Abstract
We identify ef1090 (renamed ebpR) and show its importance for the transcriptional regulation of expression of the Enterococcus faecalis pilus operon, ebpABC. An ebpR deletion (ΔebpR) mutant was found to have reduced ebpABC expression with loss of pilus production and a defect in primary adherence with, as a consequence, reduced biofilm formation.
Enterococcus faecalis is a gram-positive bacterium that is part of the normal intestinal flora of most humans but is also a major cause of opportunistic infection (9). A major regulator of virulence in E. faecalis OG1RF is the Fsr system (15). In a previous microarray analysis, we identified several Fsr-regulated genes encoding putative regulators, one of which was ef1090 (2). EF1090 contains two putative helix-turn-helix DNA-binding domains and shares 42% similarity with AcpB from Bacillus anthracis, a member of the AtxA/MgaA family (4). Upstream and in the opposite orientation of ef1090 is the ebpABC operon, which encodes pilus components shown to be important for virulence in the rat endocarditis model and the mouse urinary tract infection model (13, 17).
To test whether the putative EF1090 regulator was involved in the regulation of the pili genes, we first performed semiquantitative reverse transcription-PCR (qRT-PCR) (2). The ebpA, ebpB, and ebpC transcripts were strongly reduced in an ef1090 transposon insertion mutant (5) compared to the parent strain (data not shown). Based on these results, ef1090 was renamed ebpR for its role as an endocarditis- and biofilm-associated pilus regulator. To prevent any effects caused by the transposon insertion from complicating our analysis, we then created an unmarked in-frame ebpR deletion mutant (Table 1) of OG1RF by using the recently published PheS* system (7). The ΔebpR mutant had no growth defects compared to OG1RF in BHI, TSBG (biofilm medium), or BHI-40% horse serum (binding assay medium).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain or plasmid | Relevant characteristics or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | E. coli general cloning host | 16 |
| EC1000 | E. coli cloning host for repA-dependent plasmids; carries constitutive repA allele on chromosome; Kanr | 8 |
| E. faecalis | ||
| OG1RF | E. faecalis Fusr Rifr; p-Cl-Pher | 11 |
| CK111/pCF10-101 | E. faecalis conjugative donor strain; carries constitutive repA allele on chromosome and nontransferable pCF10 derivative; Spcr Tetr | 7 |
| DAGF27 | Tn917 transposon insertion at +581 bp from the ebpRa ATG in OG1RF; Ermr Fusr Rifr | 5 |
| TX5475 | ebpA allelic replacement aph(3′)-IIIa mutant of OG1RF; Fusr Kanr Rifr | 13 |
| TX5460 | ebpB insertion disruption mutant of OG1RF; Fusr Kanr Rifr | 13 |
| TX5448 | ebpC insertion disruption mutant of OG1RF; Fusr Kanr Rifr | 13 |
| TX5514 | ebpR in-frame deletion from bp −5 to +1337 from the ATG of ebpR of OG1RF; Fusr Rifr; p-Cl-Pher | This study |
| TX5584 | TX5514(pMSP3535); Ermr Fusr Rifr; p-Cl-Pher | This study |
| TX5582 | TX5514(pTEX5515); ebpR mutant containing ebpR gene inserted into pMSP3535; Ermr Fusr Rifr; p-Cl-Pher | This study |
| TX5590 | OG1RF(pTEX5586); OG1RF containing the ebpR promoter fused to lacZ; Ermr Fosr Rifr | This study |
| TX5591 | TX5514(pTEX5586); ebpR mutant containing the ebpR promoter fused to lacZ; Ermr | This study |
| Plasmids | ||
| pMSP3535 | Nisin-inducible expression shuttle vector with pAMβ1 and ColE1 replicons; Ermr | 3 |
| pTCV-lacZ | Shuttle vector containing promoterless lacZ; Ermr | 14 |
| pTEX5515 | pMSP3535 with ebpR from bp −20 to +1561 from the ATG; this ebpR fragment contains the full ORF and the RBS of ebpR; Ermr | This study |
| pTEX5586 | pTCV-lacZ containing 301 bp from bp 248 upstream to bp 53 downstream of the start codon of ebpR; Ermr | This study |
| pTEX5269 | pTCV-lacZ containing 103 bp from bp −110 to −8 of the start codon of fsrB; Ermr | 26 |
| Primers | ||
| ebpR deletion | Upstream fragment was amplified using GCTCTAGACAGGACCCACACTTTTCA with CGGGATCCTTTCCTCCCAAGTCCTTT, and the downstream fragment was amplified using CGGGATCCTTGAAACCAGTGTTCCAAG with GGAATTCTGGTCAGCAAACGATTTT | |
| ebpR complementation | The primers CCGAGCTCGGACTTGGGAGGAAACAATA and CGGGATCCAAGTCATTTCGACTTATGTCCT were used to amplify ebpR from 20 bp upstream of the start codon to 138 bp downstream of the stop codon | |
| ebpR promoter fusion | The primers GGAATTCAAGACTACGCCGAAAACC and CGGGATCCACACGAATGATTTCTTCCA were used to amplify from 248 bp upstream to 53 bp downstream of the start codon of ebpR. | |
| Quantitative RT-PCR primers | gyrB, ACCAACACCGTGCAAGCC and CAAGCCAAAACAGGTCGCC; ebpA, AAAAATGATTCGGCTCCAGAA and TGCCAGATTCGCTCTCAAAG; srtC, ACACATGCGGTCATTTCAGG and GCGTCTTCCCATTGACTTCG |
ORF, open reading frame; RBS, ribosome binding site. ebpR = ef1090. Kanr, kanamycin resistance; Fusr, fusidic acid resistance; Rifr, rifampin resistance; p-cl-Pher, p-chloro-phenylalanine resistance; Spcr, spectinomycin resistance; Tetr, tetracycline resistance; Ermr, erythromycin resistance. Restriction enzyme recognition sites are underlined.
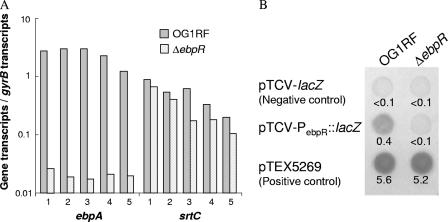
To assess transcriptional differences between OG1RF and the ΔebpR mutant, we used microarray analysis with slide preparation (1), probe labeling, hybridization, data acquisition, and statistical analysis performed as described previously (2). RNA was extracted at late log growth phase from TSBG-grown cultures (2). Within the level of detection of the microarray analysis, the ebpABC transcript was undetectable in the ΔebpR mutant compared to OG1RF, indicating that ebpR is important for ebpABC expression at the transcriptional level. srtC was not significantly altered in these conditions. We confirmed these results by using qRT-PCR (12). The amount of transcript for each gene of interest was normalized against gyrB transcripts. qRT-PCR showed that ebpA transcripts levels were decreased 119-fold (P = 0.0018) in the ebpR mutant versus the wild type, whereas srtC transcripts were only 1.7-fold decreased (P = 0.028) (Fig. 1A). The slight effect of ebpR on srtC is likely related to the presence of an independent srtC promoter and the low level of readthrough from the ebpA promoter described previously (13). Since basal ebpABC expression was detected in our ebpR mutant, our results indicate that ebpR, although important, is not essential for ebpABC expression.
FIG. 1.
ebpA, srtC, and ebpR expression. (A) Real-time transcript levels of ebpA and srtC in OG1RF and TX5514 (ΔebpR). The numbers below the x axis correspond to the five independent cultures used for this assay. (B) pTCV-PebpR::lacZ in OG1RF and in TX5514 streaked onto TSBG plates containing 10 μg of erythromycin/ml and 200 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml and incubated for 48 h at 37°C. The negative control plasmid is pTCV-lacZ, and the positive control is pTEX5269. The β-galactosidase assay results (expressed as the optical density at 420 nm/mg of protein/ml) located under the spots correspond to a representative experiment performed with samples collected at entry into the stationary growth phase of cultures grown in TSBG (performed in triplicate).
To begin examining how epbR is regulated, we mapped the transcriptional start site of OG1RF. Using the RACE (for rapid amplification of cDNA ends) kit, only one start site for ebpR was found at −21 bp. Looking at the promoter area, no canonical −10 and −35 bp sequences were detected, indicating that the expression of ebpR is likely sigma A independent. We next looked at possible ebpR autoregulation by inserting a 301-bp fragment overlapping the entire 209-bp region separating ebpA from ebpR into pTCV-lacZ to create a transcriptional lacZ fusion. The β-galactosidase activity of this construct in an ebpR background compared to the wild type was measured (6). No β-galactosidase activity was detected using the PebpR::lacZ fusion in the ebpR mutant, whereas it was detectable in the wild type (Fig. 1B). The results indicate that ebpR is autoactivated.
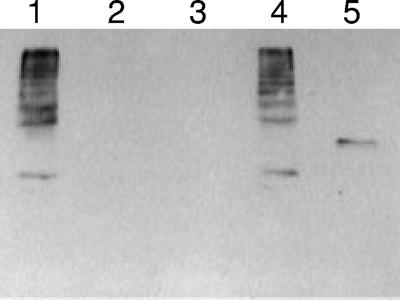
In addition to examining transcript levels, we examined pilus production in the wild type (OG1RF), the ΔebpR mutant (TX5514), and a complemented strain (TX5582) in which ebpR was cloned under the control of the nisin promoter in pMSP3535 (3). As determined by RT-PCR, the expression of ebpR in TX5582 was detected even without nisin (data not shown). In order to detect the pili, mutanolysin extracts were made from OG1RF as described previously (13). As seen before, a ladder profile was detected using antibodies to EbpA (Fig. 2, lane 1). It was not observed in extracts from the ebpR mutant (lane 2) or in extracts from the mutant carrying the empty vector (lane 3). However, it was detected in the complemented strain (Fig. 2, lane 4). A similar result was observed using EbpC antibodies (data not shown).
FIG. 2.
Western blots of OG1RF and its ΔebpR mutant (TX5514). Mutanolysin surface extract preparations were probed with anti-EbpA rabbit polyclonal immune serum. Lanes: 1, OG1RF; 2, ΔebpR mutant (TX5514); 3, ΔebpR(pMSP3535) (TX5584); 4, ΔebpR(pTEX5515) (TX5582); 5, EbpA recombinant protein with a molecular mass of 113 kDa.
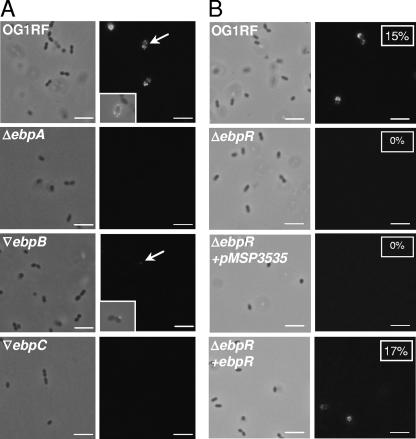
Nallapareddy et al. reported that only 10 to 20% of OG1RF cells had observable pili by electron microscopy (13). To evaluate this observation using another methodology, intact bacteria fixed with 4.4% (wt/vol) paraformaldehyde were probed with anti-EbpA or anti-EbpC rabbit antibodies by indirect immunofluorescence. Binding of anti-EbpA or anti-EbpC antibodies to the surface-exposed epitopes were detected using fluorescein isothiocyanate (FITC)-labeled anti-rabbit antibodies (Sigma, St. Louis, MO). First, we tested OG1RF and the ebpA deletion mutant for the presence of pili in cells grown in TSBG and collected in stationary phase. Using anti-EbpA, FITC-labeled cells were detected in the OG1RF population (Fig. 3A) but not in the ebpA mutant population. The fluorescence was present on the entire surface of the cells except at the septum. Since EbpA was shown to be expressed and detectable as a monomer by Western blotting in ebpB and ebpC insertion mutants (13), these two strains were also assessed by immunofluorescence. As shown in Fig. 3A, a fluorescent dot (marked by an arrow and shown at a higher resolution in the insert) was visible on a few of the ebpB insertion mutant cells, whereas no fluorescence was detected at the surface of the ebpC insertion mutant. In a subsequent experiment, OG1RF, the ebpR deletion mutant, and the complemented strain were grown under the same conditions (TSBG, stationary phase, no nisin induction). In the OG1RF population, 15% of the cells were labeled with FITC, whereas no fluorescence was detected on the surface of the ebpR mutant cells (Fig. 3B). With the complemented strain, 17% of the population was FITC labeled (Fig. 3B, boxed numbers). We can postulate that ebpR and/or ebpABC are influenced by and/or may influence other regulatory pathways, since even in the presence of a functional ebpR, the majority of the cells are not producing pili at their surfaces. However, when cloned ebpR was transferred into the wild type, the level of FITC-labeled cells increased to 70% (preliminary data), whereas only 17% were labeled when the same plasmid was introduced into the ΔebpR strain. These results indicate that the level of ebpR expression affects ebpABC expression or that the presence of the plasmid is titrating out a negative regulator.
FIG. 3.
Immunofluorescence analysis showing surface localization of EbpA. Cells were collected at stationary growth phase from a TSBG culture and assayed using anti-EbpA rabbit serum. The left panel is the phase-contrast image of the field visualized with the FITC filter (right panel). (A) Parental OG1RF. TX5475 is the ΔebpA mutant, TX5460 is the ▿ebpB mutant (▿, insertion mutant), and TX5448 is the ▿ebpC mutant. The area indicated with an arrow is shown in a box as an overlay between phase contrast and FITC visualization. (B) Parental OG1RF. TX5514 is the ΔebpR mutant; the ΔebpR mutant electroporated with vector only (pMSP3535) served as a control. The ΔebpR mutant electroporated with pMSP3535::ebpR is the complemented strain. The boxed numbers correspond to the percentage of FITC-labeled cells from at least 250 cells counted. Bar, 10 μm.
Strains containing mutations in the ebpABC operon were previously shown to have reduced biofilm formation (13). Since the ebpR mutant displays greatly reduced expression of the ebpABC operon, it was also predicted to be defective in biofilm formation. Biofilm formation and adherence to a polystyrene surface were assessed quantitatively as described previously (10). As shown in Table 2, the ebpR mutant displayed a significant reduction in biofilm formation (47% reduction, P < 0.01) (Table 2). Of interest, the complemented strain presented a statistically significant increase in biofilm formation compared to OG1RF (22% more biofilm formation, P < 0.01). The biofilm defect in the ebpA and ebpR mutants was consistent with reductions in primary adherence of 43 and 64% at 30 min and 53 and 69% reduction at 2 h, respectively (P < 0.01) (Table 2).
TABLE 2.
Biofilm and primary adherence assays
| Strain | Median (interquartile range)
|
||
|---|---|---|---|
| OD570 for biofilm assaya | Primary adherenceb at:
|
||
| 30 min | 2 h | ||
| OG1RF | 1.257 (1.184-1.313) | 267 (238-316) | 649 (614-722) |
| TX5475 (ΔebpA) | 0.817 (0.766-0.882) | 97 (89-123) | 203 (168-229) |
| TX5515 (ΔebpR) | 0.667 (0.644-0.701) | 151 (114-231) | 314 (230-385) |
| TX5584 (ΔebpR)/pMSP3535 | 0.814 (0.750-0.891) | 166 (81-202) | 293 (260-429) |
| TX5582 (ΔebpR + ebpR) | 1.534 (1.487-1.674) | 221 (181-290) | 692 (527-750) |
The optical density at 570 nm (OD570) readings for the biofilm assays are from three independent experiments (total of 24 wells).
Primary adherence was assessed after incubation for 30 min and 2 h. Bacteria in four fields from two independent plates from at least two independent experiments were subjected to light microscopy and counted.
In vivo, it has been shown that the ebpA mutant was significantly attenuated in the rat endocarditis model (13) and the murine urinary tract infection model (17). We attempted to quantify gene expression in vivo by using the rat endocarditis model. Preliminary data from vegetations collected at 48 h indicate that ebpA expression was 100 times lower in the ebpR deletion mutant, whereas srtC expression was not appreciably affected (preliminary results, data not shown). This experiment demonstrates the feasibility of detecting and quantifying gene expression in the rat endocarditis model, opening new avenues for research directed at understanding bacterial endocarditis.
In conclusion, we identified an E. faecalis transcriptional regulator, EbpR, which positively affects the expression of the endocarditis-associated pilus operon, thereby influencing biofilm formation. It will be of interest in future studies to address the signals to which ebpR is responding and whether these signals affect pilus production both in vitro and in vivo.
Acknowledgments
We are grateful to L. C. Thomson and P. Bruscella for technical assistance and T. M. Koehler and S. R. Nallapareddy for helpful discussion.
This study was supported by grant NIH R37 AI47923 from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Disease, to B.E.M. and by a new scholar award in global infectious disease to D.A.G. from the Ellison medical Foundation.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Aakra, A., H. Vebo, L. Snipen, H. Hirt, A. Aastveit, V. Kapur, G. M. Dunny, B. E. Murray, and I. F. Nes. 2005. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 49:2246-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188:2875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 4.Drysdale, M., A. Bourgogne, S. G. Hilsenbeck, and T. M. Koehler. 2004. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J. Bacteriol. 186:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garsin, D. A., J. Urbach, J. C. Huguet-Tapia, J. E. Peters, and F. M. Ausubel. 2004. Construction of an Enterococcus faecalis Tn917-mediated gene disruption library offers insight into Tn917 insertion patterns. J. Bacteriol. 186:7280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock, L. E., B. D. Shepard, and M. S. Gilmore. 2003. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J. Bacteriol. 185:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristich, C. J., J. R. Chandler, and G. M. Dunny. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabeled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 9.Malani, P. N., C. A. Kauffman, and M. J. Zervos. 2002. Enterococcal disease, epidemiology, and treatment, p. 385-408. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 10.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nallapareddy, S. R., and B. E. Murray. 2006. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect. Immun. 74:4982-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nallapareddy, S. R., K. V. Singh, J. Sillanpaa, D. A. Garsin, M. Hook, S. L. Erlandsen, and B. E. Murray. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 116:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 15.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Singh, K. V., S. R. Nallapareddy, and B. E. Murray. 2007. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 195:1671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]