Abstract
Porphyromonas gingivalis is a major oral pathogen that contributes to the development of periodontal disease. There is a significant degree of genetic variation among strains of P. gingivalis, and the population structure has been predicted to be panmictic, indicating that horizontal DNA transfer and recombination between strains are likely. The molecular events underlying this genetic exchange are not understood, although a putative type IV secretion system is present in the genome sequence of strain W83, implying that DNA conjugation may be responsible for genetic transfer in these bacteria. In this study, we provide in vitro evidence for the horizontal transfer of DNA using plasmid- and chromosome-based assays. In the plasmid assays, Bacteroides-derived shuttle vectors were tested for transfer from P. gingivalis strains into Escherichia coli. Of the eight strains tested, five were able to transfer DNA into E. coli by a mechanism most consistent with conjugation. Additionally, strains W83 and 33277 tested positive for the transfer of chromosomally integrated antibiotic resistance markers. Ten chimeras resulting from the chromosomal transfer assay were further analyzed by Southern hybridization and were shown to have exchanged DNA fragments of between 1.1 and 5.6 kb, but the overall strain identity remained intact. Chimeras showed phenotypic changes in the ability to accrete into biofilms, implying that DNA transfer events are sufficient to generate measurable changes in complex behaviors. This ability to transfer chromosomal DNA between strains may be an adaptation mechanism in the complex environment of the host oral cavity.
Porphyromonas gingivalis is a gram-negative anaerobe that colonizes plaque biofilms in the human subgingival crevice and, in cooperation with other oral pathogens, contributes to the development of periodontal disease. P. gingivalis possesses multiple virulence factors, including the gingipain proteases, fimbriae, hemagglutinins, hemolysin, iron uptake transporters, and capsule production genes (20). Additionally, P. gingivalis bacteria are able to invade and establish residence in host gingival epithelial cells, where they are protected from the immune system and can contribute to the tissue damage associated with periodontal disease (18).
Numerous studies have attempted to measure the degree of genetic variability in the P. gingivalis population by using techniques such as pulsed-field gel electrophoresis, restriction fragment length polymorphisms, insertion sequence hybridization, cross-species microarray hybridization, and multilocus sequence typing (2, 6, 7, 10, 15, 24, 27). All studies to date have pointed to significant levels of genetic variation among P. gingivalis strains, indicating that the population structure of these organisms is not strictly clonal but instead is influenced by DNA recombination between different strains. This panmictic population structure is common to human pathogens involved in chronic infections and is thought to contribute to their ability to persist in the human host in the face of changing environmental niche conditions (14, 29). Additionally, multiple alleles have been detected for several virulence factors, implying that these bacteria may have specialized adaptations for individual hosts. In the classic panmictic bacterial species, such as Helicobacter pylori and Neisseria spp., natural competence is the mechanism that facilitates DNA exchange between strains (13, 23, 33).
DNA transfer mechanisms in P. gingivalis are not well studied. P. gingivalis bacteria do not contain plasmids and are not naturally competent, although they can easily be transformed by electroporation, and many strains readily integrate Escherichia coli DNA or PCR-derived DNA into the chromosome by homologous recombination. Analysis of the genome sequence of strain W83 (9, 26) reveals one potential mediator of horizontal DNA transfer by conjugation, a distant homolog of type IV secretion systems found in other gram-negative bacteria (8). The 12-kb region encodes proteins most similar to those encoded by the DNA transfer regions (tra genes) of the Bacteroides conjugative transposons cTnDot and cTn341 (30% to 75% identity) (3, 5).
In this study, we screened eight strains of P. gingivalis for the ability to conjugate plasmid DNA, and we further show that strains ATCC 33277 and W83 are able to transfer chromosomal DNA to each other by conjugation, where it is incorporated into the genome. We postulate that this ability to transfer DNA between strains by conjugation is the underlying mechanism for allele swapping and genetic variation in the P. gingivalis population.
MATERIALS AND METHODS
Bacterial strains and cell culture.
Porphyromonas gingivalis strains (Table 1) were grown anaerobically at 37°C in supplemented Trypticase soy broth (TSB). TSB blood agar plates were made with the addition of 5% sheep's blood and 1.5% agarose. Selection for antibiotic-resistant P. gingivalis was performed with 15 μg/ml erythromycin or 1 μg/ml tetracycline. Dual resistance was selected on 5 μg/ml erythromycin and 1 μg/ml tetracycline. E. coli strains DH5α (Invitrogen) and S17-1 (32) were grown in Luria-Bertani medium supplemented as needed with ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains
| Strain | Genetic locus | Source |
|---|---|---|
| 33277 | ATCC 33277 | |
| W83 | Lab stock | |
| W50 | ATCC 53978 | |
| 381 | Lab stock | |
| 49417 | ATCC 49417 | |
| A7A1-28 | ATCC 53977 | |
| MP4-504 | 19 | |
| 5083 | 4 | |
| 33277Em | PG0653::ermF | 38 |
| 33277Tc | PG1170::tetQ | 38 |
| 33277TcEm | PG0653::ermF | |
| PG1170::tetQ | 38 | |
| W83Em | PG0653::ermF | 38 |
| W83Tc | PG1170::tetQ | This study |
| W83TcEm | PG0653::ermF | |
| PG1170::tetQ | This study | |
| A1A7-28 | traIM::ermF | This study |
P. gingivalis DNA conjugation.
Plasmids were conjugated from E. coli S17-1 donors to P. gingivalis strains by mixing log-phase cultures at a ratio of one donor to 100 recipients, pelleting the mixed cultures and resuspending the cells in a 50-μl volume, and incubating the bacterial pellets overnight on prereduced blood agar plates in a candle jar at 37°C. P. gingivalis recipients were selected by incubation for 7 to 10 days anaerobically on TSB blood agar containing 100 μg/ml gentamicin and either erythromycin or tetracycline. Plasmid conjugal transfers from P. gingivalis donors to E. coli DH5α were similar to E. coli-to-P. gingivalis transfers except that the mating pellet was incubated on prereduced TSB blood agar plates anaerobically overnight, and E. coli recipients were selected aerobically on 100 μg/ml ampicillin. P. gingivalis-to-P. gingivalis matings were performed with equal ratios of bacterial strains, and mating mixtures were incubated for 24 h anaerobically at 37°C. Conjugation efficiencies were calculated by dividing the number of transconjugants by the number of input donor cells. Controls for DNA exchange by transformation were either P. gingivalis mating mixtures resuspended in 50 μl of DNase I solution and spotted on blood agar plates for 24 h or individual recipient P. gingivalis strains resuspended in 50 μg of purified genomic DNA from the donor strain and spotted on blood agar plates. Controls for DNA exchange by bacteriophage were recipient P. gingivalis strains incubated with 0.2-μm-filtered cell culture supernatant from overnight cultures of donor strains.
Construction of P. gingivalis allelic-exchange mutants.
An allelic-exchange mutant with the region traI-traM deleted was generated through PCR amplification of approximately 2 kb flanking the gene fragments and then creating fusion PCR products with the ermF marker (17). This PCR product was cloned into the pCR4-TOPO vector (Invitrogen), and the resulting construct was digested with ScaI. The linear DNA was electroporated into P. gingivalis (34). Transformants were selected on erythromycin and confirmed by PCR and Southern hybridization. A P. gingivalis W83 double mutant with mutations in both PG0653 and PG1170 loci was created by transforming the confirmed W83 PG0653::ermF mutant with a pUC19-1170-tetQ linear construct (38).
Molecular biology.
DNA cloning, sequencing, PCR amplification, Southern blotting, E. coli plasmid purification, and other common molecular biology techniques were carried out by using standard procedures (30). Total DNA was purified from P. gingivalis using the Promega Wizard genomic DNA purification kit with further purification by phenol-chloroform extraction. For the Southern hybridizations, we employed the Invitrogen chemiluminescent DNA labeling and hybridization kit.
P. gingivalis biofilms.
Bacterial strains were grown to early log phase and labeled with 5 (and 6)-carboxyfluorescein succinimidyl ester (4 μg ml−1; Molecular Probes) as described previously (16). Labeled bacterial cells were added to 96-well cell culture plates or CultureWell chambered coverglass 16-well slides (Grace Bio Labs) and incubated anaerobically for 24 h at 37°C in 1× phosphate-buffered saline. For the 96-well plate, wells were washed and measured for green fluorescence using a Victor plate reader (Perkin-Elmer). Monobiofilms on the coverglass slides were recorded at a 40× magnification using a Zeiss fluorescent microscope (Axioplan 2) with a Spot Insight digital camera and the fluorescein isothiocyanate excitation/emission filter set. Biofilm assays were repeated independently three times with each strain in triplicate. Statistical significance was determined using an unpaired t test with Prism software (GraphPad Software).
RESULTS
Presence of type IV-related elements in P. gingivalis strains.
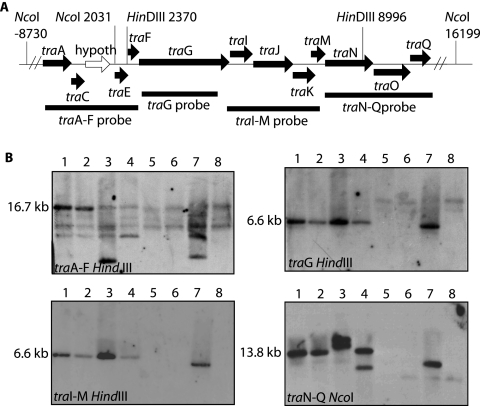
To demonstrate the presence of DNA transfer systems in P. gingivalis strains, we first analyzed eight laboratory and clinical isolates for the presence of the tra genomic loci from the W83 genome sequence (Table 1). Southern blots of HindIII- or NcoI-digested DNA were probed for the tra loci using four PCR products spanning the traA-traQ region from the W83 genome sequence (Fig. 1A). The first probe, for traA-traF, hybridized strongly to 16.7-kb bands in W83 and W50 and strongly to bands in 49417 and A1A7-28. This probe also hybridized weakly to multiple bands in all strains (Fig. 1B). For the remaining three probes, strains 33277 and 381 and clinical isolate MP4-504 either did not hybridize or did so weakly, while strains 49417 and A1A7-28 and clinical isolate 5083 showed different hybridization patterns than W83 and W50. This indicates that strains W50, 49417, 5083, and A1A7-28 have tra loci similar to those of W83 but that strains 33277, 381, and MP4-504 either do not have the traA-traQ region or the region is significantly divergent from the sequence found in W83. Attempts to PCR amplify tra genes from strains 33277, 381, and MP4-504 with W83-derived primers were unsuccessful.
FIG. 1.
(A) A putative Porphyromonas DNA transfer region. The genome region from P. gingivalis strain W83 that is similar to the Bacteroides DNA transfer genes from conjugative transposons is shown. PCR regions used as probes for Southern blots are indicated by thick horizontal bars. (B) Southern blots of P. gingivalis strains for tra genes. Results are shown for strains W83 (lanes 1), W50 (lanes 2), ATCC 49417 (lanes 3), 5083 (lanes 4), ATCC 33277 (lanes 5), 381 (lanes 6), A1A7-28 (lanes 7), and MP4-504 (lanes 8). Band sizes for strain W83 are indicated to the left of the blots.
Conjugal transfer of plasmids by P. gingivalis strains.
To determine whether the tra loci were capable of forming an active type IV secretion system, we selected two Bacteroides-E. coli shuttle vectors for testing in conjugation assays with E. coli. Plasmid pT-COW is derived from pB8-51, a promiscuous plasmid from the intestinal Bacteroides organisms (11, 31), and bears the tetracycline-selectable marker tetQ. Plasmid pFD340 is derived from the B. fragilis plasmid pBI143 (35, 36) and encodes resistance to erythromycin. Both plasmids provide ampicillin resistance for selection in E. coli. Based on growth rates in selective media, all P. gingivalis strains used in this assay maintain the plasmids at the same frequency. P. gingivalis strains containing plasmid were mated anaerobically with E. coli for 24 h, and then mating mixtures were plated aerobically on ampicillin to test for plasmid transfer from Porphyromonas strains to E. coli. Conjugation efficiencies are shown in Table 2. Controls were duplicate mating mixtures incubated in the presence of 50 μg of DNase I to confirm that DNA transfer was by conjugation and not transformation. Transfer efficiencies in the presence of DNase I were not statistically different from those not in its presence (data not shown.) Strains W83, W50, and 5083 did not transfer either plasmid at detectable levels. As these strains all contain tra-hybridizing sequences, this indicates that the tra elements in these bacteria must either be nonfunctional or recognize some DNA substrate other than those provided in this assay. In contrast, strains 49417 and A7A1-28 conjugate plasmid pT-COW at frequencies between 10−5 and 10−7, and strains 33277, 381, and MP4-504, although having no strongly hybridizing tra bands, were also able to transfer plasmid DNA at frequencies similar to those at which strains 49417 and A1A7-28 did. To determine whether the tra region is responsible for plasmid transfer in A1A7-28, a gene replacement mutant was constructed with the traI-traM open reading frames deleted and replaced with the selectable marker ermF. The pT-COW plasmid was introduced into the deletion strain and tested for the ability to conjugate into E. coli (Table 2). The mutant showed a 50-fold improvement in conjugation efficiency compared to that of the wild type, indicating that the tra region is not required for plasmid transfer but possibly interacts with plasmids in a manner that interferes with their transfer by other elements. Taken together, these data indicate the presence of a functioning DNA conjugation system in strains A1A7-28, 33277, 381, MP4-504, and 49417 that is genetically and behaviorally distinct from the tra sequences found in W83.
TABLE 2.
Mobilization of plasmids from P. gingivalis to E. coli DH5α
| Strain | Plasmid | Conjugation efficiency |
|---|---|---|
| W83 | pFD340 | <10−9 |
| pT-COW | <10−9 | |
| W50 | pT-COW | <10−9 |
| 5083 | pFD340 | <10−9 |
| pT-COW | <10−9 | |
| 49417 | pT-COW | 1.4 × 10−5 |
| 33277 | pFD340 | 9.0 × 10−9 |
| pT-COW | 3.4 × 10−6 | |
| 381 | pFD340 | 1.6 × 10−8 |
| pT-COW | 5.6 × 10−5 | |
| A1A7-28 | pT-COW | 3.8 × 10−7 |
| A1A7-28 ΔTraIM | pT-COW | 1.9 × 10−6 |
| MP4-504 | pT-COW | 1.4 × 10−5 |
Chromosomal DNA transfer.
Although the W83 tra genes were not able to conjugate plasmids pT-COW and pFD340, it is conceivable that these plasmids do not represent a functional substrate for all P. gingivalis conjugation systems. Native plasmids have not been detected in P. gingivalis, so the only potential DNA substrates for a conjugation system are integrated transposable elements or the chromosomal DNA itself. In Bacteroides spp., conjugative transposons are normally integrated into the host chromosome, from which they are able to excise, circularize, and conjugate into recipient cells (37). These transposons have also been shown to transfer chromosomal DNA by an Hfr-like mechanism (39). To determine if P. gingivalis is capable of transferring chromosomal DNA, a second conjugation assay was designed. For the chromosomal transfer assay, we utilized preexisting P. gingivalis strains 33277 and W83 containing ermF integrated at genetic locus PG0653 as well as strains with tetQ integrated at locus PG1170. Loci PG0653 and PG1170 both encode SerB phosphoserine phosphatases, and the PG0653 locus is important for P. gingivalis survival in gingival epithelial cells (38). These loci do not encode proteins predicted to be involved in DNA transfer mechanisms. For our initial trial, matings were performed between strains W83 PG0653::ermF (W83Em) and 33277 PG1170::tetQ (33277Tc), as well as between W83 PG1170::tetQ (W83Tc) and 33277 PG0653::ermF (33277Em). P. gingivalis transconjugants containing both resistance markers were obtained at an efficiency of 1 × 10−3 for both matings, a significantly higher rate than the rates from the plasmid transfer assays (Table 3). As these matings were done between different strains, it was not clear if both strains were conjugating chromosomal DNA or if one strain was acting as a donor and the other as a recipient. To clarify this point, and to assess the requirement for tra genes in the process, a second set of matings was performed between isogenic strains: W83Em was mated with W83Tc and 33277Em was mated with 33277Tc, giving mating efficiencies of 7.6 × 10−5 and 7.3 × 10−4, respectively. Thus, in contrast to the plasmid assay system, both strains are able to transfer chromosomal DNA. Moreover, as strain 33277 can transfer DNA, the presence of tra homologs is not necessary for the conjugation process. It is important to note that the resistance markers ermF and tetQ are not in regions of the chromosome associated with any mobile genetic elements; thus, their transfer must be due to an Hfr-like event. Intriguingly, the sum of these mating efficiencies (8 × 10−4) is significantly lower than the mating efficiency between different strains (P < 0.05). To confirm that the transfer of chromosomal resistance markers occurs by conjugation rather than by transfection or transduction, selected strains were incubated for 24 h with either purified chromosomal DNA or culture supernatant from filtered late-log-phase bacteria (Table 4). No DNA transfer was detected for either control condition, confirming that DNA transfer is occurring by a conjugation-like mechanism.
TABLE 3.
Mobilization of chromosomal DNA between P. gingivalis strains 33277 and W83
| Strain 1 | Chromosomal allele | Strain 2 | Chromosomal allele | DNA transfer efficiency |
|---|---|---|---|---|
| W83 | PG0653::ermF | 33277 | PG1170::tetQ | 1.0 × 10−3 |
| W83 | PG1170::tetQ | 33277 | PG0653::ermF | 1.0 × 10−3 |
| W83 | PG0653::ermF | W83 | PG1170::tetQ | 7.6 × 10−4 |
| 33277 | PG0653::ermF | 33277 | PG1170::tetQ | 7.3 × 10−4 |
TABLE 4.
Controls for mobilization of chromosomal DNA between P. gingivalis strains
| Strain 1 | Chromosomal allele | Strain and DNA source | Chromosomal allele | DNA transfer efficiency |
|---|---|---|---|---|
| W83 | PG0653::ermF | 33277 genomic DNA | PG1170::tetQ | <10−8 |
| 33277 | PG1170::tetQ | W83 genomic DNA | PG0653::ermF | <10−8 |
| W83 | PG0653::ermF | 33277 culture supernatant | PG1170::tetQ | <10−8 |
| 33277 | PG1170::tetQ | W83 culture supernatant | PG0653::ermF | <10−8 |
Genomic analysis of chimeric strains.
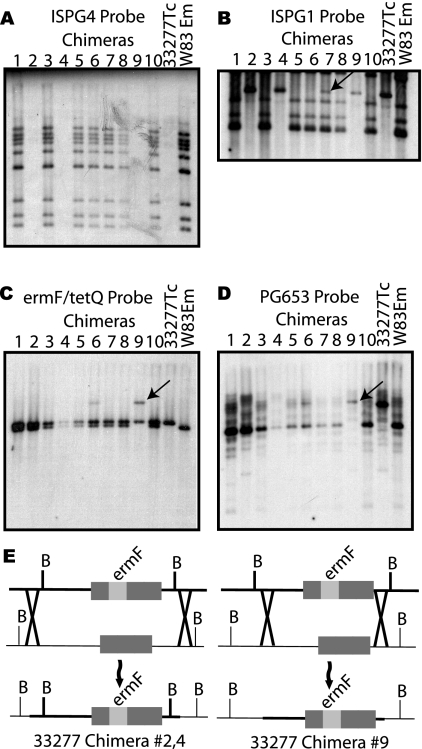
In order to produce Tcr Emr P. gingivalis transconjugants, one strain must be able to accept donated chromosomal DNA from a mating partner and integrate that DNA into its own chromosome. The extent to which chromosomal exchange occurs across the entire genome cannot be determined from the conjugation efficiency calculations. Therefore, we selected 10 transconjugants from a W83Em-33277Tc mating and analyzed these strains by Southern hybridization to determine the extent of genome chimerization. Chromosomal DNA digests were probed with ISPG4, an insertion sequence that has nine copies in W83 and none in 33277 (6). As shown in Fig. 2A, chimeras 1, 3, 5 to 8, and 10 are clearly distinguishable as W83 descendants, with no changes in the ISPG4 restriction profile. Chimeras 2, 4, and 9 are derived from strain 33277 and have no ISPG4 bands. We also probed with ISPG1, which has 22 copies in W83, and found one change between W83 and 33277, in which chimera 7 gained an additional ISPG4 band corresponding to an identically sized band in 33277 (Fig. 2B). To confirm the presence of the antibiotic markers in these chimeras, the DNA blots were simultaneously probed with ermF and tetQ (Fig. 2C). All chimeras had two bands, but chimera 9 had an ermF band that was larger in size than that in the W83 donor. The ermF marker is located in locus PG0653, which has different restriction profiles in 33277 (12.5 kb) and W83 (5.6 kb) when digested with BamHI. The chimera blot was stripped and reprobed with the PG0653 gene, which showed the ermF fragment in chimera 9 to be the same size as the PG0653 band in 33277, in contrast to those in the remainder of the chimeras, in which the ermF bands were the same size as the W83 PG0653 band (Fig. 2C). Chimeras 2, 4, and 9 are 33277 strains by ISPG4 profile and thus have acquired the ermF cassette during conjugation from W83. In the case of chimeras 2 and 4, the DNA fragment that was recombined into the 33277 genome was large enough to contain the BamHI restriction sites from the W83 donor (minimum, 5.6 kb), thus producing a restriction fragment identical in size to the donating parent (Fig. 2E). In chimera 9, the BamHI restriction sites from the donor W83 strain are not present, indicating that a smaller DNA fragment was assimilated into the genome to create this chimera. This fragment would have to be greater than 1.1 kb in size to transfer a functioning ermF gene cassette but smaller than 5.6 kb to maintain the recipient strain 33277 BamHI restriction sites. From these results, we see that widespread genome swapping is not occurring during interstrain conjugation but that the exchange of isolated regions of DNA large enough to swap alleles (such as ISPG1 and PG0653) do occur. To determine if any changes were occurring in alleles associated with pathogenicity, we utilized PCR primers to specifically amplify fimA allele 1 (1, 25) and ragB alleles 4 and 1 (12), specific for strains 33277 and W83, respectively. Each of the 10 chimeras had the correct allele for its parental type (data not shown).
FIG. 2.
Southern blot analysis of chimera strains. All blots are BamHI digests of chimera chromosomal DNA. (A) ISPG4 probe of chimeras to identify 33277- or W83-derived strains. (B) ISPG1 probe of chimera DNA. The arrow indicates the additional ISPG1 band acquired by the W83-derived chimera. (C) Location of tetQ and ermF markers in chimera genomes. The arrow indicates the ermF band in chimera 9, which shows a unique restriction profile compared to the donating parent, W83Em. (D) A PG0653 probe of the same blot as that in panel C. The PG0653 band in chimera 9 (indicated with an arrow) has the same BamHI restriction sites as the 33277Tc strain, with an additional 1.1 kb due to the presence of the ermF cassette from the donating parent, W83Em. (E) Homologous DNA recombination events that result in chimeras 2, 4, and 9. These chimeras result from the W83Em donation of ermF to 33277Tc. Bold lines represent the donated DNA, and thin lines represent the chromosomal DNA in the recipient. “B” indicates BamHI restriction sites. The bold X represents putative regions of homologous recombination.
Alterations in biofilm phenotypes in chimera strains.
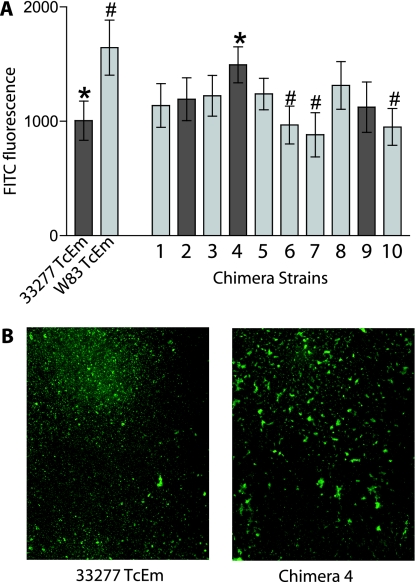
P. gingivalis strains W83 and 33277 are known to have significant genetic differences, including those in genes encoding important surface structures such as FimA, Mfa, RagB, and exopolysaccharide. It has been predicted that more than 50 different genes may vary between the two strains based on microarray analysis (7). This is likely an underestimation of the genetic differences as it does not include genes present in 33277, but not in W83, or point mutations. Therefore, many possible allele swaps may be made between these two strains, which would be difficult to detect without sequencing the 33277 and chimera genomes. Instead, we chose to measure a complex phenotype to look for differences in behavior between parent and chimera strains. Biofilm accretion is a readily measured complex phenotype which can be analyzed both qualitatively and quantitatively. Twenty-four-hour biofilms were grown in 96-well polystyrene plates or 16-well chambered coverslips as described in Materials and Methods and analyzed quantitatively by fluorescence emission or qualitatively by microscopy. The bacteria were added to wells in phosphate-buffered saline to minimize cell growth and to allow us to measure the effects on accretion to the well surface. For total biofilm accumulation, chimeras 2, 4, and 9 were compared to the nonchimeric 33277TcEm strain, while the remaining chimeras were compared to the nonchimeric W83TcEm. Chimeras 4, 6, 7, and 10 were statistically different from the nonchimeric strains, with chimera 4 showing an increase in biofilm accumulation compared to 33277TcEm, and chimeras 6, 7, and 10 showing a decrease compared to W83TcEm (Fig. 3A). Microscopic examination of the biofilms revealed that the increase in chimera 4 biofilm accumulation was due to the formation of microcolonies that were larger than those of the nonchimeric strain, implying that this strain is more self-aggregating than the parent (Fig. 3B). The biofilms of chimeras 6, 7, and 10 had overall appearances that were similar to strain W83's biofilm but had accumulated fewer bacterial cells (data not shown.) These results show that the exchange of genetic information between individual strains leads to measurable differences in the phenotypic behavior of P. gingivalis.
FIG. 3.
Quantitative and qualitative biofilm assays. (A) Quantitative analysis of biofilm accretion by P. gingivalis strains and chimeras. Chimeric strains 2, 4, and 9 (dark bars) were compared to strain 33277TcEm for statistical analysis by t test. All other chimeras (light bars) were compared to W83TcEm. Results represent three independent experiments, each done in triplicate (n = 9). Symbols * and # represent P values of <0.05. (B) Fluorescent microscopy images of 24-h biofilm formation by nonchimeric strain 33277TcEm and chimera 4. Magnification, ×40.
DISCUSSION
In periodontal disease, high numbers of P. gingivalis are found in the subgingival crevice and may compose up to 7% of the bacteria found in plaque samples (22). Individuals are most often colonized by one strain of P. gingivalis, although it is possible to be transiently coinfected by two or three strains simultaneously (21, 40). The cocolonization of strains in the same subgingival crevice could allow for DNA exchange, producing a pool of chimeric offspring that could then undergo fitness selection. Repeated cocolonization events could allow for continuing rounds of fitness improvements, and over time the numbers of bacteria present in the niche would be predicted to increase in parallel with fitness. DNA exchange between strains could therefore be contributing to the clinical development of periodontal disease, which is characterized by increasing levels of gram-negative pathogens in the subgingival crevice.
Each human mouth represents a unique ecosystem with a variety of fitness challenges facing any bacteria attempting to establish a permanent foothold. The host microbiota is highly complex, with an estimated 700 species or more capable of colonizing oral biofilms (28). Any given human host has approximately 150 bacterial species; P. gingivalis strains attempting to colonize the oral biofilm will thus have to cooperate and/or compete with a unique bacterial complement in each host. The host immune response will also present a continuing challenge, and genetic recombination between bacterial strains may facilitate antigenic variation, allowing the bacteria to evade the developing antibody response. Over time, as changes occur in the aging host's oral cavity, DNA exchange may also allow fine-tuning of bacterial fitness and contribute to persistence in the host oral niche.
In this study we show that several common laboratory strains and low-passage clinical isolates of P. gingivalis are able to transfer plasmid DNA, chromosomal DNA, or both. Based on both functional and genetic screens, P. gingivalis strains as a group are predicted to contain multiple conjugative elements. While strains W83, W50, 49417, and 5083 possessed tra homologs, these were not functional in the plasmid transfer assay adopted here. Lack of functionality could be the result of the absence of a traP gene, which is absent in the W83 sequence but has been shown to be required for plasmid transfer in Bacteroides conjugative systems (3). The element(s) present in the non-tra-hybridizing strains appears to be capable of plasmid as well as chromosomal DNA transfer. Whether there is one element responsible for the transfer of both plasmid and chromosomal DNA or there is more than one conjugative element with specialized substrates cannot be determined from these studies. The element(s) found in the non-tra-hybridizing strains might be similar to conjugative transposons in Bacteroides but is predicted to have less than 75% DNA homology to the W83 element based on a lack of high-stringency hybridization to our W83-derived Southern probes. Thus, there may be a variety of elements present in the P. gingivalis meta-genome that are capable of directing DNA transfer between strains and possibly even between species in the human flora.
Chromosomal DNA exchange between P. gingivalis strains does not appear to be extensive based on the ISPG1 and ISPG4 Southern blots, although at least 1.1 kb of ermF was transferred to create the 33277 chimeras and at least 2.7 kb to introduce tetQ into the W83 chimeras. Although we were not able to detect allele exchanges in the fimA or ragB loci by PCR, it is possible that small internal portions of genes are being exchanged, which would be detectable only by DNA sequencing or high-resolution microarray analysis.
Intriguingly, matings between different strains have better transfer efficiencies than transfers between identical strains do. Additionally, we found that chimera colonies from W83-33277 matings appear much faster on selective media (5 to 7 days) than the Tcr Emr strains from W83 and 33277 self-matings (10 to 21 days). This is counterintuitive, as identical-strain matings should be more efficient since they are not limited by restriction modification or sequence variation between donor and recipient genomes. It appears that P. gingivalis is able to detect the presence of interspecies mating pair formation or strain-specific DNA and is able to regulate its physiology to favor the uptake of this novel DNA into the genome.
These studies illustrate a previously unknown aspect of the lifestyle of an important human oral pathogen. Although the P. gingivalis population was known to be genetically diverse, the mechanisms by which this diversity was generated were unclear. Here we demonstrate that chromosomal DNA transfer between strains is a high-frequency event in vitro and contributes to important phenotypic changes in the resulting chimeric offspring. The analysis of these conjugation systems at the molecular level will provide further insight into the importance of this behavior to these oral anaerobes.
Acknowledgments
Support for this research was provided by grant DE016562 from the National Institute of Dental and Craniofacial Research to G.D.T.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664-1668. [DOI] [PubMed] [Google Scholar]
- 2.Asano, H., K. Ishihara, T. Nakagawa, S. Yamada, and K. Okuda. 2003. Relationship between transmission of Porphyromonas gingivalis and fimA type in spouses. J. Periodontol. 74:1355-1360. [DOI] [PubMed] [Google Scholar]
- 3.Bacic, M., A. C. Parker, J. Stagg, H. P. Whitley, W. G. Wells, L. A. Jacob, and C. J. Smith. 2005. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J. Bacteriol. 187:2858-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainbridge, B. W., R. C. Page, and R. P. Darveau. 1997. Serum antibodies to Porphyromonas gingivalis block the prostaglandin E2 response to lipopolysaccharide by mononuclear cells. Infect. Immun. 65:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 6.Califano, J. V., T. Arimoto, and T. Kitten. 2003. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J. Periodontal Res. 38:411-416. [DOI] [PubMed] [Google Scholar]
- 7.Chen, T., Y. Hosogi, K. Nishikawa, K. Abbey, R. D. Fleischmann, J. Walling, and M. J. Duncan. 2004. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 186:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan, M. J. 2003. Genomics of oral bacteria. Crit. Rev. Oral Biol. Med. 14:175-187. [DOI] [PubMed] [Google Scholar]
- 10.Enersen, M., I. Olsen, A. J. van Winkelhoff, and D. A. Caugant. 2006. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J. Clin. Microbiol. 44:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, R. G., J. B. Russell, D. B. Wilson, G.-R. Wang, and N. B. Shoemaker. 1996. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed β-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl. Environ. Microbiol. 62:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, L. M. C., S. C. Fawell, X. Shi, M.-C. Faray-Kele, J. Aduse-Opoku, R. A. Whiley, and M. A. Curtis. 2005. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect. Immun. 73:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton, H. L., and J. P. Dillard. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376-385. [DOI] [PubMed] [Google Scholar]
- 14.Jolley, K. A., D. J. Wilson, P. Kriz, G. McVean, and M. C. Maiden. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22:562-569. [DOI] [PubMed] [Google Scholar]
- 15.Koehler, A., H. Karch, T. Beikler, T. F. Flemmig, S. Suerbaum, and H. Schmidt. 2003. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology 149:2407-2415. [DOI] [PubMed] [Google Scholar]
- 16.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kilic, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60:121-139. [DOI] [PubMed] [Google Scholar]
- 17.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 20.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. 1999. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J. Clin. Microbiol. 37:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiden, M. C. 1993. Population genetics of a transformable bacterium: the influence of horizontal genetic exchange on the biology of Neisseria meningitidis. FEMS Microbiol. Lett. 112:243-250. [DOI] [PubMed] [Google Scholar]
- 24.Ménard, C., and C. Mouton. 1995. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Missailidis, C. G., J. E. Umeda, C. Ota-Tsuzuki, D. Anzai, and M. P. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 19:224-229. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozmeriç, N., N. R. Preus, and I. Olsen. 2000. Genetic diversity of Porphyromonas gingivalis and its possible importance to pathogenicity. Acta Odontol Scand. 58:183-187. [DOI] [PubMed] [Google Scholar]
- 28.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 29.Robinson, K., M. F. Loughlin, R. Potter, and P. J. Jenks. 2005. Host adaptation and immune modulation are mediated by homologous recombination in Helicobacter pylori. J. Infect. Dis. 191:579-587. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., V. Priefer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 33.Smeets, L. C., N. L. A. Arents, A. A. van Zwet, C. M. J. E. Vandenbroucke-Grauls, T. Verboom, W. Bitter, and J. G. Kusters. 2003. Molecular patchwork: chromosomal recombination between two Helicobacter pylori strains during natural colonization. Infect. Immun. 71:2907-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, C. J. 1995. Genetic transformation of Bacteroides spp. using electroporation. Methods Mol. Biol. 47:161-169. [DOI] [PubMed] [Google Scholar]
- 35.Smith, C. J., M. B. Rogers, and M. L. McKee. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141-154. [DOI] [PubMed] [Google Scholar]
- 36.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211-222. [DOI] [PubMed] [Google Scholar]
- 37.Smith, C. J., G. D. Tribble, and D. P. Bayley. 1998. Genetic elements of Bacteroides species: a moving story. Plasmid 40:12-29. [DOI] [PubMed] [Google Scholar]
- 38.Tribble, G. D., S. Mao, C. E. James, and R. J. Lamont. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. USA 103:11027-11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittle, G., N. Hamburger, N. B. Shoemaker, and A. A. Salyers. 2006. A Bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J. Bacteriol. 188:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. J., S. Yasui, F. Yoshimura, and I. Ishikawa. 1995. Multiple restriction fragment length polymorphism genotypes of Porphyromonas gingivalis in single periodontal pockets. Oral Microbiol. Immunol. 10:125-128. [DOI] [PubMed] [Google Scholar]