Abstract
The recent recrudescence of Mycobacterium tuberculosis infection and the emergence of multidrug-resistant strains have created an urgent need for new therapeutics against tuberculosis. The enzymes of the shikimate pathway are attractive drug targets because this route is absent in mammals and, in M. tuberculosis, it is essential for pathogen viability. This pathway leads to the biosynthesis of aromatic compounds, including aromatic amino acids, and it is found in plants, fungi, bacteria, and apicomplexan parasites. The aroB-encoded enzyme dehydroquinate synthase is the second enzyme of this pathway, and it catalyzes the cyclization of 3-deoxy-d-arabino-heptulosonate-7-phosphate in 3-dehydroquinate. Here we describe the PCR amplification and cloning of the aroB gene and the overexpression and purification of its product, dehydroquinate synthase, to homogeneity. In order to probe where the recombinant dehydroquinate synthase was active, genetic complementation studies were performed. The Escherichia coli AB2847 mutant was used to demonstrate that the plasmid construction was able to repair the mutants, allowing them to grow in minimal medium devoid of aromatic compound supplementation. In addition, homogeneous recombinant M. tuberculosis dehydroquinate synthase was active in the absence of other enzymes, showing that it is homomeric. These results will support the structural studies with M. tuberculosis dehydroquinate synthase that are essential for the rational design of antimycobacterial agents.
Tuberculosis (TB) remains the leading cause of mortality due to a bacterial pathogen, Mycobacterium tuberculosis, and infects approximately one-third of the world's population (10). The World Health Organization has estimated that 9 million people are infected per year, leading to 2 million deaths, mainly in sub-Saharan Africa and Asia (39). The discovery of the antibacterial and antituberculosis properties of streptomycin, isoniazid, and pyrazinamide led to effective chemotherapy that decreased the TB mortality rate worldwide (1). The later introduction of ethionamide, rifampin, ethambutol, and ciprofloxacin to the arsenal for TB treatment seemed to provide an adequate number of effective antimicrobial agents. The reemergence of TB is basically a consequence of anthropic factors, such as the recent human immunodeficiency virus/AIDS pandemic and the development of drug-resistant strains (stemming from inappropriate treatments and/or patient noncompliance) (13). Another contributing factor is the evolution of multidrug-resistant TB, defined as TB caused by Mycobacterium tuberculosis strains resistant to at least isoniazid and rifampin, two first-line drugs used in the standard “short-course” treatment of TB. More recently, the emergence of extensively drug resistant TB (XDR-TB), defined as TB caused by isolates resistant to isoniazid, rifampin, and at least three of the six main classes of second-line drugs, has been reported (6). XDR-TB is widespread, including occurrence in the United States, where TB had been considered under control (12). The worldwide occurrence of XDR-TB raises the prospect of virtually incurable TB (12). There is thus an urgent need for new, more effective drugs to improve the treatment of multidrug-resistant TB and XDR-TB and to shorten the duration of TB treatment.
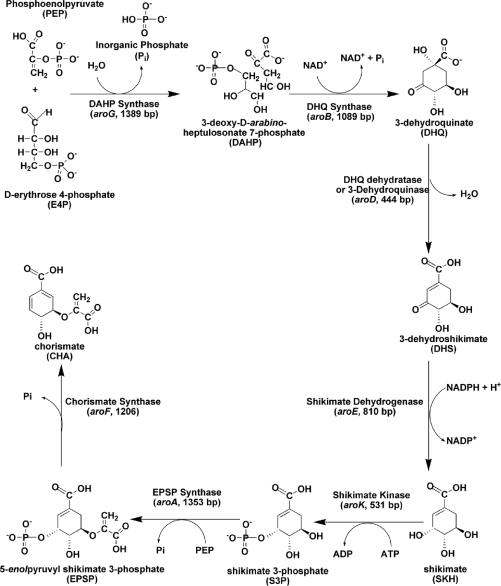
The shikimate pathway (Fig. 1) is an attractive target for the development of herbicides and antimicrobial agents because it is essential for algae, higher plants, bacteria, and fungi but is absent from mammals (4). The mycobacterial shikimate pathway leads to the biosynthesis of precursors of aromatic amino acids, naphthoquinones, menaquinones, and mycobactin (14). Homologues to the seven enzymes of the shikimate pathway have been identified in the genome sequence of M. tuberculosis (9). This pathway has been shown to be essential for the viability of M. tuberculosis (30). Accordingly, the essentiality of the mycobacterial shikimate pathway and its absence from human hosts indicate that the enzymes of this pathway represent promising targets for the development of nontoxic antimycobacterial agents.
FIG. 1.
The mycobacterial shikimate pathway (the main trunk) leads to the biosynthesis of chorismic acid, a precursor of aromatic amino acids, para-aminobenzoic acid, ubiquinone or coenzyme Q, naphthoquinones, menaquinones, and mycobactins.
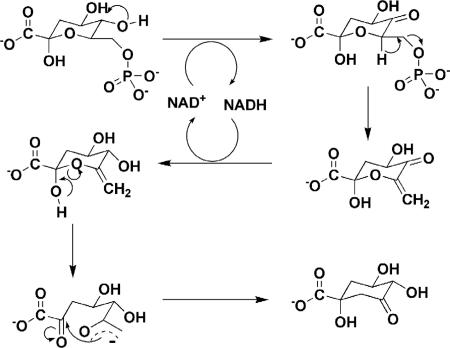
The gene (aroB; Rv2538c) encoding the dehydroquinate synthase (DHQS; EC 4.6.1.3; systematic name, 3-deoxy-arabino-heptulosonate-7-phosphate phosphate-lyase) of the shikimate pathway has been proposed to be present in M. tuberculosis by sequence homology (9). DHQS catalyzes the conversion of 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) to 3-dehydroquinate, the first cyclic compound of this pathway (Fig. 2) (34). DHQS utilizes a complex multistep mechanism that includes oxidation, β-elimination, intramolecular aldol condensation, and reduction (16). In Escherichia coli, the DHQS enzyme requires NAD+ as a cofactor, and a divalent cation, such as Co2+ or Zn2+, is also needed (3, 25). Bacillus subtilis DHQS has been purified in association with chorismate synthase and NADPH-dependent flavin reductase, and although the chorismate synthase is required for B. subtilis DHQS activity, the flavin reductase does not need to be present (20). In Neurospora crassa and Aspergillus nidulans, the DHQS activity is the first of five sequential steps of the shikimate pathway catalyzed by a pentafunctional complex, encoded by arom, whose sequence has high similarity to the five monofunctional E. coli counterparts, including DHQS (17, 35). Usually, DHQSs from bacteria are significantly smaller than their fungal enzyme analogues, and they also show significant structural divergence, especially in the amino- and carboxy-terminal sequences.
FIG. 2.
The chemical reaction catalyzed by DHQS.
To pave the way for structural and functional efforts currently under way in our laboratory on which to base the rational design of antitubercular agents, the aroB gene from M. tuberculosis strain H37Rv was PCR amplified, cloned, sequenced, and expressed, and a recombinant M. tuberculosis DHQS protein was purified to homogeneity. Electrospray ionization mass spectrometry (ESI-MS) analysis and N-terminal sequencing were carried out to unequivocally identify the recombinant M. tuberculosis DHQS protein. Genetic complementation experiments using Escherichia coli mutant strains lacking the aroB gene were carried out to confirm that recombinant M. tuberculosis DHQS was cloned in its functional form. The availability of M. tuberculosis DHQS protein in large quantities will allow enzyme kinetics and structural studies to be undertaken in order to provide a framework to guide the design of chemical compounds with antituberculosis activity.
MATERIALS AND METHODS
Amplification, cloning, and overexpression of the M. tuberculosis aroB gene.
Two oligonucleotides (5′-GGCCATATGACCGATATCGGCGCACCCG-3′ and 5′-AGGATCCTCATGGGGCGCAAACTCCGGC-3′) complementary to the amino-terminal coding and carboxy-terminal noncoding strands of the M. tuberculosis aroB gene (9) were synthesized to contain, respectively, NdeI and BamHI restriction sites (underlined). These primers were used to PCR amplify the aroB gene from M. tuberculosis H37Rv genomic DNA in the presence of 10% dimethyl sulfoxide (DMSO). The PCR product (1,089 bp) was purified by electrophoresis, digested with NdeI and BamHI (Boehringer Mannheim), and ligated into a pET23a(+) expression vector (Novagen) that had previously been digested with the same restriction enzymes. The DNA sequence of the M. tuberculosis aroB gene was determined in order to confirm the identity, integrity, and absence of PCR-introduced mutations in the cloned gene. The recombinant pET23a(+)::aroB plasmid was introduced into Escherichia coli BL21(DE3) (Novagen) electrocompetent cells and selected on LB agar plates containing 50 μg ml−1 carbenicillin. LB medium (6 liters) containing carbenicillin was inoculated with single colonies, which were grown for 20 h at 180 rpm and 37°C without isopropyl-β-d-thiogalactopyranoside (IPTG) induction. Cells were harvested by centrifugation at 4,000 × g for 30 min at 4°C and were stored at −20°C. Soluble and insoluble fractions were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22).
Purification of M. tuberculosis DHQS.
Approximately 30 g of cells was suspended in 140 ml of 50 mM Tris·HCl buffer, pH 7.8 (buffer A), containing protease inhibitors (Complete protease inhibitor cocktail tablets; Boehringer Mannheim) and lysozyme (0.2 mg ml−1). Cells were disrupted by sonication, and cell debris was removed by centrifugation (at 48,000 × g for 30 min). The supernatant was incubated with 1% (wt/vol) streptomycin sulfate and centrifuged at 48,000 × g for 30 min. The resulting supernatant was dialyzed against buffer A and loaded onto a Q-Sepharose Fast Flow column (GE Healthcare) preequilibrated with buffer A, and the absorbed material was eluted with a linear gradient from 0 to 0.5 M NaCl. Fractions containing M. tuberculosis DHQS were pooled and loaded onto a HiLoad 16/10 phenyl-Sepharose (GE Healthcare) column, and protein elution was achieved with a linear gradient of 1 to 0 M ammonium sulfate. The fractions containing M. tuberculosis DHQS were pooled and concentrated down to 10 ml using an Amicon ultrafiltration cell (molecular weight cutoff, 30,000). The sample was loaded onto a Sephacryl S-200 HR column (GE Healthcare) and eluted with buffer A. The homogeneous recombinant protein was stored in a saturated ammonium sulfate solution. Protein expression and all purification steps were analyzed by SDS-PAGE, and protein concentrations were determined by the method of Bradford (5) using the Bio-Rad Laboratories protein assay kit.
N-terminal amino acid sequencing.
The N-terminal amino acid residues of homogeneous recombinant M. tuberculosis DHQS were identified by automated Edman degradation sequencing using a PPSQ 21A gas-phase sequencer (Shimadzu).
Mass spectrometry analysis.
The homogeneity of the protein preparation was assessed by ESI-MS with some adaptations (8). Samples were analyzed on a triple quadrupole mass spectrometer (model QUATTRO II) equipped with a standard ESI probe (Micromass, Altrincham, United Kingdom) and adjusted to a flow rate of ca. 250 μl min−1. The source temperature (80°C) and needle voltage (3.6 kV) were maintained constant throughout the collection of experimental data, applying a drying gas (nitrogen) flow of 200 liters h−1 and a nebulizer gas flow of 20 liters h−1. The mass spectrometer was calibrated with intact horse heart myoglobin and its typical cone voltage-induced fragments. The molecular mass of the recombinant M. tuberculosis DHQS subunit was determined by ESI-MS, by adjusting the mass spectrometer to give a peak with a half-height of 1 mass unit, and the sampling cone-to-skimmer lens voltage controlling the transfer of ions to the mass analyzer was set to 38 V. About 50 pmol (10 μl) of each sample was injected into the electrospray transport solvent. The ESI spectrum was obtained in the multichannel acquisition mode, with scanning from 500 to 1,800 m/z at a scan time of 7 s. The mass spectrometer is equipped with MassLynx and Transform software for data acquisition and spectrum handling.
Genetic complementation.
The EcoRI and HindIII (Boehringer Mannheim) restriction sites were introduced into the aroB gene through a new round of PCR amplification. The amplified fragment was cloned into the pKK223-3 expression vector (NCCB 3190). Genetic complementation was performed using the mutant E. coli strain AB2847 (Genetic Stock Center), whose aroB gene has been knocked out. Two batches of solid medium were prepared, one lacking phenylalanine, tyrosine, and tryptophan and the other supplemented with these aromatic compounds (Sigma) at final concentrations of 40 μg ml−1 each. The E. coli mutant strain AB2847 was transformed with the recombinant plasmid, grown on minimal medium agar plates (11) with 100 μg ml−1 ampicillin, either with or without aromatic amino acid supplementation, at 37°C. For control experiments, the E. coli mutant strain AB2847 was transformed with the pKK223-3 vector lacking the M. tuberculosis aroB gene.
M. tuberculosis DHQS activity assay.
Recombinant M. tuberculosis DHQS protein was assayed in the forward direction in 50 mM Tris-HCl, pH 7.6, at 25°C. Enzyme activity was measured by estimating the rate of inorganic phosphate (Pi) release in solution using a continuous spectrophotometric coupled assay with human purine nucleoside phosphorylase (PNP) and 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG). This assay is based on the maximum difference in absorbance at 360 nm between MESG and the purine base product (2-amino-6-mercapto-7-methylpurine) produced by PNP-catalyzed phosphorolysis of MESG (37). The PNP-coupled assay has been utilized to continuously monitor Pi production catalyzed by a number of enzymes (27, 28, 29). Typically, a 500-μl assay mixture contained 0.5 mM MESG, 0.5 U PNP, 0.250 mM DAHP (Toronto Research Chemicals Inc.), and 0.250 mM NAD+ (Sigma). All components were mixed and incubated for 10 min to consume any endogenous Pi that might be present in the DAHP solution, and reactions were initiated by addition of M. tuberculosis DHQS (stock solution, 1.7 mg ml−1).
RESULTS AND DISCUSSION
The M. tuberculosis genome has a high content of cytosine and guanine bases, approximately 65.6% (9). Although higher temperatures are usually used for DNA denaturation of these types of genomes, the aroB gene was amplified using standard PCR conditions. However, amplification of the aroB gene from M. tuberculosis H37Rv genomic DNA was achieved only in the presence of 10% DMSO in the reaction mixture. DMSO is a cosolvent that improves the denaturation of GC-rich DNA (38) and facilitates PCR extension by DNA polymerase through DNA secondary structures that may hinder amplification (31). The PCR fragment was inserted into the pET23a(+) expression vector between the NdeI and BamHI restriction sites, and DNA sequencing of the entire aroB structural gene confirmed the identity of the gene and the absence of PCR-introduced mutations.
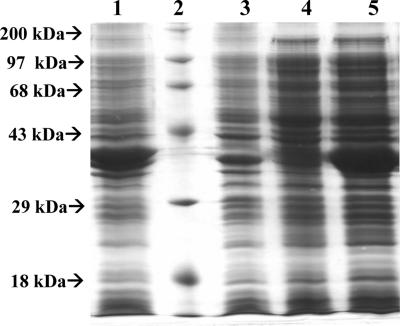
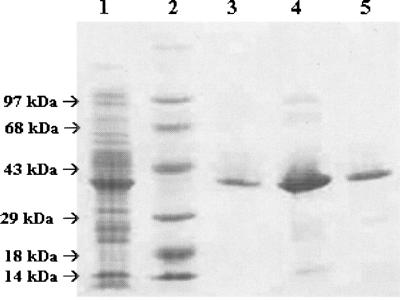
Recombinant M. tuberculosis DHQS was overexpressed in its soluble form in E. coli BL21(DE3) host cells in the absence of IPTG induction (Fig. 3). It has been shown previously that lac-controlled systems, including the pET system, could have high-level protein expression in the absence of inducer. It has been proposed that leaky protein expression is a property of the lac-controlled system as cells approach stationary phase in complex medium and that cyclic AMP, acetate, and low pH are required to achieve high-level expression in the absence of IPTG induction, which may be part of a general cellular response to nutrient limitation (19). Accordingly, a number of reports have described similar results (24, 28, 32, 33). SDS-PAGE showed expression of a soluble protein with a molecular size consistent with that expected for M. tuberculosis DHQS (38.1 kDa) (Fig. 3). M. tuberculosis DHQS was purified to homogeneity from a crude extract by using a protocol including anionic exchange, followed by hydrophobic interaction and elution on gel filtration columns, yielding 5 mg of recombinant protein per liter of cell culture. No contaminants were detected by SDS-PAGE analysis (Fig. 4). Homogeneous recombinant protein was stored in an 85% (NH4)2SO4 saturated solution.
FIG. 3.
Analysis of DHQS expression by SDS-PAGE. Lanes: 1, insoluble fraction of E. coli BL21(DE3) transformed with pET23a(+)::aroB; 2, protein molecular size standards (200, 97, 68, 43, 29, and 18 kDa) (Gibco); 3, insoluble fraction of E. coli BL21(DE3) transformed with pET23a(+) (control); 4, soluble fraction of E. coli BL21(DE3) transformed with pET23a(+) (control); 5, soluble fraction of E. coli BL21(DE3) transformed with pET23a(+)::aroB.
FIG. 4.
Analysis of purification steps of DHQS by SDS-PAGE. Lanes: 1, crude extract after dialysis; 2, protein molecular size standards (97, 68, 43, 29, 18, and 14 kDa) (Gibco); 3 through 5, DHQS after elution on Q-Sepharose Fast Flow, phenyl-Sepharose, and Sephacryl S-200 HR columns, respectively.
The molecular mass of the M. tuberculosis DHQS subunit was determined by ESI-MS to be 38,135.70 Da, consistent with the posttranslational removal of an N-terminal methionine residue from the full-length gene product (predicted mass, 38,266.00 Da). The ESI-MS result revealed no peak at the expected mass for E. coli DHQS (38,880.90 Da). The first 18 amino-terminal amino acid residues of the purified recombinant protein were determined to be TDIGAPVTVQVAVDPPYP by Edman degradation, thereby unambiguously identifying M. tuberculosis DHQS and confirming removal of the N-terminal methionine residue from it.
To confirm the correct assignment to the structural gene encoding M. tuberculosis DHQS, the biological activity of recombinant M. tuberculosis DHQS was probed by genetic complementation using E. coli mutants lacking the aroB gene (E. coli AB2847). Although strain AB2847 is not able to grow in minimal medium due to the lack of the DHQS-encoding gene, supplementation of minimal medium with phenylalanine, tyrosine, and tryptophan enables the cells to grow. On the other hand, the expression of the target protein in pKK233-3 is not controlled by T7 RNA polymerase as in the pET system. Therefore, since E. coli strain AB2847 does not have a copy of T7 RNA polymerase in its genome, a different plasmid must be used to express M. tuberculosis DHQS in this E. coli mutant. Accordingly, PCR amplification was performed using the recombinant plasmid pET23a(+)::aroB as a template to subclone the M. tuberculosis aroB fragment into the pKK233-3 expression vector, containing EcoRI and HindIII restriction sites. Transformed E. coli AB2847 harboring the recombinant pKK223-3::aroB plasmid was able to grow in minimal medium without aromatic amino acid supplements, whereas E. coli AB2847 transformed with the pKK223-3 vector lacking the M. tuberculosis aroB gene was able to grow only in minimal medium containing aromatic amino acid supplements (Fig. 5). These results demonstrate that the M. tuberculosis aroB gene encodes an active DHQS enzyme that is responsible for the auxotrophic phenotype and consequently probes the functionality of the recombinant protein, which also functions in E. coli. It has been reported previously that the M. tuberculosis aroB gene codes for a DHQS activity in E. coli (18). However, Garbe et al. transformed an E. coli strain lacking the aroB gene with a recombinant plasmid containing a 3.4-kb DNA fragment that encompassed aroD (named aroQ by Garbe et al.) and aroB genes as well as sequences of unknown function (18). Accordingly, to the best of our knowledge, here we describe the first experimental evidence for the correct assignment to the open reading frame of the aroB coding sequence.
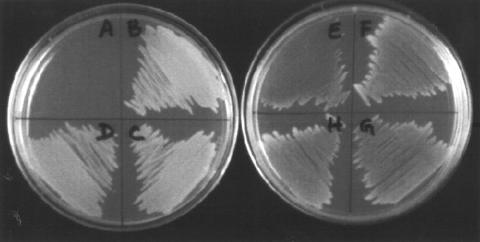
FIG. 5.
Analysis of functional complementation. (Left) Minimal medium agar plates. Sector A, AB2847 transformed with pKK223-3 (negative control); sectors B, C, and D, AB2847 transformed with pkk223-3::aroB. (Right) Minimal medium supplemented with phenylalanine, tyrosine, and tryptophan. Sector E, AB2847 transformed with pKK223-3 (negative control); sectors F, G, and H, AB2847 transformed with pkk223-3::aroB.
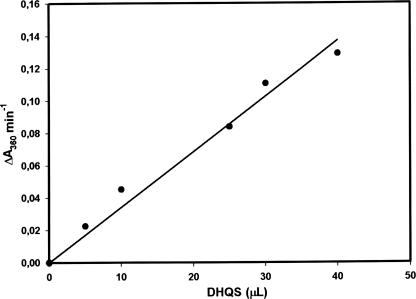
In B. subtilis, DHQS has been purified in association with chorismate synthase and NADPH-dependent flavin reductase. The enzyme was active only when associated with chorismate synthase, whereas the flavin reductase could be separated from the complex with retention of DHQS activity (20). DHQS, chorismate synthase, and flavin reductase form a trifunctional enzyme complex, and chorismate synthase is required for DHQS activity. The complex can be dissociated into flavin reductase and chorismate synthase with the loss of DHQS activity (20). Interestingly, in the work reported here, M. tuberculosis DHQS was purified as a single protein with no other associated protein, which suggests that the quaternary structure is homomeric, like that of E. coli DHQS (3). However, it is also conceivable that M. tuberculosis DHQS requires other enzymes for activity that were not observed in the homogeneous recombinant protein because they were not present at concentrations comparable to that of the overexpressed protein. Accordingly, increasing amounts of homogeneous M. tuberculosis DHQS were added to the reaction mixture, and enzyme activity was measured. The M. tuberculosis DHQS enzyme activity was linearly dependent on the volume of sample added to the reaction mixture (Fig. 6), thereby showing that M. tuberculosis DHQS does not require other enzymes for activity and is thus homomeric. These results also show that true initial velocities are being measured.
FIG. 6.
Linear dependence of M. tuberculosis DHQS activity on homogeneous recombinant protein volume. The rates of Pi release due to M. tuberculosis DHQS enzyme activity were followed in the forward direction by continuously monitoring the increase in the concentration of 2-amino-6-mercapto-7-methylpurine at 360 nm produced by phosphorolysis of MESG catalyzed by PNP in a coupled assay.
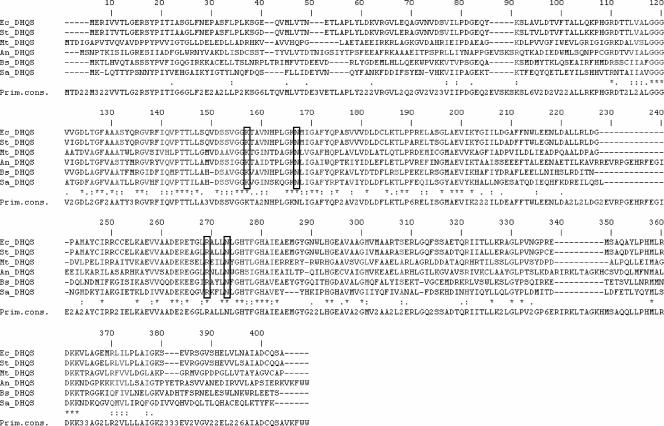
The amino acid sequence comparison between M. tuberculosis DHQS and DHQSs from other organisms showed highly conserved residues (Fig. 7), which may be involved in protein function and activity, either directly or indirectly, by maintenance of an appropriate conformation (15). Even though bacterial DHQSs are smaller than their eukaryotic counterparts, the degree of homology does not diverge among prokaryotes (either gram-positive or gram-negative bacteria) and eukaryotes. In eukaryotic organisms such as N. crassa, A. nidulans, and Saccharomyces cerevisiae, the DHQS reaction is the first of five sequential steps in the shikimate pathway that are catalyzed by the arom enzyme complex (21). The multifunctional complex enzymes have spatially distinct catalytic sites for DHQS and the four subsequent enzymes. Moreover, eukaryotic DHQSs have a nonconserved insertion (of different lengths in different species) in a downstream domain, which may be important for their assembly as a multifunctional complex. M. tuberculosis DHQS shows 25 to 37% overall identity and 52 to 69% similarity with DHQSs from E. coli, Salmonella enterica serovar Typhimurium, B. subtilis, A. nidulans, and Staphylococcus aureus. Amino acid sequence comparison (Fig. 7) indicates the conservation of Lys157, Asn167, Arg269, and Asn273 (according to M. tuberculosis aroB gene numbering). These conserved amino acid residues have been shown to be involved in conformational changes upon substrate binding to the enzyme active site in S. aureus DHQS and A. nidulans DHQS (26).
FIG. 7.
Comparison of prokaryotic and eukaryotic DHQS amino acid sequences. Shown are DHQS sequences for Escherichia coli (Ec) (362 residues), Salmonella serovar Typhimurium (St) (362 residues), Mycobacterium tuberculosis (Mt) (362 residues), Aspergillus nidulans (An) (403 residues), Bacillus subtilis (Bs) (362 residues), and Staphylococcus aureus (Sa) (354 residues). The conserved amino acid residues Lys157, Asn167, Arg269, and Asn273 in M. tuberculosis DHQS (corresponding to amino acid residues Lys136, Asn146, Arg235, and Asn239 in S. aureus DHQS [26]) are boxed.
The chemical reaction catalyzed by DHQS (Fig. 2) is mechanistically unusually diverse for a single enzyme and includes (i) the oxidation of the secondary alcohol at C-5 of DAHP, (ii) the β-elimination of inorganic phosphate across C-6 and C-7, (iii) the reduction of the resulting eneone at C-5, (iv) the ring opening of the enol pyranose, and (v) the final intramolecular aldol-like reaction that produces 3-dehydroquinate (3, 25). Despite the fact that the bioavailability of Zn2+ in nature is much greater than that of Co2+, the Co2+ form of the enzyme has been reported to be more stable and has a higher specific activity than the Zn2+ form (23). However, the mode of action of M. tuberculosis DHQS is still unknown, and the availability of a homogeneous enzyme will allow mechanistic and structural studies to be carried out. Expression of functional proteins in soluble form has been identified as an important bottleneck in efforts to determine the biological activity and crystal structure of M. tuberculosis proteins (36). Moreover, protein purification has become an important asset for any research group, since demand for homogeneous proteins has been increasing (7).
In this report, we present the cloning, purification, and genetic complementation of the aroB gene from M. tuberculosis. The availability of functional homogeneous M. tuberculosis DHQS will provide protein in quantities necessary for both X-ray crystal structure determination and studies on the mode of action of the enzyme by steady-state and pre-steady-state kinetics to allow the rational design of antimycobacterial agents. Moreover, the availability of homogeneous M. tuberculosis DHQS will allow immobilization on a solid support in order to screen for new chemical entities from plant-derived chemical compound libraries to identify antituberculosis agents as described elsewhere (2).
Acknowledgments
Financial support for this work was provided by Millennium Initiative Program MCT-CNPq, Ministry of Health—Department of Science and Technology and PRONEX/CNPq/FAPERGS (Brazil), to D.S.S. and L.A.B. D.S.S (CNPq, 304051/1975-06), L.A.B. (CNPq, 520182/99-5), and J.F. (CNPq, 301131/2003-1) are research career awardees from the National Council for Scientific and Technological Development of Brazil.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Basso, L. A., and D. S. Santos. 2005. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis—an update. Med. Chem. Rev. 2:393-413. [DOI] [PubMed] [Google Scholar]
- 2.Basso, L. A., L. H. P. da Silva, A. G. Fett-Neto, W. F. de Azevedo, I. S. Moreira, M. S. Palma, J. B. Calixto, S. Astolfi Filho, R. R. dos Santos, M. B. P. Soares, and D. S. Santos. 2005. The use of biodiversity as source of new chemical entities against defined molecular targets for treatment of malaria, tuberculosis, and T-cell mediated diseases—a review. Mem. Inst. Oswaldo Cruz 100:475-506. [DOI] [PubMed] [Google Scholar]
- 3.Bender, S. L., S. Mehdi, and J. R. Knowles. 1989. Dehydroquinate synthase: the role of divalent metal cations and of nicotinamide adenine dinucleotide in catalysis. Biochemistry 28:7555-7560. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, R. 1990. The shikimate pathway—a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25:307-384. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2006. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs worldwide. Morb. Mortal. Wkly. Rep. 55:301-305. [PubMed] [Google Scholar]
- 7.Chapman, T. 2005. Pure but not simple. Nature 434:795-798. [DOI] [PubMed] [Google Scholar]
- 8.Chassaigne, H., and R. Lobinski. 1998. Characterization of horse kidney metalothionein isoforms by electrospray MS and reversed-phase HPLC-electrospray MS. Analyst 123:2125-2130. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 11.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman, S. E., and R. E. Chaisson. 2007. From magic bullets back to the Magic Mountain: the rise of extensively drug-resistant tuberculosis. Nat. Med. 13:295-298. [DOI] [PubMed] [Google Scholar]
- 13.Ducati, R. G., A. Ruffino-Netto, L. A. Basso, and D. S. Santos. 2006. The resumption of consumption—a review on tuberculosis. Mem. Inst. Oswaldo Cruz 101:697-714. [DOI] [PubMed] [Google Scholar]
- 14.Ducati, R. G., L. A. Basso, and D. S. Santos. 2007. Mycobacterial shikimate pathway enzymes as targets for drug design. Curr. Drug Targets 8:423-435. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, K., R. M. Edwards, and J. R. Coggins. 1987. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem. J. 246:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost, J. W., J. L. Bender, J. T. Kadonaga, and J. R. Knowles. 1984. Dehydroquinate synthase from Escherichia coli: purification, cloning and construction of overproducers of the enzyme. Biochemistry 23:4470-4475. [DOI] [PubMed] [Google Scholar]
- 17.Gaertner, F. W., and K. W. Cole. 1977. A cluster-gene: evidence for one gene, one polypeptide, five enzymes. Biochem. Biophys. Res. Commun. 75:259-264. [DOI] [PubMed] [Google Scholar]
- 18.Garbe, T., S. Servos, A. Hawkins, G. Dimitriadis, D. Young, G. Dougan, and I. Charles. 1991. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol. Gen. Genet. 228:385-392. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, T. H., E. S. Kawaski, S. R. Punreddy, and M. S. Osburne. 1998. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene 209:95-103. [DOI] [PubMed] [Google Scholar]
- 20.Hasan, N., and E. W. Nester. 1978. Dehydroquinate synthase in Bacillus subtilis. An enzyme associated with chorismate synthase and flavin reductase. J. Biol. Chem. 253:4999-5004. [PubMed] [Google Scholar]
- 21.Hawkins, A. R., H. K. Lamb, J. D. Moore, I. G. Charles, and C. F. Roberts. 1993. The pre-chorismate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects. J. Gen. Microbiol. 139:2891-2899. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lambert, J. M., M. R. Boocock, and J. R. Coggins. 1985. The 3-dehydroquinate synthase activity of the pentafunctional arom enzyme complex of Neurospora crassa is Zn2+-dependent. Biochem. J. 226:817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magalhães, M. L. B., C. P. Pereira, L. A. Basso, and D. S. Santos. 2002. Cloning and expression of functional shikimate dehydrogenase (EC 1.1.1.25) from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 26:59-64. [DOI] [PubMed] [Google Scholar]
- 25.Moore, J. D., M. A. Skinner, D. R. Swatman, A. R. Hawkins, and K. A. Brown. 1998. Reactivation of 3-dehydroquinate synthase by lanthanide cations. J. Am. Chem. Soc. 120:7105-7106. [Google Scholar]
- 26.Nichols, C. E., J. Ren, K. Leslie, B. Dhaliwal, M. Lockyer, I. Charles, A. R. Hawkins, and D. K. Stammers. 2004. Comparison of ligand-induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases. J. Mol. Biol. 343:533-546. [DOI] [PubMed] [Google Scholar]
- 27.Nixon, A. E., J. L. Hunter, G. Bonifacio, J. F. Eccleston, and M. R. Webb. 1998. Purine nucleoside phosphorylase: its use in a spectroscopic assay for inorganic phosphate and for removing inorganic phosphate with the aid of phosphodeoxyribomutase. Anal. Biochem. 265:299-307. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, J. S., C. A. Pinto, L. A. Basso, and D. S. Santos. 2001. Cloning and overexpression in soluble form of functional shikimate kinase and 5-enolpyruvylshikimate 3-phosphate synthase enzymes from Mycobacterium tuberculosis. Protein Expr. Purif. 22:430-435. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira, J. S., M. A. Mendes, M. S. Palma, L. A. Basso, and D. S. Santos. 2003. One-step purification of 5-enolpyruvylshikimate-3-phosphate synthase enzyme from Mycobacterium tuberculosis. Protein Expr. Purif. 28:287-292. [DOI] [PubMed] [Google Scholar]
- 30.Parish, T., and N. G. Stoker. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148:3069-3077. [DOI] [PubMed] [Google Scholar]
- 31.Pomp, D., and J. F. Medrano. 1991. Organic solvents as facilitators of a polymerase chain reaction. BioTechniques 10:58-59. [PubMed] [Google Scholar]
- 32.Rizzi, C., J. Frazzon, F. Ely, P. G. Weber, I. O. Fonseca, M. Gallas, J. S. Oliveira, M. A. Mendes, B. M. Souza, M. S. Palma, D. S. Santos, and L. A. Basso. 2005. DAHP synthase from Mycobacterium tuberculosis H37Rv: cloning, expression, and purification of functional enzyme. Protein Expr. Purif. 40:23-30. [DOI] [PubMed] [Google Scholar]
- 33.Silva, R. G., L. P. S. Carvalho, J. S. Oliveira, C. A. Pinto, M. A. Mendes, M. S. Palma, L. A. Basso, and D. S. Santos. 2003. Cloning, overexpression, and purification of functional human purine nucleoside phosphorylase. Protein Expr. Purif. 27:158-164. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan, P. R., J. Rothschild, and D. B. Sprinson. 1963. The enzymic conversion of 3-deoxy-d-arabino-heptulosonic acid 7-phosphate to 5-dehydroquinate. J. Biol. Chem. 238:3176-3182. [PubMed] [Google Scholar]
- 35.van den Hombergh, J. P. T. W., J. D. Moore, I. G. Charles, and A. R. Hawkins. 1992. Overproduction in Escherichia coli of the dehydroquinate synthase domain of the Aspergillus nidulans pentafunctional AROM protein. Biochem. J. 284:861-867. [PMC free article] [PubMed] [Google Scholar]
- 36.Vincentelli, R., C. Bignon, A. Gruez, S. Canaan, G. Sulzenbacher, M. Tegoni, V. Campanacci, and C. Cambillau. 2003. Medium-scale structural genomics: strategies for protein expression and crystallization. Acc. Chem. Res. 36:165-172. [DOI] [PubMed] [Google Scholar]
- 37.Webb, M. R. 1992. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA 89:4884-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winship, P. R. 1989. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 17:1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization. 2006. Global tuberculosis control: surveillance, planning, financing. WHO report 2006. World Health Organization, Geneva, Switzerland.