Abstract
Comparison of nadA and nadB in 14 Shigella strains and enteroinvasive Escherichia coli versus E. coli showed that at least one locus is altered in all strains. These observations explain the characteristic nicotinic acid auxotrophy of Shigella organisms and are consistent with the previously identified antivirulence nature of these genes for these pathogens.
The evolution of bacterial pathogens from their commensal ancestors, allowing them to colonize a new ecological niche, is believed to occur by the acquisition of genes via horizontal transfer (6). The presence of virulence genes in pathogenicity islands reflects this form of gene acquisition (12). This first step of evolution is followed by progressive adaptation of the bacteria to their new niche (the host) by mutation and selection for improved fitness (23). An important element of this process of pathoadaptive evolution is the selection of “black holes” in pathogen genomes, that is, the inactivation or loss of genes that are incompatible with, and even antagonistic to, the new pathogenic lifestyle (14). These incompatible genes, which we define as antivirulence loci (AVL), are present in the genome of a nonpathogenic ancestor but absent or inactive in the pathogen because expression of AVL is detrimental to the expression of some virulence phenotypes.
Bacteria of the genus Shigella are the causative agents of bacillary dysentery. We previously demonstrated the contribution of black holes in the evolution of Shigella from the nonpathogenic gut commensal Escherichia coli (15). The cadA gene encoding lysine decarboxylase in E. coli K-12 was shown to be part of a large region deleted or altered in all Shigella spp., and its expression proved to be incompatible with Shigella virulence (3, 16).
The genetic similarities between Shigella and E. coli are strong enough to justify grouping them in the same genus. Shigella spp. and enteroinvasive E. coli (EIEC) are even more closely related, with the latter causing a disease very similar to dysentery. A recent study focusing both on housekeeping and virulence genes classified both species as a single pathovar of E. coli (9). However, there are several well-known auxotrophic requirements of Shigella that are not found among most isolates of E. coli. Most Shigella strains require nicotinic acid supplementation for growth on minimal medium (1). The nicotinic acid requirement of Shigella flexneri 2a strain 2457T is due to mutations in two unlinked loci, nadA and nadB (5), that encode enzymes in the l-aspartate-dihydroxyacetone pathway leading to de novo synthesis of NAD. In S. flexneri 5a strain M90T, Mantis et al. showed that nadB inactivation is responsible for this phenotype since nicotinic acid prototrophy can be restored by transformation with the cloned nadB gene from Salmonella enterica serovar Typhimurium (13). The nadA gene encodes the quinolinate synthetase A, while nadB encodes the l-aspartate oxydase (or quinolinate synthetase B). Both function together as a multienzyme complex catalyzing the oxidation and condensation of l-aspartate to quinolinate (QUIN). QUIN is subsequently ribosylated to nicotinic acid mononucleotide and converted to NAD by other enzymes in the pathway. In the absence of functional nadA and/or nadB, exogenous nicotinic acid can be used instead of QUIN to produce nicotinic acid mononucleotide, thus bypassing the need for nadA and nadB functionality and synthesis of QUIN.
We recently reported that QUIN is a strong and specific inhibitor of several virulence phenotypes of Shigella (19). We also found that complementation of strain M90T with functional copies of nadA and nadB from E. coli K-12, while conferring nicotinic acid prototrophy to the strain, strongly impaired its virulence. These results define nadA and nadB as AVL in Shigella. The model of pathoadaptive evolution predicts that strong selective pressure against expression of AVL will lead to their inactivation by mutation. In this study, we wanted to test this prediction for the Shigella AVL nadA and nadB by analyzing these loci in other strains of Shigella and EIEC.
We first analyzed the nadA and nadB loci in strain M90T. Table 1 shows the overlapping primers used for amplification and sequencing of both loci. PCR amplification for sequencing, cloning, and plasmid screening purposes utilized the Taq DNA polymerase (QIAGEN). For sequencing PCR-generated products, samples were prepared with an ABI Prism Dye Terminator Cycle Sequencing Core Kit (Applied Biosystems) and analyzed using an ABI Prism 377 DNA sequencer (Applied Biosystems). Clone Manager (SciEd software) was used to perform DNA translation to protein and comparative alignments. We found that nadA and nadB sequences from M90T were identical to those from the published sequence for S. flexneri 5b strain 8401 (18). Both genes contained point mutations that result in 2- and 8-amino-acid substitutions compared to the functional NadA and NadB of E. coli K-12, respectively (Table 2). In addition, the nadB gene of M90T contained a single base change that created a stop codon at amino acid 354, resulting in truncation of the C-terminal third of the nadB product. Gemski et al. originally reported that nadA and nadB of S. flexneri 2a strain 2457T are both mutated (5). Mantis et al. reported defects limited to nadB in M90T, and since the nicotinic acid auxotrophy of M90T could be satisfied by introduction of the nadB gene from S. enterica serovar Typhimurium, they suggested that its nadA gene was functional (13). In order to confirm these latter results, the nadAPF/nadAM and nadBPromF/nadBM primers (Table 1) were used to amplify the full nadA and nadB loci and their promoters from M90T. The fragments obtained were cloned into pGEM-T (Promega) and tested for their ability to complement nadA or nadB mutants of E. coli K-12 on M9 glucose minimal medium (MM). Nicotinic acid (Sigma) at a concentration of 10 μg/ml was used as a supplement in control plates. Table 3 shows that neither clone from M90T was able to complement the respective mutant strains of E. coli K-12. In reciprocal experiments, the wild-type nadA and nadB genes of E. coli K-12 were cloned into pBluescript (Stratagene) and pGEM-T, respectively, and the constructs obtained were transformed into M90T, generating strains BS830 and BS831, respectively. Neither transformant showed nicotinic acid independence. However, when both genes cloned together on pBluescript were transformed into M90T, the resulting strain (BS813) grew on MM without nicotinic acid (Table 3). These data extend the initial work of Gemski et al. (5) to another S. flexneri serotype and suggest that, contrary to the observations of Mantis et al. (13), the nicotinic acid auxotrophy of M90T is due to loss of function of both nadA and nadB. Bergthorsson and Roth (2) identified an A111V substitution in NadA as responsible for the nicotinic acid auxotrophy of the cattle pathogen S. enterica serovar Dublin, indicating that this residue is essential for the functionality of the protein. The same change was found in M90T (Table 2), supporting our conclusion that NadA is not functional in this strain. Regarding NadB, while a nonsense mutation alone usually leads to a mutant phenotype, one or more of the other amino acid changes in this protein must also lead to a null phenotype since we were unable to select spontaneous nadB+ revertants of BS830 on MM. The reciprocal experiment using BS831 failed to yield spontaneous nadA+ revertants (Table 3). These results suggest that multiple point mutations altered essential amino acid residues in both loci, emphasizing a strong selective pressure for their loss of functionality.
TABLE 1.
Primers used in this study
| Gene | Primer name (direction) | Primer sequence (5′ to 3′) | Positiona |
|---|---|---|---|
| nadA | nadAPF (sense)b | CAAGCAACTCTATGTCGGTGG | −184 |
| nadAPR (antisense) | CTTTGCACCGAAGCGCGCCAT | +231 | |
| nadA2R (antisense) | GTCTTAAATTCATCATGCAC | +623 | |
| nadA2 (sense) | CCACCAGTCAACTGATCGCT | +725 | |
| nadAP (sense) | AAAGAACGTAATGCGGTGATGG | +112 | |
| nadAM (antisense)b | TGGCAAGGCCAATACACAGC | +1202 | |
| nadB | nadBPromF (sense)b | CAAAGGGTTAGAGTGTCTCG | −332 |
| nadBPF (sense) | CTGCCTGAAGAGTAACCCAAC | −191 | |
| nadBPR (antisense) | ATCAAACACGGCGGCAATACC | +174 | |
| nadB2 (sense) | GAAACCTGCCACGCAAAAGC | +577 | |
| nadB3R (antisense) | CACGCCATAGCAATGCCAT | +686 | |
| nadB2R (antisense) | CGCCCATGATCATCAACCA | +1091 | |
| nadB3F (sense) | AGCCGCGTTGAGAACCCTGAC | +1273 | |
| nadBP (sense) | CAAAGAAAATGAATACTCTCCCTGA | −8 | |
| nadBM (antisense)b | CGTGGGCCAGACCAGAACTATTCC | +1676 |
Base numbering relative to ATG.
Primer used for cloning.
TABLE 2.
NadA/B sequence changes in various Shigella and EIEC strains compared to E. coli K-12
| Strain | Serotype | Clustera | Changes in NadAb | Accession no.c | Changes in NadBb | Accession no.c | Reference or sourced |
|---|---|---|---|---|---|---|---|
| Shigella strains | |||||||
| S. boydii BS511 | 14 | 1 | Q271* | EF473660 | (I108V, E141Q, T142S, L149Q), A226D, R234Q, (Y412D, D415G, I416V), T468N | EF473669 | CDC |
| S. boydii BS512 | 18 | 1 | A148V | NZ_AAKA00000000 | IS600 between aa 178 and 179 | NZ_AAKA00000000 | CDC |
| S. boydii 227 | 4 | 1 | G304D | NC_007613 | IS600 between aa 178 and 179 | NC_007613 | 25 |
| S. dysenteriae 1012 | 4 | 1 | R35H, V45A, K77R, R134H, 1-bp insertion frameshift at position 442 | NZ_AAMJ00000000 | R138K, (V167I), W185*, (Y412D, D415G, I416V), T468N | NZ_AAMJ00000000 | 24 |
| S. boydii BS686 | 17 | 2 | R35H, V45A, R134H, 1-bp insertion frameshift at position 442 | EF473663 | A19T, V67E, V102G, E114Q, D134Y, R138K, (V167I, Y412D, D415G, I416V), T457K, T468N | EF473672 | CDC |
| S. dysenteriae BS507 | 2 | 2 | R35H, V45A, K77R, R134H, 1-bp insertion frameshift at position 442 | EF473658 | A75V, R138K, (V167I), W185*, (Y412D, D415G, I416V), T468N | EF473667 | CDC |
| S. flexneri 2457T | 2a | 3 | A111V, C128Y, T252A, Q271R, G304D | AF403415 | R80H, Q95P, (I108V, E141Q, T142S, L149Q), C354*, (D415G, I416V) | AF403416 | 4 |
| S. flexneri BS510 | 3a | 3 | None | EF473659 | A73S, (I108V, E141Q, T142S, L149Q), C354*, (D415G, I416V) | EF473668 | CDC |
| S. flexneri M90T | 5a | 3 | A111V, T252A | EF473666 | R80H, Q95P, (I108V E141Q, T142S, L149Q), C354*, (D415G, I416V) | EF473657 | 21 |
| S. dysenteriae 197 | 1 | Outlier | R134H, (G191A), P219L, R257W | NC_007606 | (V167I), D218N, (D415G, I416V) | NC_007606 | 25 |
| S. dysenteriae BS681 | 8 | Outlier | none | EF473662 | G44V, (I108V, E141Q, L149Q), V180I, (Y412D, D415G, I416V) | EF473671 | CDC |
| S. sonnei BS513 | NAe | Outlier | IS21 between aa 292 and 293 | EF473661 | IS600 in codon 233 | EF473670 | CDC |
| E. coli (EIEC) strains | |||||||
| EDL1284 | O124 | NA | G198S, V260G | EF473665 | Portion of IS600 after aa 52; deletion aa 53 to 192 | EF473674 | 7 |
| E. coli (EIEC) strain 1 | O136 | NA | G198S | EF473664 | IS600 between aa 52 and 53 | EF473673 | L. Trabulsi |
Classification according to Pupo et al. (20).
Changes are based on the comparison of each sequence to the translated E. coli K-12 sequences for nadA (gene identifier 945351) and nadB (gene identifier 947049). Nonsense mutations in DNA leading to the replacement of an amino acid by a stop codon are represented by an asterisk. Changes in boldface correspond to amino acids conserved among E. coli K-12 and four other bacterial species (Fig. 1). Parentheses indicate polymorphic changes also found in other E. coli strains. Total sequence length is 347 amino acids for NadA and 540 amino acids for NadB. aa, amino acids.
The National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) was used to collect the available sequence data. NZ and NC accession numbers correspond to the whole genome or whole genome project. The remaining accession numbers correspond to the specific gene sequences determined for this study. The analysis was performed in October 2006.
Centers for Disease Control and Prevention, Nancy Strockbine.
NA, not applicable.
TABLE 3.
Results of nadA/B complementation experiments
| Straina | Relevant characteristicsb | Growth on MM |
|---|---|---|
| E. coli strains | ||
| K-12 MC4100 | nadAECnadBEC | + |
| K-12 CAG12147 | nadA57::Tn10 | − |
| K-12 CAG18480 | nadB51::Tn10 | − |
| ATM876 | CAG12147 + pBluescript/nadAEC | + |
| ATM839 | CAG12147 + pGEM-T/nadASF | − |
| ATM877 | CAG18480 + pGEM-T/nadBEC | + |
| ATM878 | CAG18480 + pGEM-T/nadBSF | − |
| S. flexneri 5a strains | ||
| M90T | nadASFnadBSF | − |
| BS830 | M90T + pBluescript/nadAEC | − |
| BS831 | M90T + pGEM-T/nadBEC | − |
| BS813 | M90T + pBluescript/nadAEC-nadBEC | + |
We next compiled the available nadA and nadB sequences of Shigella species from the databases and complemented these data by sequencing nadA and nadB in representative isolates of each species and in two EIEC strains that are also nicotinic acid auxotrophs. All the strains studied displayed changes in at least one of the loci compared to E. coli K-12 (Table 2). The EIEC strains, two of the Shigella boydii strains, and a Shigella sonnei strain showed a significant disruption of nadB by insertion of insertion sequence (IS) elements. EIEC O124 contained an insertion of a portion of IS600 after codon 52, and the gene also showed a large deletion of the coding region for amino acids 53 to 192. Interestingly, EIEC O136 also had an IS600 insertion at codon 52, but neither the IS element nor the surrounding open reading frame showed any rearrangements. Both strains also displayed the same amino acid change in NadA at codon 198 compared to E. coli K-12, with an additional amino acid change in EIEC O124. These results strongly suggest that both strains are related and that they share a common ancestor, even though they correspond to different serotypes. The nadB sequences of S. boydii serotypes 4 and 18 were both disrupted by insertion of IS600 after codon 178. This result also suggests a common ancestor for these strains. Finally, both the S. sonnei nadA and nadB loci showed disruption by IS21 and IS600, respectively. Their sequences are identical to the nadA and nadB loci of S. sonnei 46, the sequenced strain from the databases. These results are consistent with the well-known clonal nature of S. sonnei strains (8). In all of these strains, it is very likely that the IS-related disruptions of nadA and/or nadB loci are responsible for the nicotinic acid auxotrophy by preventing the production of functional proteins.
Numerous amino acid changes were found among all the Shigella and EIEC strains studied compared to E. coli K-12. In order to determine if some of these changes were due to polymorphisms of the nadA and nadB loci, we compared the deduced NadA and NadB protein sequences from the 17 DNA sequences of E. coli available in the databases. This analysis revealed 12 and 33 polymorphic positions in NadA and NadB, respectively. Several of these changes were also found in some of our strains and are indicated in parentheses in Table 2. However, all the Shigella strains studied were found to contain additional and specific changes in at least one locus compared to E. coli. Several strains had nonsense mutations or frameshifts, which likely result in production of nonfunctional protein(s). Overall, among the 14 strains studied, only two (Shigella dysenteriae 1 and 8) contain no more than a few specific amino acid substitutions compared to E. coli K-12 and are still likely to produce full-sized NadA and NadB proteins. To determine whether these mutations might be in essential residues, we performed alignments of NadA and NadB sequences for E. coli K-12, S. enterica CT18, Yersinia enterocolitica 8081, Vibrio cholerae N16961, and Pseudomonas aeruginosa PAO1, all of which contain functional alleles of nadA and nadB (Fig. 1). The P219L substitution in NadA found in S. dysenteriae 1, as well as the D218N and G44V substitutions in NadB found in S. dysenteriae 1 and 8, respectively, represent alterations in residues that are conserved among the five different species. This observation strongly suggests that these residues are important for the function of the protein(s). Therefore, it is likely that some or all of these amino acid changes in the S. dysenteriae loci lead to loss of protein function. Additionally, among the 12 other Shigella and EIEC strains studied, eight contained one or more changes in conserved amino acids in NadA and/or NadB (Table 2; Fig. 1). This observation provides further evidence of a strong selective pressure toward the loss of function of both proteins in Shigella species. Finally, only two strains, S. flexneri 3a and S. dysenteriae 8, displayed no changes in NadA compared to E. coli. In both strains, the ability to grow on MM was restored by the reintroduction of a functional E. coli copy of nadB, confirming that mutations in nadB were responsible for their nicotinic acid auxotrophy (data not shown).
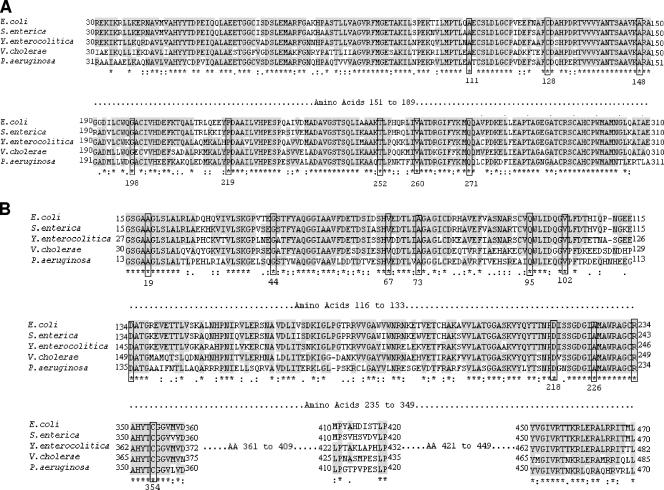
FIG. 1.
Partial alignments of NadA and NadB sequences from various bacterial species. The National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) was used to collect the protein sequence data. The accession number is indicated for each sequence. (A) Partial alignment of NadA from E. coli K-12 (NP_415271), S. enterica CT18 (NP_455307), Y. enterocolitica 8081 (YP_001007117), V. cholerae N16961 (NP_231467), and P. aeruginosa PAO1 (NP_249695). (B) Partial alignment of NadB from E. coli K-12 (NP_417069), S. enterica CT18 (NP_457117), Y. enterocolitica 8081 (YP_001005342), V. cholerae N16961 (NP_232098), and P. aeruginosa PAO1 (NP_249452). Both alignments were obtained via ClustalW (http://www.ebi.ac.uk/clustalw/) and edited using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Conserved amino acids among three or more of the five sequences are shaded. An asterisk indicates a conserved amino acid among all five species, a colon indicates conservative amino acid substitutions, and a period indicates semiconservative substitutions. Boxes show conserved amino acids that were found to be mutated in one or more of the Shigella and EIEC strains studied, with E. coli numbering indicated underneath. The analysis was performed in February 2007.
The phylogenetic relationships of Shigella strains to each other and to E. coli strains have been analyzed by sequencing two virulence plasmid genes and eight housekeeping genes in four separate regions around the chromosome (9-11, 20). These data show that the majority of Shigella strains fall into three main clusters within E. coli and that EIEC and S. sonnei strains, as well as S. dysenteriae serotypes 1, 8, and 10, lie outside these three clusters. Our results show a good correlation of nadA and nadB mutations with these clusters (Table 2). Only a few of the amino acid substitutions were common to strains from several clusters and even outliers (i.e., R35H, V45A, K77R, R134H, and G304D in NadA and R138K and T468N in NadB) while the majority of the amino acid substitutions were either unique to strains from a single cluster (i.e., A111V and T252A in NadA and R80H, Q95P, and C354* in NadB for cluster 3) or even unique to one single strain. In summary, most of the substitutions we found were likely selected for because they affected essential residues of the proteins (Table 2), and very few may represent polymorphisms due to their position in a particular “hot spot” for mutation within the gene. Overall, these results support the idea that a strong selective pressure toward the loss of nadA and nadB function occurred in the evolution of Shigella species.
Inactivation of either nadA or nadB would satisfy this selective pressure to block the synthesis of QUIN, but mutations would tend to subsequently accumulate in both genes because of the absence of selective pressure to maintain gene function. It is difficult to definitely distinguish which gene is the preferred target for inactivation. However, several of our observations suggest that mutation of NadB was the first event in the evolution of Shigella species toward nicotinic acid auxotrophy. First, two strains, S. dysenteriae 8 and S. flexneri 3a, had no alterations of nadA and produced functional NadA protein. Their defect in NAD synthesis is solely due to alterations in NadB (missense mutations in conserved residues and/or a nonsense mutation that truncates one-third of the protein). Second, in the two EIEC strains, as well as in the two S. boydii serotypes 4 and 18, the nadB gene sustained a major disruption by IS elements while nadA showed only point mutations compared to the E. coli K-12 sequence. The latter could be the result of genetic drift of nadA after inactivation of nadB as a result of the loss of the selective pressure to maintain a functional nadA gene. In general, we also found that the majority of the strains studied (except the S. sonnei strain) show a higher number of modifications in NadB than in NadA. This phenomenon could be explained by the larger size of the nadB locus relative to nadA, which makes it a more frequent target for random mutation during the selection process for nicotinic acid auxotrophs.
In summary, we have found that among 14 strains of Shigella and EIEC, all contained alterations in at least one of the nadA or nadB loci that could explain their nicotinic acid auxotrophy. These observations are consistent with our previous report that the nadA and nadB genes are AVL in Shigella (19). Additionally, the distinct nature of mutations defining the nadA and nadB loci of the four different species of Shigella suggests their acquisition and accumulation after Shigella speciation. This conclusion is in accord with the model that clones of Shigella evolved multiple times from different lines of E. coli ancestor strains (20). Our findings confirm and extend the concept that bacterial pathogen evolution proceeds through convergent evolution toward removal or inactivation of AVL by whatever alteration (IS, deletion, or point mutation) leads to a more virulent phenotype or to improved fitness for survival in the host (14).
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Ahmed, Z. U., M. R. Sarker, and D. A. Sack. 1988. Nutritional requirements of Shigella for growth in a minimal medium. Infect. Immun. 56:1007-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergthorsson, U., and J. R. Roth. 2005. Natural isolates of Salmonella enterica serovar Dublin carry a single nadA missense mutation. J. Bacteriol. 187:400-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day, W. A., Jr., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Formal, S. B., G. J. Dammin, E. H. Labrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemski, P., S. B. Formal, and L. S. Baron. 1971. Identification of two widely separated loci conferring nicotinic acid dependence on wild-type Shigella flexneri 2a. Infect. Immun. 3:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 7.Harris, J. R., I. K. Wachsmuth, B. R. Davis, and M. L. Cohen. 1982. High-molecular-weight plasmid correlates with Escherichia coli enteroinvasiveness. Infect. Immun. 37:1295-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaolis, D. K., R. Lan, and P. R. Reeves. 1994. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J. Clin. Microbiol. 32:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan, R., M. C. Alles, K. Donohoe, M. B. Martinez, and P. R. Reeves. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72:5080-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan, R., B. Lumb, D. Ryan, and P. R. Reeves. 2001. Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli. Infect. Immun. 69:6303-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes. Infect. 4:1125-1132. [DOI] [PubMed] [Google Scholar]
- 12.Lee, C. A. 1996. Pathogenicity islands and the evolution of bacterial pathogens. Infect. Agents Dis. 5:1-7. [PubMed] [Google Scholar]
- 13.Mantis, N. J., and P. J. Sansonetti. 1996. The nadB gene of Salmonella typhimurium complements the nicotinic acid auxotrophy of Shigella flexneri. Mol. Gen. Genet. 252:626-629. [DOI] [PubMed] [Google Scholar]
- 14.Maurelli, A. T. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick, B. A., M. I. Fernandez, A. M. Siber, and A. T. Maurelli. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell Microbiol. 1:143-155. [DOI] [PubMed] [Google Scholar]
- 17.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie, H., F. Yang, X. Zhang, J. Yang, L. Chen, J. Wang, Z. Xiong, J. Peng, L. Sun, J. Dong, Y. Xue, X. Xu, S. Chen, Z. Yao, Y. Shen, and Q. Jin. 2006. Complete genome sequence of Shigella flexneri 5b and comparison with Shigella flexneri 2a. BMC Genomics 7:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prunier, A. L., R. Schuch, R. E. Fernandez, K. L. Mumy, H. Kohler, B. A. McCormick, and A. T. Maurelli. 2007. nadA and nadB of S. flexneri 5a are anti-virulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of Shigella pathogenicity. Microbiology 153:2363-2372. [DOI] [PubMed] [Google Scholar]
- 20.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 24.Talukder, K. A., B. K. Khajanchi, D. K. Dutta, Z. Islam, M. A. Islam, M. S. Iqbal, G. B. Nair, and D. A. Sack. 2005. An unusual cluster of dysentery due to Shigella dysenteriae type 4 in Dhaka, Bangladesh. J. Med. Microbiol. 54:511-513. [DOI] [PubMed] [Google Scholar]
- 25.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]