Abstract
Regulation of pyrimidine biosynthetic (pyr) genes by a transcription attenuation mechanism that is mediated by the PyrR mRNA-binding regulatory protein has been demonstrated for numerous gram-positive bacteria. Mycobacterial genomes specify pyrR genes and contain obvious PyrR-binding sequences in the initially transcribed regions of their pyr operons, but transcription antiterminator and attenuation terminator sequences are absent from their pyr 5′ leader regions. This work demonstrates that repression of pyr operon expression in Mycobacterium smegmatis by exogenous uracil requires the pyrR gene and the pyr leader RNA sequence for binding of PyrR. Plasmids containing the M. smegmatis pyr promoter-leader region translationally fused to lacZ also displayed pyrR-dependent repression, but transcriptional fusions of the same sequences to a lacZ gene that retained the lacZ ribosome-binding site were not regulated by PyrR plus uracil. We propose that PyrR regulates pyr expression in M. smegmatis, other mycobacteria, and probably in numerous other bacteria by a translational repression mechanism in which nucleotide-regulated binding of PyrR occludes the first ribosome-binding site of the pyr operon.
The genes of de novo pyrimidine nucleotide biosynthesis (pyr genes) are regulated in many diverse bacteria by the pyr mRNA-binding regulatory protein PyrR (33). The mechanism by which PyrR regulates the expression of pyr genes has been studied in Bacillus (17-19, 35, 36), Enterococcus (13), Lactococcus (20, 21), and Lactobacillus (4, 12, 25) species. In these organisms PyrR acts by mediating a nucleotide-regulated transcription attenuation mechanism, which has been well characterized in genetic and biochemical studies with Bacillus subtilis (33) and Bacillus caldolyticus (8, 14). PyrR binds to pyr mRNA at a site which lies upstream of the genes that it regulates. This site, called the binding loop, consists of an RNA stem-loop with a purine-rich internal loop in the 5′ strand that contains two segments of highly conserved nucleotide sequence (6). Thus, PyrR-binding loops are readily identified by examination of the DNA sequences at the 5′ untranslated regions of pyr genes. Furthermore, the attenuation mechanism requires the presence of two additional RNA secondary structures that lie immediately downstream from the binding loop. These are the attenuator, which is a typical bacterial factor-independent terminator, and an upstream antiterminator, a stem-loop structure whose formation prevents formation of the attenuator stem-loop. Binding of PyrR to the binding loop occludes the 5′ strand of the antiterminator, permitting the formation of the attenuator, which results in premature termination of transcription and reduced expression of the downstream pyr genes. Thus, binding of PyrR, which is stimulated by uridine nucleotides, leads to repression, whereas the antagonism of binding by guanosine nucleotides leads to cross-regulatory derepression of pyr gene expression (8).
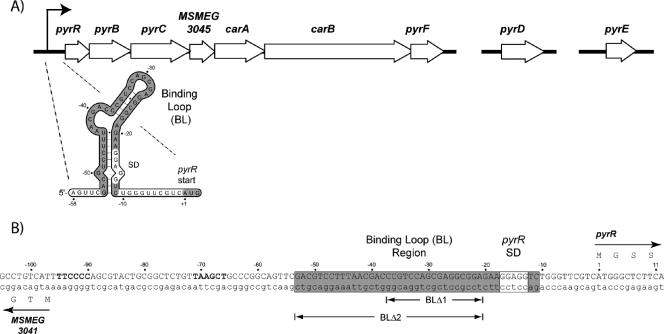
Sequencing of the genomes of a number of Mycobacterium species has revealed that most of their pyr genes are organized into an operon in which the first gene encodes a PyrR homolog and an obvious binding loop sequence lies immediately upstream of the pyrR open reading frame (7, 9, 32) (GenBank accession no. CP000480 for the M. smegmatis genome) (Fig. 1A). However, sequences encoding attenuator (terminator) and antiterminator structures are not found in the pyr leader region (Fig. 1B), which makes it highly unlikely that mycobacterial pyr genes are regulated by the transcription attenuation mechanism described above. If PyrR participates in the regulation of pyr genes in mycobacteria, an indication as to how it might do so comes from the observation that the binding loop sequence overlaps the Shine-Dalgarno sequence of a putative ribosome-binding site for the pyrR gene (Fig. 1A). Binding of PyrR to the binding loop might prevent binding of ribosomes and thus inhibit translation of the gene. If the binding of PyrR to the binding loop is in turn regulated by nucleotides as in other bacteria, a means would be provided for regulation of translation of pyrR and, via translational coupling, of downstream pyr genes.
FIG. 1.
A. Schematic diagram of the chromosomal organization of the pyrimidine biosynthetic (pyr) genes in M. smegmatis. The pyrRBC-orf3045-carAB-pyrF operon is shown at the left; the pyrD and pyrE genes are unlinked and located away from the pyr operon. MSMEG3045 denotes an open reading frame of unknown function. The bent arrow indicates the start of transcription of the operon. A portion of the promoter-leader region of the transcript shown below illustrates the predicted secondary fold of the PyrR-binding loop (BL; shaded) in the RNA, the Shine-Dalgarno sequence of a putative ribosome binding site (SD), and start codon (shaded AUG) for pyrR, the first gene of the operon. B. DNA sequence on the pyr operon promoter-leader region; the nontemplate strand is shown in capital letters. The sequence is numbered with the first base of the pyrR start codon as +1. The sequence specifying the PyrR-binding loop is enclosed in a box and shaded, except for a box designating the putative Shine-Dalgarno sequence. Putative −35 and −10 promoter sequences are shown in boldface. MSMEG3041 indicates an open reading frame of unknown function (annotated as a member of the thiopurine S-methyltransferase family) that is transcribed in an opposite direction to the pyr operon. Regions deleted from the binding loop in pyr′-lacZ fusion plasmids pFS20 and pFS22 are denoted by Δ1; those deleted in pyr′-lacZ fusion plasmids pFS21 and pFS23 are denoted by Δ2.
In this work evidence is presented that (i) the expression of pyr genes is repressed by exogenous uracil in Mycobacterium smegmatis, (ii) repression of pyr genes requires the presence of both the pyrR gene and the binding loop RNA sequence to which PyrR binds, and (iii) PyrR-dependent regulation is mediated by a translational repression mechanism. We propose that these generalizations apply to other mycobacterium species as well.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. smegmatis and Escherichia coli strains used are listed in Table 1. M. smegmatis cultures were maintained on Middlebrook 7H9 broth medium and 7H10 solid medium (Difco) and were supplemented with ADC supplement (1× final concentration; Difco); liquid medium was supplemented with 0.05% Tween 20 to inhibit cell clumping. Where noted, the following supplements and antibiotics were added (final concentrations in parentheses): kanamycin (50 μg/ml for both E. coli and M. smegmatis), gentamicin (10 μg/ml for both E. coli and M. smegmatis), ampicillin (100 μg/ml for E. coli only), uracil (100 μg/ml), uridine (200 μg/ml), l-arginine (50 μg/ml), sucrose (10%), 5-bromo-4-chloro-3-indoyl-β-d-galactoside (X-Gal; 50 μg/ml). When β-galactosidase and aspartate transcarbamylase activities were measured, the cultures were grown in 25 ml of Middlebrook medium in 125-ml sidearm flasks with aeration (250 rpm in a New Brunswick shaker). Growth was monitored using a Klett-Summerson meter with a no. 54 filter. Cells were harvested by centrifugation at early to mid-log phase at 60 to 75 Klett units; cell pellets were quickly frozen in liquid N2 and stored at −20°C prior to analysis.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| M. smegmatis strains | ||
| mc2155 | Parent strain; ATCC 700084 | 31 |
| CF1 | Derivative of mc2155; ΔpyrD::gm Gmr | This work |
| CF22 | Derivative of mc2155; ΔpyrR | This work |
| CF24 | Derivative of CF1; ΔpyrR ΔpyrD::gm Gmr | This work |
| E. coli strain | ||
| DH5α | recA1 endA1 gyrA96 thi-1 relA1 hsdR17(rK− mK+) supE44 φ80lacZΔM15 Δ(lacZYA-argF)U169 | 28 |
| Plasmids | ||
| pJEM13 | E. coli-Mycobacterium translational lacZ reporter shuttle vector; Kmr | 34 |
| pJEM15 | E. coli-Mycobacterium transcriptional lacZ reporter shuttle vector; Kmr | 34 |
| pZErO2.1 | Cloning vector; Kmr | Invitrogen |
| p2NIL | Cloning vector; Kmr | 26 |
| pGOAL19 | Plasmid carrying lacZ, sacB, and hyg genes in a PacI cassette; Apr Hygr | 26 |
| pGMΩ1 | Plasmid carrying the aacC1 omega interposon in a SmaI cassette; Gmr | 30 |
| pFS1 | M. smegmatis pyrR in pZERO2.1; Kmr | This work |
| pFS3 | M. smegmatis pyrD in pZERO2.1; Kmr | This work |
| pFS3Δ1 | pyrD in-frame deletion in pZERO2.1; Kmr | This work |
| pFS3Δ2 | pFS3Δ1 pyrD in-frame deletion with omega interposon; Kmr Gmr | This work |
| pFS4 | pJEM13 with −488 to +9b of pyrR gene; Kmr | This workc |
| pFS5 | pJEM13 with −233 to +9 of pyrR gene; Kmr | This workc |
| pFS6 | pJEM13 with −102 to +9 of pyrR gene; Kmr | This workc |
| pFS8 | pJEM15 with −488 to +9 of pyrR gene; Kmr | This workc |
| pFS9 | pJEM15 with −233 to +9 of pyrR gene; Kmr | This workc |
| pFS10 | pJEM15 with −102 to +9 of pyrR gene; Kmr | This workc |
| pFS11 | pZErO2.1 with −488 to +9 of pyrR gene; Kmr | This work |
| pFS14 | pFS1 derivative, pyrR in-frame deletion; Kmr | This work |
| pFS16 | pFS11 derivative, binding loop deletion (BLΔ1); Kmr | This work |
| pFS18 | pFS11 derivative, binding loop deletion (BLΔ2); Kmr | This work |
| pFS19 | pyrR in-frame deletion cloned in p2NIL; Kmr | This work |
| pFS20 | pJEM13 with −233 to +9 of pyrR gene, BLΔ1); Kmr | This workd |
| pFS21 | pJEM13 with −233 to +9 of pyrR gene, BLΔ2); Kmr | This worke |
| pFS22 | pJEM15 with −233 to +9 of pyrR gene, BLΔ1; Kmr | This workd |
| pFS23 | pJEM15 with −233 to +9 of pyrR gene, BLΔ2; Kmr | This worke |
| pFS24 | pFS19 with pGOAL19 PacI cassette; Kmr Hygr | This work |
| pFS28 | pJEM15 with −488 to +579 of pyrR gene (includes intact pyrR); Kmr | This workc |
| pFS30 | pJEM15 with MSMEG_3041 promoter; Kmr | This workc |
| pFS31 | pJEM15 with Ag80 promoter; Kmr | This workf |
| pFS32 | pJEM15 with groEL2 (hsp60) promoter; Kmr | This workf |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Gmr, gentamicin resistance; Hygr, hygromycin resistance.
Coordinates are relative to the pyrR translational start codon.
PCR template was pFS1.
PCR template was pFS16.
PCR template was pFS18.
PCR template was pGOAL19.
General techniques.
All PCRs were performed using Platinum Pfx DNA polymerase (Invitrogen) and chemically synthesized primers. Plasmid clones were verified by sequencing (University of Illinois Core Sequencing Facility). The identities of plasmids transformed into M. smegmatis strains were verified by breaking a sample of the isolated clones using a Mini-BeadBeater (BioSpec, Inc.) and transforming E. coli DH5α with 5 to 10 μl of the supernatant; DNA isolated from E. coli transformants was then sequenced for verification.
Rapid screening of mycobacterial colonies for chromosomal rearrangements was performed using a modified colony PCR method (28). A small sample of a representative isolated colony was picked using a sterile pipette tip and added to 50 μl of distilled H2O, vortexing briefly. The resuspended cells were incubated at 95°C for 5 min, followed by a second round of vortexing and incubation at 95°C. Cell debris was centrifuged for 10 min at 14,000 × g, and 2 to 5 μl of the resulting supernatant was used directly in the amplification reaction mixture.
Preliminary M. smegmatis genome sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. The GenBank accession number for the genome sequence is CP000480.
Construction of lacZ transcriptional and translational fusion plasmids.
All lacZ fusion plasmids were constructed as follows and as detailed in Table 1. Primers flanking the region of interest were used to amplify DNA from the indicated template DNA (Table 1). Amplified DNA was gel purified, cut using ApaI and KpnI, and cloned into pJEM13 (translational fusion), pJEM15 (transcriptional fusions), or both to generate the indicated plasmid fusions. Plasmids were transformed into M. smegmatis by electroporation (15).
Construction of plasmids with PyrR-binding loop deletions BLΔ1 and BLΔ2.
A fragment of 513 bp specifying the entire M. smegmatis pyr promoter region and DNA upstream of it was amplified by PCR and cloned into pZErO2.1, generating pFS11. Two deletion variants were constructed by inverse PCR. Amplified DNA was gel purified, phosphorylated using T4 polynucleotide kinase (Promega), and self-ligated to generate pFS16 (BLΔ1) and pFS18 (BLΔ2) (Table 1). The structures of the resulting deletion regions were verified by sequencing, and they were subcloned as described above into pJEM13 and pJEM15 (generating pFS20 to pFS23) (Table 1).
Construction of M. smegmatis ΔpyrD mutant strain CF1.
The M. smegmatis pyrD gene was cloned by PCR amplification of 2,972 bp from the chromosomal pyrD region. The PCR fragment was digested with XbaI and HindIII and cloned into similarly cut pZeRO2.1 to generate plasmid pFS3 (Table 1). A pyrD in-frame deletion plasmid was generated by digesting pFS3 with AgeI and BlpI (New England BioLabs) followed by a fill-in reaction using Klenow fragment of E. coli DNA polymerase (Promega), which regenerated the AgeI restriction site. This fragment was self-ligated to generate pFS3Δ1 (Table 1). Subsequent attempts at creating a markerless chromosomal pyrD deletion using sacB-based counterselection methods (16) were unsuccessful (data not shown). In order to create a marked allele for easier isolation of a pyrD mutant, a SmaI fragment from pGMΩ1 containing a gentamicin resistance gene in an omega interposon was ligated to AgeI-digested, blunt-ended pFS3Δ1 to generate pFS3Δ2. This plasmid is both kanamycin and gentamicin resistant and unable to replicate in M. smegmatis.
Allelic exchange experiments using pFS3Δ2 were conducted by electrotransformation of M. smegmatis mc2155 using 2 μg of plasmid DNA treated with 100 kJ/cm2 of UV irradiation to increase the frequency of recombination, as described previously (15). Single-crossover integrants from each transformation were selected on 7H10 plates supplemented with uracil, gentamicin, and kanamycin. Several independent single-crossover integrants were grown overnight under nonselective conditions in 7H9 broth with uracil and plated on 7H10 plates supplemented with uracil and gentamicin. Several hundred colonies were screened for potential double crossovers by kanamycin sensitivity via replica plating. From those, 20 colonies demonstrated the expected antibiotic sensitivity; these were subsequently replica plated to 7H10 plates with and without uracil. From that pool, one colony demonstrated a requirement for uracil for growth. This strain was designated CF1 (Table 1) and was used for all subsequent experiments.
Construction of M. smegmatis ΔpyrR strain CF22 and ΔpyrR ΔpyrD strain CF24.
Plasmid pFS1 (Table 1), containing the wild-type M. smegmatis pyrR gene, was constructed by PCR amplification of 1,984 bp of the chromosomal pyr gene region. The PCR amplicon was gel purified, cut with HindIII and XbaI, and ligated into similarly cut pZErO2.1 (Table 1) to generate pFS1. An in-frame deletion of the M. smegmatis pyrR gene was constructed by digesting plasmid pFS1 with BbsI and PshAI followed by a fill-in reaction using Klenow fragment and subsequent religation, resulting in plasmid pFS14. After verifying the in-frame deletions by sequencing, the fragment was cloned into HindIII- and ScaI-digested p2NIL (26) to generate plasmid pFS19. A PacI cassette containing sacB, lacZ, and hygromycin resistance genes was cloned from pGOAL19 (26) into pFS19 to generate the final knockout vector, pFS24 (Table 1).
pFS24 DNA (5 μg) was UV treated and electrotransformed into M. smegmatis strains mc2155 and CF1. Single-crossover plasmid integrants were selected on 7H10 medium containing uracil, arginine, X-Gal, and kanamycin. Several candidate single-crossover integrants (blue colonies) were screened by colony PCR to verify the presence of both pyrR alleles. Once verified, integrants were grown overnight in 7H9 broth with uracil and arginine, and dilutions were plated onto 7H10 plates containing 10% sucrose and uracil. After 3 to 5 days of growth, colonies were replica plated onto 7H10 plates containing sucrose, uracil, and X-Gal. Double-crossover candidates (white colonies) were further analyzed by colony PCR for the presence of the deletion allele only, using both bacterial strains (mc2155 and CF1) and plasmids (pFS1 and pFS24) as controls. Aspartate transcarbamylase (ATCase) assays were performed as a secondary screen to detect altered pyr gene expression. Strains CF22 (parent strain mc2155) and CF24 (parent strain CF1) were shown by colony PCR to contain the expected deletion in pyrR and by assay to display elevated ATCase activity when grown with uracil (Table 1).
Assays.
ATCase activity was monitored using a variation of the 14C-based radioassay (27) under assay conditions similar to those described for previous studies on the M. smegmatis ATCase (22). Frozen cell pellets were thawed on ice and washed three times with 50 mM Tris-acetate (pH 7.5), resuspended in the same buffer with 0.1-mm glass zirconium beads, and then disrupted by a single 90-s pulse using a Mini-BeadBeater. Beads and cell debris were removed by a 10-min centrifugation (14,000 × g), and the supernatant was used in assays. ATCase assays were performed at 30°C in a 100-μl final volume containing the following: 50 mM Tris-acetate (pH 7.5), 10 mM [14C]aspartate (final specific activity, 50 μCi/mmol; Amersham Biosciences), and 5 mM carbamoyl-phosphate (Sigma). Reactions were initiated by addition of enzyme and stopped with 900 μl 0.2 N acetic acid after 30 min. A 950-μl portion of each reaction mixture was added to a column containing Dowex AG-50W-X8 (200 to 400 mesh hydrogen form; Bio-Rad) to retain unreacted aspartate. Columns were washed four times with 400 μl 0.2 N acetic acid with the eluate collected in scintillation vials. Aquasol-2 (Packard Research) was added to the collected eluate, and total counts were determined using liquid scintillation counting.
For β-galactosidase assays the cells were grown and treated as for ATCase assays. In experiments where only β-galactosidase activity was determined, the cells were resuspended in Z buffer lacking 2-mercaptoethanol (24) prior to breaking in the Mini-BeadBeater as described above; in experiments where both ATCase and β-galactosidase were determined, 50 mM Tris-acetate was used as the breaking buffer to avoid possible phosphate inhibition of ATCase activity. β-Galactosidase activity was determined as described previously (34) using Z buffer containing 40 mM β-mercaptoethanol.
Protein concentrations were determined using the BCA protein assay kit (Pierce), using bovine serum albumin as the protein standard.
RESULTS
Repression of M. smegmatis pyr genes by exogenous uracil requires the pyrR gene.
ATCase is encoded by pyrB, the second gene in the M. smegmatis pyr operon (Fig. 1A). Assays of ATCase activity were used to measure the level of pyr operon expression in M. smegmatis cells grown under repressing and derepressing conditions. The repressive effects of exogenous uracil on ATCase levels in the parent strain, mc2155, were modest, only about twofold (Table 2). When a pyrimidine auxotroph derived from mc2155, strain CF1 (ΔpyrD), was grown with uridine as the pyrimidine source, growth was very slow (doubling time of 18 h compared to 4.7 h when this strain was grown with uracil supplementation), which indicates very poor utilization of exogenous uridine by M. smegmatis with consequent starvation for pyrimidines. With strain CF1, pyrimidine-limited growth led to a 14-fold derepression of ATCase, relative to the repressed level in cells grown with uracil (Table 2). Thus, expression of the pyr operon is clearly regulated by the supply of exogenous pyrimidines, presumably acting via effects on the intracellular concentration of pyrimidine nucleotides.
TABLE 2.
Repression of M. smegmatis pyrB (ATCase) by uracil and derepression by pyrimidine-limiting growth on uridine require the pyrR gene
| Function | Strain | Strain genotype | Plasmid genotype | Chromosomally encoded ATCase
|
Plasmid-encoded β-galactosidase
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Sp act (nmol/min/mg)
|
Ratio | Sp act (nmol/min/mg)
|
Ratio | ||||||
| Uracil | NE or uridinea | Uracil | NEa or uridine | ||||||
| Repression | mc2155 | Wild type | NAb | 7.7 ± 0.2 | 19 ± 1.6 | 2.5 | − | − | − |
| CF22 | ΔpyrR | NA | 64 ± 2.7 | 66 ± 4.1 | 1.0 | − | − | − | |
| CF22/pJEM15c | ΔpyrR | None | 64.5 ± 4.6 | 64.5 ± 2.9 | 1.0 | 3.5 ± 0.4 | 4.2 ± 1.9 | 1.2 | |
| CF22/pFS28d | ΔpyrR | pyrR-pyrB′-lacZ | 11 ± 0.8 | 20 ± 2.0 | 1.8 | 500 ± 27 | 540 ± 6.9 | 1.1 | |
| Derepression | CF1 | ΔpyrD | NA | 9 ± 1.4 | 130 ± 6.2 | 14 | − | − | − |
| CF24 | ΔpyrD ΔpyrR | NA | 69 ± 1.9 | 150 ± 7.8 | 2.2 | − | − | − | |
| CF24/pJEM15 | ΔpyrD ΔpyrR | None | 60 ± 1.4 | 180 ± 15 | 3.0 | 1.3 ± 0.1 | 6.8 ± 0.5 | 5.2 | |
| CF24/pFS28 | ΔpyrD ΔpyrR | pyrR-pyrB′-lacZ | 9.1 ± 0.6 | 130 ± 21 | 14 | 470 ± 19 | 6,000 ± 260 | 13 | |
NE, no effector (for strains mc2155 and CF22); uridine was used for derepression of strains CF1 and CF24.
NA, not applicable.
pJEM15 is the vector plasmid (promoterless lacZ).
pFS28 is derived from pJEM15 and contains the pyr promoter region, pyrR, and the 5′ end of pyrB fused transcriptionally to lacZ (see text for details).
Repression of pyr expression was dependent on the presence of an intact pyrR gene. This was demonstrated by determination of the levels of ATCase activity in strain CF22, a derivative of strain mc2155 containing an in-frame deletion of the pyrR gene. ATCase specific activity was eightfold higher in CF22 cells than in the repressed pyrR+ parent strain and was not repressed by growth of the cells in the presence of uracil (Table 2). The ATCase level in strain CF24, a ΔpyrR derivative of the ΔpyrD strain CF1, was also eight times higher than in the parent CF1 strain when both were grown in the presence of repressing levels of uracil. When both ΔpyrD strains were starved for pyrimidines by growth in the presence of uridine, the level of ATCase in the ΔpyrD ΔpyrR strain CF24 was similar to, although about 15% higher than, the ATCase level in the ΔpyrD pyrR+ strain CF1 (Table 2). These findings document a requirement of the pyrR gene for repression of the pyr operon in pyrimidine-starved cells. It is noteworthy that the level of ATCase formed in the ΔpyrD strains CF1 and CF24 during pyrimidine-limited growth on uridine was about twice as high as that observed in cells containing the ΔpyrR mutations alone. This indicates that a small portion of the total derepression seen during slow growth caused by pyrimidine limitation is pyrR independent. Additional evidence for a component of pyrR-independent derepression in pyrimidine-starved cells is seen in experiments presented in Table 3, below.
TABLE 3.
Regulation of pyr′-lacZ transcriptional and translational fusion plasmids by pyrimidines
| Strain | Relevant genotype | Plasmid | Promoter regiona | β-Galactosidase
|
||
|---|---|---|---|---|---|---|
| Sp act (nmol/min/mg)
|
Ratio | |||||
| Uracilb | Uridineb | |||||
| Translational fusions | ||||||
| CF1 | ΔpyrD | pFS4 | −488 to +9 | 400 ± 20 | 7,900 ± 330 | 20 |
| CF1 | ΔpyrD | pFS5 | −233 to +9 | 68 ± 4.1 | 3,200 ± 160 | 47 |
| CF1 | ΔpyrD | pFS6 | −102 to +9 | 6.7 ± 1.2 | 280 ± 33 | 42 |
| CF1 | ΔpyrD | pJEM13 | NAc | 1.1 ± 0.44 | 0.76 ± 0.27 | 0.7 |
| CF24 | ΔpyrD ΔpyrR | pFS5 | −233 to +9 | 780 ± 99 | 3,500 ± 440 | 4.5 |
| CF1 | ΔpyrD | pFS20 | −233 to +9, with BLΔ1d | 380 ± 21 | 2,000 ± 180 | 5.3 |
| CF1 | ΔpyrD | pFS21 | −233 to +9, with BLΔ2e | 1,300 ± 55 | 6,300 ± 430 | 4.8 |
| CF24 | ΔpyrD ΔpyrR | pFS20 | −233 to +9, with BLΔ1 | 360 ± 25 | 1,300 ± 41 | 3.6 |
| CF24 | ΔpyrD ΔpyrR | pFS21 | −233 to +9, with BLΔ2 | 1,300 ± 140 | 4,900 ± 550 | 3.8 |
| Transcriptional fusions | ||||||
| CF1 | ΔpyrD | pFS8 | −488 to +9 | 6,300 ± 300 | 15,000 ± 1,100 | 2.4 |
| CF1 | ΔpyrD | pFS9 | −233 to +9 | 4,400 ± 180 | 8,800 ± 640 | 2.0 |
| CF1 | ΔpyrD | pFS10 | −102 to +9 | 550 ± 59 | 1,000 ± 190 | 1.8 |
| CF1 | ΔpyrD | pJEM15 | NA | 2.3 ± 0.20 | 8.6 ± 1.6 | 3.7 |
| CF24 | ΔpyrD ΔpyrR | pFS9 | −233 to +9 | 1,000 ± 77 | 6,500 ± 650 | 6.5 |
| CF1 | ΔpyrD | pFS22 | −233 to +9, with BLΔ1 | 1,100 ± 15 | 6,900 ± 500 | 6.3 |
| CF1 | ΔpyrD | pFS23 | −233 to +9, with BLΔ2 | 1,800 ± 75 | 11,000 ± 1,300 | 6.1 |
| CF24 | ΔpyrD ΔpyrR | pFS22 | −233 to +9, with BLΔ1 | 870 ± 30 | 4,900 ± 330 | 5.6 |
| CF24 | ΔpyrD ΔpyrR | pFS23 | −233 to +9, with BLΔ2 | 1,600 ± 63 | 8,700 ± 890 | 5.4 |
Coordinates are relative to the start codon of pyrR as +1.
Concentration in medium: uracil, 100 μg/ml; uridine 200 μg/ml.
NA, not applicable.
BL region with nucleotides −21 to −37 deleted.
BL region with nucleotides −21 to −53 deleted.
Complementation of the ΔpyrR mutation in strains CF22 and CF24 by transformation with a pyrR-bearing plasmid, pFS28, restored ATCase regulation to the same pattern as observed in the pyrR+ parent strains mc2155 and CF1, respectively (Table 2). When the same strains were transformed with the vector plasmid pJEM15, they displayed the same phenotype as the ΔpyrR host strains. Complementation of the ΔpyrR chromosomal mutation by plasmid-borne pyrR demonstrated that the defects in regulation of pyrB in the ΔpyrR strains did not result from an unexpected cis-acting effect of constructing the deletions.
The pJEM15 plasmid used to construct the pFS28 pyrR complementation plasmid contained a promoterless lacZ gene and expressed β-galactosidase at only very low levels (Table 2). However, in the pFS28 plasmid the lacZ gene lies downstream from the pyrR insert and is under the control of the pyr promoter (Fig. 2). When pFS28 was expressed in strain CF22, β-galactosidase expression was elevated and was only slightly repressed by uracil (Table 2). When pFS28 was expressed under pyrimidine-limiting growth conditions in the CF24 host, β-galactosidase activity was expressed at very high levels and was repressed 13-fold by growth with uracil supplementation (Table 2). PyrR-dependent regulation of the downstream lacZ gene by uracil in pFS28 provides evidence for translational coupling between the upstream pyrR gene, whose ribosome-binding site is the putative site of PyrR action, and downstream genes (see Discussion, below).
FIG. 2.
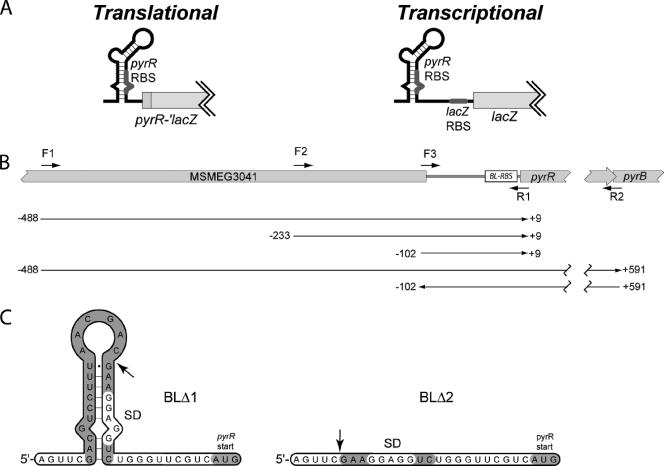
Construction of pyr′-lacZ fusion plasmids for characterization of regulation. A. Schematic diagrams showing the set of pJEM13-derived translational fusions in which the pyr leader, including the PyrR-binding loop region, pyrR ribosome-binding site (RBS), and first three codons of pyrR were fused in frame via a five-codon linker (Gly-Thr-Lys-Leu-Ala) to lacZ at codon 13 (left), and the set of pJEM15-derived transcriptional fusions in which the same pyr segments were fused to 56 bp of the lacZ leader so that the lacZ ribosome-binding site lies at the normal distance from the lacZ open reading frame (right). B. Diagram showing the locations of PCR primers (F1, F2, F3, R1, and R2) and segments of M. smegmatis DNA that were inserted upstream of lacZ to generate the plasmids described in the text and in Tables 2 and 3. Numbering of the sequence is as in Fig. 1B. C. Proposed secondary structures of the pyr leader RNA regions in the pyr′-lacZ fusion plasmids from which segments of the PyrR-binding loop were deleted (Table 3). Arrows denote the site of deletion. SD denotes the Shine-Dalgarno sequence of the presumed pyr ribosome-binding site.
Repression of the pyr operon in M. smegmatis is exerted at the level of mRNA translation.
To analyze the mechanism of PyrR involvement in repression of M. smegmatis pyr gene expression by uracil, plasmids were constructed that contained the pyr promoter-leader region fused to a lacZ reporter gene in two ways. In the translational fusions the M. smegmatis pyr promoter and leader, which includes the putative PyrR RNA binding loop sequences and the ribosome-binding site for pyrR followed by the first three codons of pyrR, were fused in frame to codon eight of lacZ in pJEM13 (Fig. 2A, left). In the transcriptional fusions, the pyr promoter, leader, and first three codons of the pyrR coding sequence were fused via a 30-nucleotide linker to the native lacZ ribosome-binding site, followed by the complete lacZ gene in pJEM15 (Fig. 2A, right). Since the precise location of the pyr promoter was not known, in both types of fusions segments of M. smegmatis DNA containing 111, 242, and 497 bp directly 5′ to codon 4 of the pyrR open reading frame were fused to lacZ (Fig. 2B). This yielded three translational and three transcriptional pyr-lacZ fusion plasmids that differed only in the length of upstream DNA fused to lacZ. M. smegmatis strain CF1 (ΔpyrD) was transformed with all six plasmids, the transformants were grown with uracil (repressing conditions) and with uridine (pyrimidine-limiting conditions), and β-galactosidase levels were assayed to determine pyr promoter-directed expression (Table 3). For the translational fusions (Table 3), the derepressed β-galactosidase expression was much higher with the fusion containing a 241-bp insert than in the fusion containing the 111-bp insert, but only increased by another 2.5-fold in the fusion containing the 497-bp insert. This observation indicates that most or all of the DNA sequences needed for a fully functional M. smegmatis pyr promoter lie on the 241-bp segment upstream of pyrR. Since about 55 bp of the 3′ portion of this segment specifies the binding loop RNA, the pyrR ribosome-binding site, and the first three codons of pyrR, we suggest that the complete pyr promoter lies on a 180-bp segment in the 5′ portion of the 242-bp insert. A functional, but much weaker, promoter sequence lies in the 49-bp segment at the 5′ end of the 111-bp insert (see Discussion, below).
Significantly, all three inserts specified uracil-repressible activity in the translational fusion plasmids (Table 3). However, the corresponding inserts in the transcriptional fusion plasmids were repressed only about twofold by uracil (Table 3). It is shown below that this residual repression by uracil is independent of the pyrR gene. Thus, we conclude that the transcriptional fusion plasmids, which include the lacZ ribosome-binding site that is not present in the translational fusion plasmids, escape PyrR-dependent repression by uracil. Since both types of fusion plasmids contained the identical pyr promoter-leader region, we conclude that uracil and PyrR exert their repressive actions by inhibition of translation at the pyrR ribosome-binding site, rather than acting to inhibit initiation of transcription. The most obvious mechanism for inhibition of translation is for PyrR to bind to the conserved RNA-binding loop sequences that overlap the pyrR ribosome-binding site. The transcriptional fusions escape repression because PyrR cannot bind to the lacZ ribosome-binding site.
Strain CF24 (ΔpyrDΔpyrR) was transformed with the translational and transcriptional plasmids containing the 242-bp insert (pFS5 and pFS9, respectively), and β-galactosidase levels were determined in cells grown in the presence of uracil and uridine to assess the role of the pyrR gene in regulation of expression of the pyr′-lacZ plasmids. The ability of uracil to repress expression of the pyr′-lacZ fusion translation was largely abolished in the ΔpyrR background, as shown by the 11-fold increase in β-galactosidase level formed in CF24 cells compared to that produced from the same plasmid in pyrR+ cells (Table 3, CF1 with pFS5 grown with uracil versus CF24 with pFS5). On the other hand, β-galactosidase expression, which was elevated and poorly repressed by uracil in the transcriptional pyr′-lacZ fusion, was also poorly regulated in the ΔpyrR host (Table 3, CF1 with pFS9 versus CF24 with pFS9). These observations confirm that regulation of expression of the pyr′-lacZ fusion by uracil was largely dependent on the pyrR gene. However, there was a pyrR-independent component to the increased expression of β-galactosidase seen with these plasmids during pyrimidine-limited growth, as indicated by the 4.5- to 6.5-fold higher levels seen in CF24 cells grown with uridine versus CF24 cells grown with uracil.
Repression of the pyr operon in M. smegmatis requires an intact PyrR-binding site adjacent to the pyrR ribosome-binding site in the pyr transcript.
If PyrR acts to repress pyr operon expression by binding to the conserved RNA-binding loop sequences in the pyr leader, as we have proposed, pyr′-lacZ translational fusion plasmids from which DNA specifying portions of the binding loop RNA has been deleted should no longer be repressed by uracil. Effects of uracil on expression of transcriptional fusions, from which the same binding loop sequences are deleted but which still contain the lacZ ribosome-binding site, should be small and similar to those seen with the corresponding plasmids from which the binding loop sequences are not deleted. These predictions were tested with translational fusion plasmids pFS20 and pFS21 (Table 3) and transcriptional fusion plasmids pFS22 and pFS23 (Table 3). In pFS20 and pFS22, a 17-bp segment specifying nucleotide −37 to −21 of the binding loop RNA (BLΔ1) was deleted; in pFS21 and pFS23 the entire binding loop upstream of the putative pyrR ribosome-binding site (−53 to −21; BLΔ2) was deleted (Fig. 1B and 2C). In all of the plasmids the putative pyrR Shine-Dalgarno sequence (−13 to −17) was retained. All four binding loop deletion plasmids were introduced separately into strain CF1 (ΔpyrD), and β-galactosidase levels were determined in cells grown with uracil and uridine as pyrimidine sources. Deletion of the binding loop sequences sharply reduced the ability of uracil to repress expression in the translational fusion plasmids pFS20 and pFS21 (about fivefold repression) (Table 3) compared to the corresponding translational fusion plasmid pFS5 (47-fold) (Table 3) from which the binding loop was not deleted. Effects of uracil and uridine on expression of the transcriptional fusion plasmids containing binding loop deletions, pFS22 and pFS23 (Table 3), were similar to those seen with the parent plasmid pFS9 (Table 3), although repression ratios were actually somewhat higher (sixfold versus twofold) with the transcriptional fusion plasmids with binding loop deletions. These results confirm a requirement for the PyrR-binding loop sequence for strong repression by uracil. Curiously, the expression of all of the plasmids was elevated by five- to sixfold in uridine-grown cells, which is greater than the two- to threefold elevation of ATCase expression observed in pyrimidine-starved ΔpyrR cells (Table 2). This might suggest that some residual PyrR-dependent regulation occurs even when the binding loop is deleted. However, the experiments in Table 3 using strain CF24 (ΔpyrD ΔpyrR) transformed with pFS20, pFS21, pFS23, and pFS23 demonstrated that the four- to sixfold derepression observed in cells grown under pyrimidine-limiting conditions was independent of the pyrR gene.
Effects of pyrimidine-limited growth on expression controlled by other M. smegmatis promoters.
The highest expression of the M. smegmatis pyr genes was obtained in our studies when ΔpyrD host cells were starved for pyrimidines by slow growth on uridine. From the data in Table 2 it is seen that most of the derepression of pyrB (ATCase) under these conditions requires the pyrR gene, but two- to threefold derepression occurs in ΔpyrR strains. In studies with pyr′-lacZ fusion plasmids, the pyrR-independent component of derepression in uridine-grown cells was usually higher, generally four- to sixfold (Table 3). Does the pyrR-independent component of derepression represent a second mode of regulation of pyr genes by uracil, or does it result from a general, nonspecific effect of slow growth during pyrimidine starvation? We attempted to address this question by examining the effects of pyrimidine starvation on expression of three plasmids in which the hsp60 (groEL2) (10, 26), Ag85 (fbpA) (2, 26), and putative MSMEG3041 (see Materials and Methods) promoters were transcriptionally fused to lacZ in pJEM15. We reasoned that a second mechanism that was specific for regulation of pyr gene expression, if such a mechanism existed, would act on the pyr promoter but not on promoters that have no function in nucleotide metabolism. Strain CF1 and CF24 cells were transformed with the new promoter-lacZ fusion plasmids, the cells were grown with uracil and uridine as in the experiments of Tables 2 and 3, and β-galactosidase expression was assayed. Expression of two of the promoter fusion plasmids was elevated by about threefold in the cells grown under pyrimidine limitation (Table 4), whether or not the host strain contained a functional pyrR gene. Expression of the Ag85-lacZ fusion plasmid was also elevated during pyrimidine-limited growth, but to a lesser extent. We conclude that most of the pyrR-independent derepression previously observed with the pyr′-lacZ fusion plasmids was a nonspecific effect of slow growth or depleted intracellular pyrimidine nucleotide levels (see Discussion).
TABLE 4.
Expression of alternative lacZ transcriptional fusion plasmids under pyrimidine-limiting conditions
| Strain | Relevant genotype | Plasmid | Promoter | β-Galactosidase
|
||
|---|---|---|---|---|---|---|
| Sp act (nmol/min/mg)
|
Ratio | |||||
| Uracila | Uridinea | |||||
| CF1 | ΔpyrD | pFS30 | MSMEG3041 | 380 ± 14 | 1,100 ± 100 | 2.9 |
| CF1 | ΔpyrD | pFS31 | Ag85A (fbpA) | 77 ± 6.6 | 120 ± 21 | 1.6 |
| CF1 | ΔpyrD | pFS32 | hsp60 (groEL2) | 8,300 ± 420 | 25,000 ± 2,200 | 3.0 |
| CF24 | ΔpyrD ΔpyrR | pFS30 | MSMEG3041 | 450 ± 30 | 1,000 ± 170 | 2.2 |
| CF24 | ΔpyrD ΔpyrR | pFS31 | Ag85A (fbpA) | 57 ± 3.5 | 77 ± 8.3 | 1.4 |
| CF24 | ΔpyrD ΔpyrR | pFS32 | hsp60 (groEL2) | 7,500 ± 180 | 21,000 ± 1,700 | 2.8 |
Concentrations in medium: uracil, 100 μg/ml; uridine, 200 μg/ml.
DISCUSSION
Location and properties of the M. smegmatis pyr promoter.
Although the site of transcription initiation has not been mapped precisely for the M. smegmatis pyr operon by primer extension, our experiments with pyr′-lacZ fusion plasmids demonstrated that a functional pyrimidine-regulated pyr promoter lies in a 72-bp segment of DNA lying from 55 to 102 bp 5′ to the start of pyrR translation (Fig. 1B). Within this region a consensus sequence (TANNNT) for the −10 region mycobacterial promoters (1) is found at −66 to −71 (TAAGCT), with numbering from the start of translation. Separated by 18 bp upstream at −90 to −95, the sequence TTCCCC is found, which matches the mycobacterial −35 consensus sequence (TTGCGA) (1) in three of six positions. These sequences would predict a likely start for pyr transcription 7 or 8 bp downstream from the −10 sequence at A-58 or G-57, just 4 or 5 bp before the sequence that specifies the beginning of the PyrR-binding loop RNA. Possibly as a result of the relatively weak −35 sequence, the core promoter sequence located in the 111-bp pyr′-lacZ fusion plasmid pFS6 supported 10- and 20-fold lower expression, respectively, than the 242-bp (pFS5) and 497-bp (pFS4) pyr′-lacZ fusion plasmids (Table 3). This suggests that upstream elements, while not part of the core promoter, serve to activate pyr transcription in M. smegmatis. The activation of the M. smegmatis rrnB operon by upstream elements provides precedence for such a phenomenon (3), but the specific sequences involved in rrnB activation are not found upstream of the pyr operon.
A model for regulation of the M. smegmatis pyr operon by PyrR-dependent translational repression.
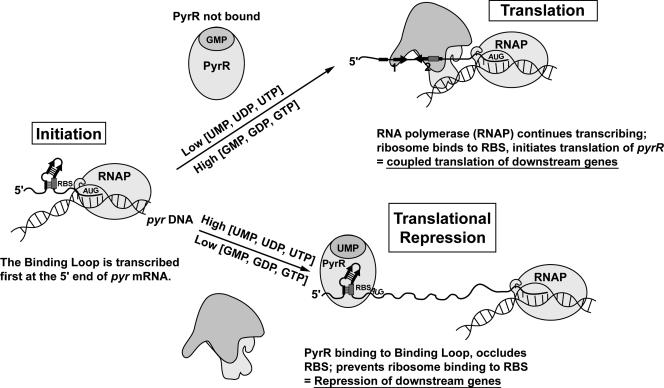
Our experiments demonstrated that most of the repressive effects of exogenous uracil on pyr expression in M. smegmatis required both the pyrR gene and the presence of the PyrR-binding loop RNA sequence that lies in the untranslated leader upstream of the pyrR gene. Furthermore, this regulatory effect was largely abolished if a second ribosome-binding site that does not overlap the PyrR binding sequence was provided for the lacZ reporter gene. We propose that a translational repression model for PyrR action in M. smegmatis accounts for these observations (Fig. 3). According to this model, PyrR binding to its specific binding site in the pyr leader is stimulated by high levels of uridine nucleotides and antagonized by high levels of guanosine nucleotides, as has been demonstrated for the Bacillus PyrR (8). We have also demonstrated these effects of nucleotides on the binding of purified PyrR from M. tuberculosis to M. tuberculosis binding loop RNA (C. J. Fields and R. L. Switzer, unpublished data). When PyrR is not bound, ribosomes are able to bind to a site just upstream of the pyrR open reading frame and initiate translation. However, the PyrR-binding site and the Shine-Dalgarno sequence of the pyrR ribosome-binding site overlap, and so when PyrR is bound at high uridine nucleotide concentrations, binding of ribosomes is inhibited and translation is repressed. Although competition between ribosomes and PyrR would occur only at a site upstream of pyrR, the first cistron of the pyr operon, translation of all of the downstream pyr genes might also be inhibited because of translational coupling. Evidence that such translational coupling occurs is seen from the efficient regulation of the expression of pyrB, the second cistron of the operon by exogenous uracil (Table 2), and by similar regulation of lacZ when this gene lies artificially downstream from pyrR in the pyrR complementation plasmid pFS28 (Table 2). Transcriptional polarity of the polycistronic pyr mRNA could also affect expression of the downstream genes. Further testing of the proposed model by biochemical experiments is in progress in our laboratory.
FIG. 3.
Proposed mechanism of PyrR-mediated regulation of the M. smegmatis pyr operon by translational repression.
The nature of PyrR-independent derepression during growth under pyrimidine-limiting conditions.
Addition of uracil to defined pyrimidine-free medium gave only a twofold decrease in pyrB expression in M. smegmatis, and so it was necessary to grow a pyrimidine auxotroph on uridine, which resulted in slow growth and consequent derepression of pyrB, to demonstrate the full extent of repression by uracil. Such a device is commonly used to study regulation of bacterial biosynthetic genes. Full derepression of the B. subtilis pyr operon (19) and pyrG (23) required growth of auxotrophs on orotate, which supports slow pyrimidine-limited growth in that organism. Most of the derepression of pyrB or pyr′-lacZ fusion plasmids seen in M. smegmatis grown under pyrimidine-limiting conditions was dependent on the pyrR gene, but a residual portion, ranging from two- to sixfold, was observed in ΔpyrR strains (Tables 2 and 3). The experiments reported in Table 4 document that threefold derepression of two other M. smegmatis promoters, whose functions are unrelated to pyrimidine metabolism, was observed under the same conditions of pyrimidine-limited growth. Thus, this effect reflects, largely if not entirely, a generalized and nonspecific effect of such growth conditions on gene expression and is not the reflection of a second PyrR-independent regulatory mechanism that specifically governs pyr gene expression. The biochemical basis of this effect is not clear. It could represent a generalized stress response induced by slow growth, or it could be mediated by the low intracellular concentrations of pyrimidine nucleotides under these conditions. It is noteworthy that a similar two- to threefold effect of slow growth of B. subtilis pyrimidine auxotrophs on orotate was observed for the pyr operon in a ΔpyrR strain (19) and for the pyrG gene in a strain in which the 5′ leader attenuator was deleted (23).
Translational repression of pyr genes by PyrR in other species.
The pyr genes of M. tuberculosis (9) and other mycobacteria (7, 32) are organized into operons in which the first cistron is pyrR and the 5′ leader sequences include a PyrR-binding loop that overlaps a putative ribosome-binding site in the same way as seen in M. smegmatis. It seems very likely, therefore, that the pyr operons in these species are regulated by a translational repression mechanism like that described here.
A review of published genome sequences indicated that pyrR genes and PyrR-binding sequences that overlap the ribosome-binding sites for pyr genes are quite widespread in other bacterial species. Often when this arrangement is found, attenuator terminator and antiterminator sequences are not present downstream from the PyrR-binding sequence, which argues against a transcription attenuation regulatory mechanism for such genes. Our findings in this work provide support for the idea that such arrangements indicate likely regulation of these genes by translational repression, although they do not, of course, demonstrate that this is the case. C. J. Fields (unpublished) has identified a number of species in which open reading frames for putative pyrP (uraA) genes encoding uracil transporters and/or another putative transport protein of the major facilitator protein superfamily fit this pattern. In some species (e.g., Clostridium spp.) this arrangement coexists with pyr genes that are regulated by PyrR-mediated transcriptional attenuation. In other cases (e.g., Haemophilus influenzae, Pasteurella multocida), translational repression of the genes for transport proteins and/or carA appears to be the only mode of PyrR regulation. A regulatory mechanism involving both PyrR-dependent translational repression and transcription attenuation has been proposed for Thermus strain ZO5 and related Thermus species (37). In Thermus strain ZO5 an open reading frame for short leader polypeptide precedes the pyrR gene and other downstream genes of a pyr operon. The ribosome-binding site in the mRNA for the leader polypeptide overlaps a consensus PyrR-binding sequence so that translational repression by PyrR is likely. The RNA encoding the leader polypeptide is also capable of forming a transcription terminator. Van de Casteele et al. (37) suggested that attenuation at this terminator is regulated by the rate of translation of the leader polypeptide, which is in turn responsive to pyrimidines via PyrR-mediated translational repression. Furthermore, 6 of 28 codons in the leader polypeptide encode arginine, which might account for stimulation of pyr gene expression in Thermus strain ZO5 by this amino acid. This interesting model has not yet been tested by genetic or biochemical means, however. Altogether, these observations indicate that translational repression by PyrR may be as widespread among bacteria as PyrR-dependent transcription attenuation and may have preceded it in evolutionary development.
Translational repression is a widespread regulatory mechanism in prokaryotes.
Numerous examples of the regulation of genes by translational repression by various mechanisms have been reported (29). Of these examples, translational repression of the B. subtilis trpG (11), trpP (39), and ycbK (38) genes by the trp RNA-binding attenuation protein TRAP (5) presents a close analogy to the mechanism we have proposed for regulation of pyr genes by M. smegmatis PyrR. Like PyrR, TRAP also regulates gene expression by a transcription attenuation mechanism (5). However, in the cases cited, strong evidence has been presented that TRAP acts to inhibit translation initiation by binding to mRNA at the ribosome-binding site. Remarkably, TRAP can also inhibit translation of the trpE gene by binding to a site that does not overlap the ribosome-binding site but instead favors an alternative fold of trpE RNA that occludes the ribosome-binding site in a hairpin structure (5). We suspect that numerous additional examples and mechanistic variations of translational repression mechanisms remain to be discovered.
Acknowledgments
This research was supported by National Institutes of Health grant GM47112.
We gratefully acknowledge the following persons for providing plasmids used in this research: Brigette Gicquel for pJEM13 and pJEM15, Tanya Parish for pGOAL19 and p2NIL, and Herbert Schweizer for pGMΩ1. Preliminary M. smegmatis genome sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of M. smegmatis was accomplished with support from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 29 June 2007.
REFERENCES
- 1.Agarwal, N., and A. K. Tyagi. 2006. Mycobacterial transcription signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res. 34:4245-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitige, L. Y., C. Jagannath, A. R. Wanger, and S. J. Norris. 2000. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect. Immun. 68:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnvig, K. B., B. Gopal, K. G. Papavinasasundaram, R. A. Cox, and M. J. Colston. 2005. The mechanism of upstream activation in the rrnB operon of Mycobacterium smegmatis is different from the Escherichia coli paradigm. Microbiology 151:467-473. [DOI] [PubMed] [Google Scholar]
- 4.Arsene-Ploetz, F., V. Kugler, J. Martinussen, and F. Bringel. 2006. Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine regulator PyrR1. J. Bacteriol. 188:8607-8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 183:5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosch, R., S. V. Gordon, T. Garnier, et al. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Nat. Acad. Sci. USA 104:5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chander, P., K. M. Halbig, J. K. Miller, C. J. Fields, H. K. S. Bonner, G. K. Grabner, R. L. Switzer, and J. L. Smith. 2005. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J. Bacteriol. 187:1773-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Dellagostin, O. A., G. Esposito, L. J. Eales, J. W. Dale, and J. McFadden. 1995. Activity of mycobacterial promoters during intracellular and extracellular growth. Microbiology 141:1785-1792. [DOI] [PubMed] [Google Scholar]
- 11.Du, H., R. Tarpey, and P. Babitzke. 1997. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 179:2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elagöz, A., A. Abdi, J.-C. Hubert, and B. Kammerer. 1996. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene 182:37-43. [DOI] [PubMed] [Google Scholar]
- 13.Ghim, S.-Y., C. C. Kim, E. R. Bonner, J. N. D'Elia, G. K. Grabner, and R. L. Switzer. 1999. The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 181:1324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghim, S.-Y., and J. Neuhard. 1994. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J. Bacteriol. 176:3698-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds, J., E. Mahenthiralingam, K. E. Kempsell, K. Duncan, R. W. Stokes, T. Parish, and N. G. Stoker. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519-527. [DOI] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 17.Lu, Y., and R. L. Switzer. 1996. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol. 178:7206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, Y., R. J. Turner, and R. L. Switzer. 1996. Function of the RNA secondary structures in the regulation of the Bacillus subtilis pyr operon expression. Proc. Natl. Acad. Sci. USA 93:14462-14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, Y., R. J. Turner, and R. L. Switzer. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol. 177:1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinussen, J., and K. Hammer. 1998. The carB gene encoding the large subunit of carbamoylphosphate synthetase from Lactococcus lactis is transcribed monocistronically. J. Bacteriol. 180:4380-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masood, R., and T. A. Venkitasubramanian. 1988. Purification and properties of aspartate transcarbamylase from Mycobacterium smegmatis. Biochim. Biophys. Acta 953:106-113. [DOI] [PubMed] [Google Scholar]
- 23.Meng, Q., and R. L. Switzer. 2001. Regulation of transcription of the Bacillus subtilis pyrG gene encoding cytidine triphosphate synthetase. J. Bacteriol. 183:5513-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Nicoloff, H., A. Elagoz, F. Arsene-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 187:2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 27.Perbal, B., and G. Herve. 1972. Biosynthesis of Escherichia coli aspartate transcarbamylase I. Parameters of gene expression and sequential biosynthesis of the subunits. J. Mol. Biol. 70:511-529. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schlax, P. J., and D. J. Worhunsky. 2003. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 48:1157-1169. [DOI] [PubMed] [Google Scholar]
- 30.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 31.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 32.Stinear, T. P., T. Seemann, S. Pidot, et al. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 34.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, R. J., E. R. Bonner, G. K. Grabner, and R. L. Switzer. 1998. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J. Biol. Chem. 273:5932-5938. [DOI] [PubMed] [Google Scholar]
- 36.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Casteele, M., P. Chen, M. Roovers, C. Legrain, and N. Glansdorff. 1997. Structure and expression of a pyrimidine gene cluster from the extreme thermophile Thermus strain ZO5. J. Bacteriol. 179:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yakhnin, H., A. V. Yakhnin, and P. Babitzke. 2006. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol. 61:1252-1266. [DOI] [PubMed] [Google Scholar]
- 39.Yakhnin, H., H. Zhang, A. V. Yakhnin, and P. Babitzke. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 186:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]