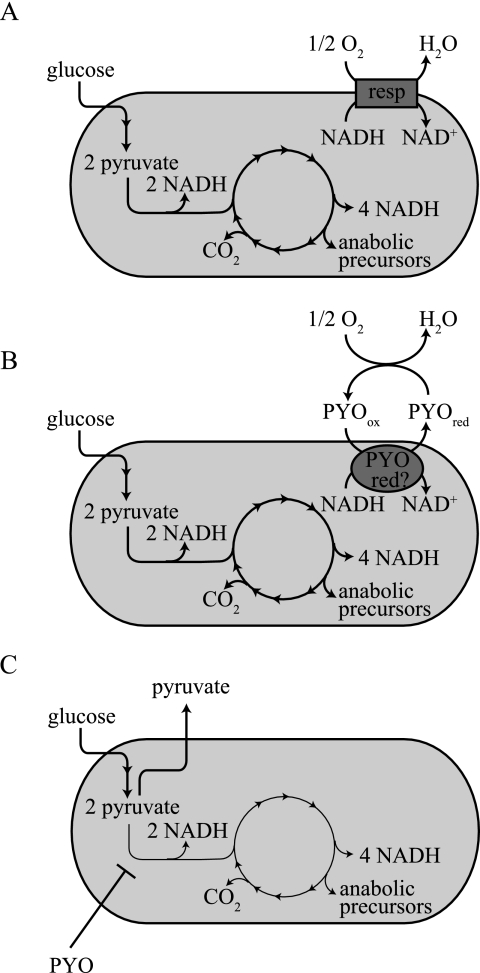
Given that the reduction of pyocyanin by NADH is a thermodynamically favorable reaction, one could attribute the observed inverse relationship between intracellular [NADH] and pyocyanin production (Fig. 4C) to a model such as that shown in Fig. 7. In this model, stationary-phase cells accumulate NADH as a consequence of oxygen limitation (Fig. 7A) and transfer electrons to pyocyanin when it becomes available (Fig. 7B). Pyocyanin can subsequently be reoxidized through abiotic electron transfer to oxygen. Differences in central metabolism depending on the presence or absence of synthetic and environmental electron shuttles have been reported for fermentative bacteria (5, 19, 20) and apparently arise from the effects of extracellular electron shuttling on the intracellular redox state. If the mechanism of redox balancing depicted in Fig. 7B were operational, we might expect a phenazine-null mutant (lacking the naturally produced pseudomonad electron shuttle) to be defective in complete oxidation of its carbon source, since flux through the citric acid cycle would be inhibited by a relatively reduced NAD(H) pool. Instead, we observed that the wild-type strain appears to be affected in its ability to mineralize its carbon source, based on the excretion of pyruvate in late stationary phase (Fig. 5 and 6). This phenomenon thus suggests an alternative mechanism for lowering the NADH/NAD+ ratio, in which NADH accumulation is avoided in the wild type by excretion of pyruvate before it can enter the citric acid cycle and reduce NAD+ (Fig. 7C). Because stationary-phase P. aeruginosa cells both catalyze pyocyanin reduction and excrete pyruvate, maintenance of redox homeostasis in stationary-phase P. aeruginosa PA14 could be due to a combination of the mechanisms shown in Fig. 7B and C.
FIG. 7.
Model for how pyocyanin reduction allows P. aeruginosa PA14 to maintain redox homeostasis under oxygen-limited conditions. When sufficient oxygen is available for growth (A), the aerobic respiratory chain (“resp”) can catalyze the reoxidation of NADH. Under conditions in which terminal electron acceptors for respiration are limiting (B), P. aeruginosa can couple the reoxidation of NADH to the reduction of pyocyanin, either directly or through an enzyme-mediated reaction as represented by “PYO red,” a putative phenazine reductase. The electrons could be transferred from pyocyanin to oxygen through an abiotic extracellular reaction. (C) Also under conditions of oxygen limitation, the NADH/NAD+ ratio could be balanced through inactivation of the pyruvate dehydrogenase complex by pyocyanin. NAD+ reduction (and therefore NADH production) would be avoided because pyruvate would be excreted without further oxidation. PYO, pyocyanin.
Although pyruvate excretion has been observed in cultures of other bacteria, such as Aerobacter aerogenes and Photobacterium fischeri (since reclassified as members of Enterobacter and Vibrio, respectively), the mechanisms underlying its regulation have not been elucidated (57, 68). An understanding of the pyruvate oxidation machinery in pseudomonads provides insight into the potential mechanisms whereby this reaction may be inhibited in pyocyanin-producing cells. The conversion of pyruvate to acetyl coenzyme A (acetyl-CoA) is catalyzed by pyruvate dehydrogenase, a large multienzyme complex prevalent during aerobic growth in bacteria and eukarya, though there is some evidence for its occurrence in archaea as well (34). All pyruvate dehydrogenase multienzyme complexes require dihydrolipoamide, a cofactor which is covalently bound to the E2 subunit, to transfer an acyl group derived from pyruvate to CoA and produce acetyl-CoA. Dihydrolipoamide contains a disulfide bond that is broken and reformed during the three-step pyruvate decarboxylation and oxidation mechanism (14, 17). The one-electron reduction of each suflhydryl group of the lipoamide cofactor on the E2 subunit, catalyzed by superoxide (8, 64), is thought to inactivate the enzyme. The generation of superoxide by pyocyanin, or even the formation of pyocyanin radical itself (29), may therefore inhibit the pyruvate dehydrogenase complex, leading to accumulation of pyruvate.
Regardless of whether the excretion of pyruvate by wild-type P. aeruginosa results from an apparently toxic side reaction, it may be beneficial under pyocyanin-producing conditions. Excreted pyruvate that remains in the immediate environment could potentially be accessed later during a “last gasp” under conditions of extreme energy starvation. While this scenario is unlikely to occur in the soil, where other microorganisms might consume the pyruvate before it was metabolized by the producer, it may be relevant for conditions encountered by P. aeruginosa during chronic infection of the lung. Individuals with chronic P. aeruginosa infections resulting from impaired lung function harbor monocultures of this bacterium at cell densities of as high as 107 CFU/g sputum (43). It is thought that bacteria in the lung cavity experience steep gradients of electron acceptor availability (74), and the ability to reserve a pool of substrate or transfer it to an energy-starved neighbor may contribute to the ability of P. aeruginosa populations to persist throughout the lifetime of the individual host (59). Pseudomonad phenazines as well as other high-cell-density signals have been detected in the sputa of chronically infected patients (60, 72). An interesting avenue for future research, therefore, is to determine whether the physiological effects of pyocyanin on P. aeruginosa contribute to its long-term survival during chronic colonization of the lung.
Supplementary Material
Acknowledgments
We acknowledge all of the members of the Newman laboratory, particularly Tracy K. Teal, for their technical advice and for comments and suggestions regarding the manuscript.
This work was supported by NIH training grant 5T32GM07616 (to A.P.-W.), an EMBO long-term fellowship (to L.E.P.D.), and grants from the Packard Foundation and Howard Hughes Medical Institute (to D.K.N.).
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aslund, F., K. D. Berndt, and A. Holmgren. 1997. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 272:30780-30786. [DOI] [PubMed] [Google Scholar]
- 2.Baron, S. S., and J. J. Rowe. 1981. Antibiotic action of pyocyanin. Antimicrob. Agents Chemother. 20:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, S. S., G. Terranova, and J. J. Rowe. 1989. Molecular mechanism of the antimicrobial action of pyocyanin. Curr. Microbiol. 18:223-230. [Google Scholar]
- 4.Beifuss, U., and M. Tietze. 2005. Methanophenazine and other natural biologically active phenazines. Top. Curr. Chem. 244:77-113. [Google Scholar]
- 5.Benz, M., B. Schink, and A. Brune. 1998. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl. Environ. Microbiol. 64:4507-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernofsky, C., and M. Swan. 1973. Improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 53:452-458. [DOI] [PubMed] [Google Scholar]
- 7.Bessette, P. H., F. Aslund, J. Beckwith, and G. Georgiou. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA 96:13703-13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunik, V. I., and C. Sievers. 2002. Inactivation of the 2-oxo acid dehydrogenase complexes upon generation of intrinsic radical species. Eur. J. Biochem. 269:5004-5015. [DOI] [PubMed] [Google Scholar]
- 9.Byng, G. S., D. C. Eustice, and R. A. Jensen. 1979. Biosynthesis of phenazine pigments in mutant and wild-type cultures of Pseudomonas aeruginosa. J. Bacteriol. 138:846-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, P. C., and A. C. Blackwood. 1969. Simultaneous production of three phenazine pigments by Pseudomonas aeruginosa Mac 436. Can. J. Microbiol. 15:439-444. [DOI] [PubMed] [Google Scholar]
- 11.Clark, L. C., Jr., R. Wolf, D. Granger, and Z. Taylor. 1953. Continuous recording of blood oxygen tensions by polarography. J. Appl. Physiol. 6:189-193. [DOI] [PubMed] [Google Scholar]
- 12.Conway, T. 1992. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 103:1-28. [DOI] [PubMed] [Google Scholar]
- 13.Cox, C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cronan, J. E., X. Zhao, and Y. Jiang. 2005. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 50:103-146. [DOI] [PubMed] [Google Scholar]
- 15.Davis, G., and P. J. Thornalley. 1983. Free radical production from the aerobic oxidation of reduced pyridine nucleotides catalyzed by phenazine derivatives. Biochim. Biophys. Acta 724:456-464. [DOI] [PubMed] [Google Scholar]
- 16.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. T. de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Kok, A., A. F. Hengeveld, A. Martin, and A. H. Westphal. 1998. The pyruvate dehydrogenase complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385:353-366. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich, L. E. P., A. Price-Whelan, A. Petersen, M. Whiteley, and D. K. Newman. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308-1321. [DOI] [PubMed] [Google Scholar]
- 19.Emde, R., and B. Schink. 1990. Oxidation of glycerol, lactate, and propionate by Propionibacterium freudenreichii in a poised-potential amperometric culture system. Arch. Microbiol. 153:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emde, R., A. Swain, and B. Schink. 1989. Anaerobic oxidation of glycerol by Escherichia coli in an amperometric poised-potential culture system. Appl. Microbiol. Biotechnol. 32:170-175. [Google Scholar]
- 21.Eschbach, M., K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186:4596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedheim, E., and L. Michaelis. 1931. Potentiometric study of pyocyanine. J. Biol. Chem. 91:355-368. [Google Scholar]
- 23.Friedheim, E. A. H. 1931. Pyocyanine, an accessory respiratory pigment. J. Exp. Med. 54:207-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fultz, M. L., and R. A. Durst. 1982. Mediator compounds for the electrochemical study of biological redox systems: a compilation. Anal. Chim. Acta 140:1-18. [Google Scholar]
- 26.Gardner, P. R. 1996. Superoxide production by the mycobacterial and pseudomonad quinoid pigments phthiocol and pyocyanine in human lung cells. Arch. Biochem. Biophys. 333:267-274. [DOI] [PubMed] [Google Scholar]
- 27.Hassan, H. M., and I. Fridovich. 1979. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch. Biochem. Biophys. 192:385-395. [DOI] [PubMed] [Google Scholar]
- 28.Hassan, H. M., and I. Fridovich. 1980. Mechanism of the antibiotic action of pyocyanine. J. Bacteriol. 141:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassett, D. J., L. Charniga, K. Bean, D. E. Ohman, and M. S. Cohen. 1992. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect. Immun. 60:328-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassett, D. J., H. P. Schweizer, and D. E. Ohman. 1995. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 177:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez, M. E., A. Kappler, and D. K. Newman. 2004. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70:921-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez, M. E., and D. K. Newman. 2001. Extracellular electron transfer. Cell. Mol. Life Sci. 58:1562-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingledew, W. M., and J. J. Campbell. 1969. A new resuspension medium for pyocyanine production. Can. J. Microbiol. 15:595-598. [DOI] [PubMed] [Google Scholar]
- 34.Jolley, K. A., D. G. Maddocks, S. L. Gyles, Z. Mullan, S. L. Tang, M. L. Dyall-Smith, D. W. Hough, and M. J. Danson. 2000. 2-Oxoacid dehydrogenase multienzyme complexes in the halophilic Archaea? Gene sequences and protein structural predictions. Microbiology 146:1061-1069. [DOI] [PubMed] [Google Scholar]
- 35.Kerr, J. R., G. W. Taylor, A. Rutman, N. Hoiby, P. J. Cole, and R. Wilson. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52:385-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kito, N., Y. Ohnishi, M. Nagami, and A. Ohno. 1974. Reduction by a model of NAD(P)H: construction of electron bridges. Chem. Lett. 1974:353-356. [Google Scholar]
- 37.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 38.Lau, G. W., D. J. Hassett, H. Ran, and F. Kong. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599-606. [DOI] [PubMed] [Google Scholar]
- 39.Learoyd, S. A., R. G. Kroll, and C. F. Thurston. 1992. An investigation of dye reduction by food-borne bacteria. J. Appl. Bacteriol. 72:479-485. [DOI] [PubMed] [Google Scholar]
- 40.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, W. Gang, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lies, D. P., M. E. Hernandez, A. Kappler, R. E. Mielke, J. A. Gralnick, and D. K. Newman. 2005. Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl. Environ. Microbiol. 71:4414-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Look, D. C., L. L. Stoll, S. A. Romig, A. Humlicek, B. E. Britigan, and G. M. Denning. 2005. Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. J. Immunol. 175:4017-4023. [DOI] [PubMed] [Google Scholar]
- 43.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavrodi, D., W. Blankenfeldt, and L. S. Thomashow. 2006. Phenazine compounds in fluorescent Pseudomonas spp.: biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417-445. [DOI] [PubMed] [Google Scholar]
- 45.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinlay, J. B., and J. G. Zeikus. 2004. Extracellular iron reduction is mediated in part by neutral red and hydrogenase in Escherichia coli. Appl. Environ. Microbiol. 70:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mossner, E., M. Huber-Wunderlich, A. Rietsch, J. Beckwith, R. Glockshuber, and F. Aslund. 1999. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from Escherichia coli. J. Biol. Chem. 274:25254-25259. [DOI] [PubMed] [Google Scholar]
- 48.O'Malley, Y. Q., K. J. Reszka, G. T. Rasmussen, M. Y. Abdalla, G. M. Denning, and B. E. Britigan. 2003. The Pseudomonas secretory product pyocyanin inhibits catalase activity in human lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L1077-L1086. [DOI] [PubMed] [Google Scholar]
- 49.O'Malley, Y. Q., K. J. Reszka, D. R. Spitz, G. M. Denning, and B. E. Britigan. 2004. Pseudomonas aeruginosa pyocyanin directly oxidizes glutathione and decreases its levels in airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 287:L94-L103. [DOI] [PubMed] [Google Scholar]
- 50.Palmer, K. L., L. M. Mashburn, P. K. Singh, and M. Whiteley. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierson, L. S., III, V. D. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price-Whelan, A., L. E. P. Dietrich, and D. K. Newman. 2006. Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71-78. [DOI] [PubMed] [Google Scholar]
- 53.Rabaey, K., N. Boon, M. Hofte, and W. Verstraete. 2005. Microbial phenazine production enhances electron transfer in biofuel cells. Environ. Sci. Technol. 39:3401-3408. [DOI] [PubMed] [Google Scholar]
- 54.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 55.Ran, H., D. J. Hassett, and G. W. Lau. 2003. Human targets of Pseudomonas aeruginosa pyocyanin. Proc. Natl. Acad. Sci. USA 100:14315-14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reszka, K. J., Y. O'Malley, M. L. McCormick, G. M. Denning, and B. E. Britigan. 2004. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Biol. Med. 36:1448-1459. [DOI] [PubMed] [Google Scholar]
- 57.Ruby, E. G., and K. H. Nealson. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.San, K. Y., G. N. Bennett, S. J. Berrios-Rivera, R. V. Vadali, Y. T. Yang, E. Horton, F. B. Rudolph, B. Sariyar, and K. Blackwood. 2002. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4:182-192. [DOI] [PubMed] [Google Scholar]
- 59.Schreiber, K., N. Boes, M. Eschbach, L. Jaensch, J. Wehland, T. Bjarnsholt, M. Givskov, M. Hentzer, and M. Schobert. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188:659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 61.Stams, A. J. M., F. A. M. de Bok, C. M. Plugge, M. H. A. van Eekert, J. Dolfing, and G. Schraa. 2006. Exocellular electron transfer in anaerobic microbial communities. Environ. Microbiol. 8:371-382. [DOI] [PubMed] [Google Scholar]
- 62.Stewart-Tull, D. E. S., and A. V. Armstrong. 1971. The effect of 1-hydroxyphenazine and pyocyanin from Pseudomonas aeruginosa on mammalian cell respiration. J. Med. Microbiol. 5:67-73. [DOI] [PubMed] [Google Scholar]
- 63.Sweet, W. J., and J. A. Peterson. 1978. Changes in cytochrome content and electron transport patterns in Pseudomonas putida as a function of growth phase. J. Bacteriol. 133:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabatabaie, T., J. D. Potts, and R. A. Floyd. 1996. Reactive oxygen species-mediated inactivation of pyruvate dehydrogenase. Arch. Biochem. Biophys. 336:290-296. [DOI] [PubMed] [Google Scholar]
- 65.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trutko, S. M. 1989. The physiological role of phenazine pigments synthesized by the bacteria Pseudomonas aureofaciens. Biochemistry (Moscow) 54:1092-1098. [Google Scholar]
- 67.Trutko, S. M., A. D. Garagulya, E. A. Kiprianova, and V. K. Akimenko. 1989. Physiological role of pyocyanine synthesized by Pseudomonas aeruginosa. Microbiologya 57:957-964. [PubMed] [Google Scholar]
- 68.Webb, M. 1968. Pyruvate accumulation in growth-inhibited cultures of Aerobacter aerogenes. Biochem. J. 106:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams, D. R., J. J. Rowe, P. Romero, and R. G. Eagon. 1978. Denitrifying Pseudomonas aeruginosa: some parameters of growth and active transport. Appl. Environ. Microbiol. 36:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams, H. D., J. E. A. Zlosnik, and B. Ryall. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52:1-71. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, R., D. A. Sykes, D. Watson, A. Rutman, G. W. Taylor, and P. J. Cole. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wimpenny, J. W. T., and A. Firth. 1972. Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J. Bacteriol. 111:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu, K. D., P. S. Stewart, F. Xia, C. T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.