Abstract
c-type cytochromes are located partially or completely in the periplasm of gram-negative bacteria, and the heme prosthetic group is covalently bound to the protein. The cytochrome c maturation (Ccm) multiprotein system is required for transport of heme to the periplasm and its covalent linkage to the peptide. Other cytochromes and hemoglobins contain a noncovalently bound heme and do not require accessory proteins for assembly. Here we show that Bradyrhizobium japonicum cytochrome c550 polypeptide accumulation in Escherichia coli was heme dependent, with very low levels found in heme-deficient cells. However, apoproteins of the periplasmic E. coli cytochrome b562 or the cytosolic Vitreoscilla hemoglobin (Vhb) accumulated independently of the heme status. Mutation of the heme-binding cysteines of cytochrome c550 or the absence of Ccm also resulted in a low apoprotein level. These levels were restored in a degP mutant strain, showing that apocytochrome c550 is degraded by the periplasmic protease DegP. Introduction of the cytochrome c heme-binding motif CXXCH into cytochrome b562 (c-b562) resulted in a c-type cytochrome covalently bound to heme in a Ccm-dependent manner. This variant polypeptide was stable in heme-deficient cells but was degraded by DegP in the absence of Ccm. Furthermore, a Vhb variant containing a periplasmic signal peptide and a CXXCH motif did not form a c-type cytochrome, but accumulation was Ccm dependent nonetheless. The data show that the cytochrome c heme-binding motif is an instability element and that stabilization by Ccm does not require ligation of the heme moiety to the protein.
Cytochromes are heme proteins that participate in respiration, photosynthesis, and other aspects of oxygen metabolism. Unlike most other heme proteins, c-type cytochromes have the heme moiety covalently bound to the protein via thioether linkages with vicinal cysteines in the context CXXCH (reviewed in references 15 and 20). Cytochromes c are located outside the cytoplasmic membrane, which corresponds to the periplasmic space in gram-negative bacteria. They may be completely soluble within that space or anchored to the cytoplasmic membrane at the N terminus. Thus, cytochrome c biosynthesis requires the translocation of both the apoprotein and heme moiety from the cytoplasmic side of the membrane to the periplasm and the covalent attachment of heme to the protein. Apocytochrome c is translocated to the periplasm by the Sec pathway (29), followed by heme attachment and folding of the protein into its native confirmation. Heme translocation and its covalent attachment to the apoprotein are carried out by the so-called system I cytochrome c maturation (Ccm) pathway in Escherichia coli, encoded by the ccmABCDEFGH operon. The system II pathway found in other bacteria contains different and fewer proteins, but it likely carries out the same function as the type I system (15, 16). System I requires less cellular heme for cytochrome c formation than does system II, probably because the latter system has a low-affinity heme-binding site (24). System III is composed of a heme lyase and is found in mitochondria.
Whereas the cytochrome polypeptide is a primary translation product, heme is the end product of a biosynthetic pathway. Thus, heme and apoprotein syntheses are presumably coordinated for the formation of heme proteins. Cytochrome c1 apoprotein accumulation depends on heme in Bradyrhizobium japonicum, as concluded from the observation that little polypeptide is found in heme-deficient cells (11). Heme has little effect on B. japonicum cytochrome c1 transcripts, but mutations in the heme-binding site of the protein abrogate polypeptide accumulation (11). Thus, heme likely exerts its effect primarily at the level of protein stability. Because B. japonicum contains multiple copies of genes encoding putative periplasmic proteases (based on the genome sequence [13]) and expresses the ccm genes constitutively, it is a difficult system with which to further pursue studies of heme-dependent control of c-type cytochromes in bacteria. However, the expression of heterologous bacterial genes encoding soluble, periplasmic c-type cytochromes in E. coli has been used extensively as a model for studying cytochrome c biogenesis (3, 9, 24, 30, 32). The ccm genes are expressed only anaerobically in E. coli (30), but the cloned genes can be expressed aerobically from a plasmid under the control of a constitutive promoter (3). In the present study, we use this system to study the control of cytochrome c by heme and to identify features of the cytochrome c maturation process that contribute to stability.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α was used for propagation of plasmids. Cells containing pBluescript II-based plasmids were grown in LB medium containing 200 μg/ml ampicillin. Mutant strains S905 (17), KS474 (27), and S906 are defective in hemA, degP, and both hemA and degP, respectively. Transfer of the degP allele from KS474 to S905 was achieved using P1vir-mediated generalized transduction (18). The Tn5-containing strains KS474 and S906 were grown in medium containing 50 μg/ml kanamycin. Media were supplemented with 60 μM or 600 μM δ-aminolevulinic acid (ALA) for either low- or high-ALA conditions, respectively. Strains containing plasmid pEC86, which contains ccmABCDEFGH expressed from a constitutive promoter (3), was grown in LB medium containing 25 μg/ml chloramphenicol.
Analyses of cytochrome c550, cytochrome b562, and Vhb.
Polypeptides of cytochrome c550, cytochrome b562, and the Vitreoscilla hemoglobin (Vhb) apo- or holocytochrome were measured by immunoblot analysis. To prepare whole-cell extracts, cells were harvested by centrifugation. The cell pellets were washed once with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4) and then resuspended in 200 μl of PBS. The cell solutions were mixed with equal volumes of 2× sodium dodecyl sulfate (SDS) sample buffer (20 mM sodium phosphate, 20% glycerol, 4% SDS, 0.2 M dithiothreitol, 0.001% bromophenol blue) and boiled for 5 min, followed by centrifugation at 13,000 × g for 1 min. Aliquots of the supernatant were loaded onto 15% SDS-polyacrylamide gels and analyzed by immunoblotting using anti-tetra-His antibody (QIAGEN, Valencia, CA). Protein concentrations were determined using a bicinchoninic acid assay (Sigma). Bovine serum albumin was used as a standard.
Analysis of holoproteins by absorption spectroscopy.
Heme proteins were expressed in an E. coli hemA strain grown with either 60 μM or 600 μM of ALA. Cells expressing E. coli cytochrome c550, E. coli cytochrome b562, or Vhb were broken by two passages through a French pressure cell (18,000 lb/in2), followed by centrifugation twice at 13,000 × g for 10 min. The resulting supernatant was centrifuged at 140,000 × g for 90 min. A sample of the supernatant fraction (12 mg/ml) was reduced with sodium dithionite, and the spectrum was determined with a DW-2000 UV-Vis spectrophotometer against a PBS reference. A pyridine hemochromogen assay was done as described previously (26).
Construction of plasmids encoding His-tagged cytochrome c550, cytochrome b562, and Vhb.
pRJ3290, which carries the B. japonicum cytochrome c550 coding region with codons for a C-terminal six-His tag, was a gift from L. Thöny-Meyer (30). The E. coli cytochrome b562 coding region was amplified by PCR, using E. coli strain BL21 genomic DNA as the template and the following forward and reverse primers: 5′-TATCATATGCGTAAAAGCCTGTTAGC-3′ and 5′-TATGAATTCTTAGTGGTGGTGGTGGTGGTGACGATACTTCTGGTGATAGG-3′, respectively. The forward primer includes an NdeI site which contains an ATG initiation codon. The reverse primer contains an EcoRI cloning site and six consecutive histidine codons. The PCR product was ligated into the EcoRV site of pBluescript KS(+). The resultant plasmid was digested with NdeI and EcoRI, whose sites originated with the PCR primers. The resultant fragment, which contained the coding sequence for E. coli cytochrome b562 with a C-terminal six-His tag, was ligated with NdeI/EcoRI-linearized pISC2, which contains an arabinose-inducible promoter and a ribosome binding site. The plasmid pVhbHis, which contains the upstream promoter region, ribosome binding site, and coding region of Vhb with a C-terminal six-His tag in pUC8, was constructed on the basis of pVhb (6), whereby the six-His tag was added to the C terminus of Vhb to facilitate Western blot assay.
Construction of leader peptide plus Vhb by PCR technique using splicing with overlap extension.
A leader signal sequence was added to the N-terminal part of the Vhb coding region by a PCR technique using splicing with overlap extension (12). The 93-nucleotide (nt) primer A (5′-CATATGACAAAACTGACTTTCGGCGCGCTAGTTGCCCTCGCCATGACTGCCGCAGCGTCCACCGCCAT GTCTTCCAAAGCGATGGCGCAGGAC-3′), which encodes an NdeI site as well as the 30-amino-acid N-terminal signal sequence of B. japonicum cytochrome c550, was synthesized by Integrated DNA Technologies (Coralville, IA). The coding region of Vhb was PCR amplified using primer B (5′-CCAAAGCGATGGCGCAGGACTTAGACCAGCAAACCATTAA-3′), which contains the last 20 nt of primer A and the first 20 nt of the Vhb coding region, and primer C (5′-TATGAATTCTTAGTGGTGGTGGTGGTGGTGACGATACTTCTGGTGATAGG-3′), which contains an EcoRI cloning site and six consecutive histidine codons. The PCR product was mixed with primer A and PCR amplified again using the primer 5′-CACCATATGACAAAACTGACTTTCGGCG-3′ and primer C. The resultant PCR product was cloned into pBluescript KS(+) to generate the leader peptide plus Vhb (SPVhb), which was then either subcloned into pUC8 immediately downstream of the Vhb promoter or used for further mutagenesis.
Mutagenesis of the genes encoding cytochrome c550, cytochrome b562, and the leader peptide plus Vhb.
Mutagenesis of the genes encoding cytochrome c550, cytochrome b562, and the leader peptide plus Vhb was carried out using a QuikChange kit (Stratagene, La Jolla, CA) as described previously (33).
Staining of covalently attached heme.
Heme stains were performed as described previously (8), with modifications. Cells expressing E. coli cytochrome b562 or cytochrome b562 with c-type heme-binding cysteines were broken by two passages through a French pressure cell (18,000 lb/in2), followed by centrifugation twice at 13,000 × g for 10 min. The resulting supernatant was centrifuged at 140,000 × g for 90 min. The resultant supernatant was mixed with an equal volume of SDS sample buffer without the addition of either dithiothreitol or β-mercaptoethanol to avoid the loss of heme binding (20 mM sodium phosphate, 20% glycerol, 4% SDS, 0.2 M dithiothreitol, 0.001% bromophenol blue) and boiled for 5 min, followed by centrifugation at 13,000 × g for 1 min. Aliquots of the supernatant were loaded onto 15% SDS-polyacrylamide gels and analyzed by heme staining using a Supersignal West Femto maximum sensitivity substrate kit (Pierce, Rockford, IL). The chemiluminescence of heme stains was detected by exposing and developing them on F-BX810 blue X-ray films (Phenix, Hayward, CA). Protein concentrations were determined using a bicinchoninic acid assay (Sigma). Bovine serum albumin was used as a standard.
RESULTS
Heme regulates cytochrome c550 peptide accumulation.
In a previous study, we showed that a defect in heme synthesis or in a heme-binding site abrogated the accumulation of cytochrome c1 protein in B. japonicum, presumably due to protein degradation (11). We chose to further address this system in E. coli, which offers several experimental advantages. In addition, we used cytochrome c550 from B. japonicum because it is a soluble periplasmic protein in E. coli and in B. japonicum (30). The genes encoding the Ccm system are not normally expressed in aerobic E. coli cells but can be expressed readily from a constitutive promoter on a plasmid in those cells.
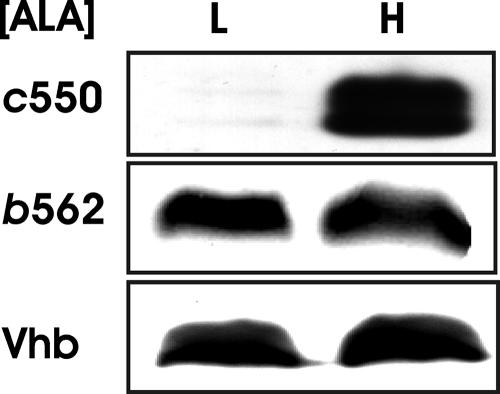
The hemA (gtrA) gene of E. coli encodes the heme biosynthetic enzyme glutamyl-tRNA reductase. Thus, a hemA mutant is heme defective but can be rescued by the addition of the heme precursor ALA to the growth medium. We examined the effects of heme on cytochrome c550 expression in the hemA mutant strain grown with low and high concentrations of ALA (Fig. 1). Cytochrome c550 accumulated in cells grown in ALA-containing medium, as determined by Western blot analysis. The peptide often appeared as a doublet, which was probably due to both processed and unprocessed peptide. The protein was almost undetectable in cells grown in low-ALA medium, showing that the peptide level was heme dependent. This is similar to what was observed previously for cytochrome c1 in B. japonicum cells (11).
FIG. 1.
Heme-dependent expression of heme protein polypeptides. B. japonicum cytochrome c550, E. coli cytochrome b562, and Vhb were expressed from a plasmid as His-tagged proteins in E. coli hemA strain S905 grown in medium containing either 60 μM (L) or 600 μM (H) ALA. Cells that expressed B. japonicum cytochrome c550 also harbored the ccm genes on pEC86. Cell extracts were analyzed by immunoblotting using antibodies against the His tag. Thirty micrograms of protein was loaded per lane.
Accumulation of b-type heme protein peptides is independent of heme.
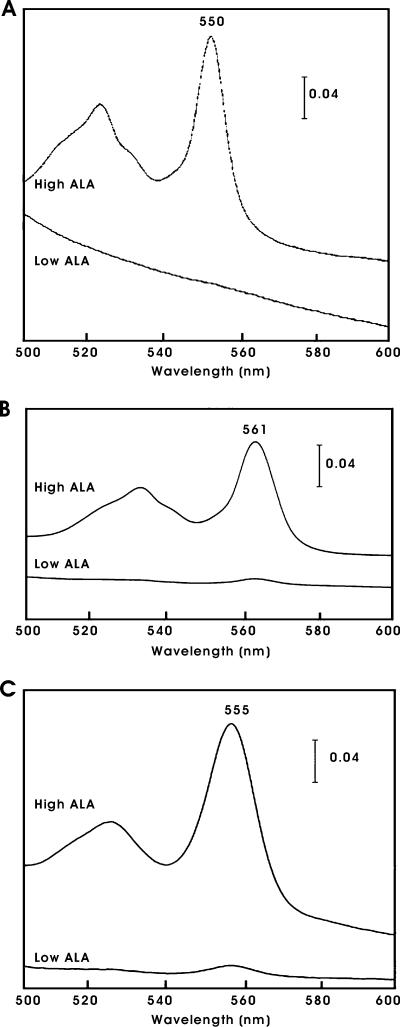
Whereas heme is linked covalently to the protein in c-type cytochromes, cytochromes b and hemoglobins have protoheme (b-type heme) associated noncovalently. We examined the effects of heme on protein accumulation of E. coli cytochrome b562 and Vhb. Cytochrome b562 is a periplasmic protein, whereas Vhb is found in the cytoplasm (4, 23). Both proteins accumulated in the hemA mutant grown in high- or low-ALA medium (Fig. 1). Thus, unlike the case for cytochrome c550, accumulation of cytochrome b562 and Vhb is independent of heme. To confirm that the proteins grown in low-ALA medium were heme deficient, we monitored the heme content of each of the heme proteins. The reduced state of a heme protein has a characteristic absorption spectrum. As expected, cytochromes c550 and b562 from cells grown in medium supplemented with ALA showed absorption peaks at 550 nm and 561 nm, respectively (Fig. 2A and B). However, for low-ALA medium, the spectral features were nearly absent, showing very low levels of heme. Vhb does not show a single sharp peak in the 500- to 600-nm region. Thus, we extracted the heme and took a spectrum of the pyridine hemochromagen (Fig. 2C). As observed with the other proteins, Vhb heme was detected in cells grown in high-ALA medium but was very low in cells grown in low-ALA medium. These data confirm that cells grown in low-ALA medium are heme deficient and that the apoproteins of cytochrome b562 and Vhb accumulate in heme-deficient cells.
FIG. 2.
Absorption spectra of heme proteins in heme-sufficient or heme-deficient cells. B. japonicum cytochrome c550 (A), E. coli cytochrome b562 (B), and Vhb (C) were overexpressed in E. coli hemA strain S905 grown in medium containing either 60 μM (low) or 600 μM (high) ALA. Cells that expressed B. japonicum cytochrome c550 also harbored the ccm genes on pEC86. The soluble cell fraction containing cytochrome c550 or cytochrome b562 was reduced with sodium dithionite (A and B). The dithionite-reduced spectrum of the pyridine hemochromagen of Vhb was taken (C).
Apocytochrome c550 is degraded in a heme-deficient strain or Ccm-deficient strain by the periplasmic protease DegP.
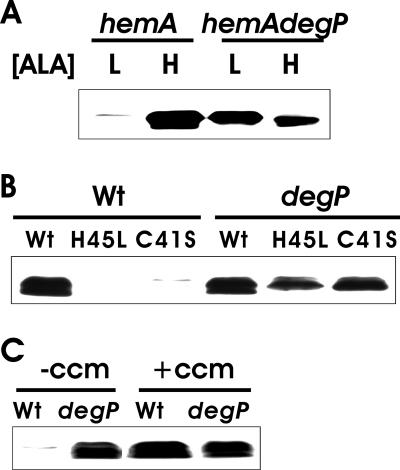
Heme could affect the accumulation of apocytochrome c550 at some step in the synthesis or degradation of the protein. To address the latter possibility, we examined cytochrome c550 polypeptide accumulation in a DegP-deficient strain. We chose DegP because it is located in the periplasm, as are cytochromes c, and it has a very broad substrate specificity (14). Cytochrome c550 apoprotein was measured by Western blot analysis of hemA and hemA degP strains. In heme-deficient cells (cells grown in low-ALA medium), the protein was not found for the hemA strain, but it was easily detected in the hemA degP double mutant growing as both heme-deficient and -sufficient cells (Fig. 3A). Thus, heme deficiency affects cytochrome c550 at the level of protein turnover, and DegP is required for this degradation. These data also show that the apoprotein is degraded in the periplasm and that heme is not required for its synthesis or targeting.
FIG. 3.
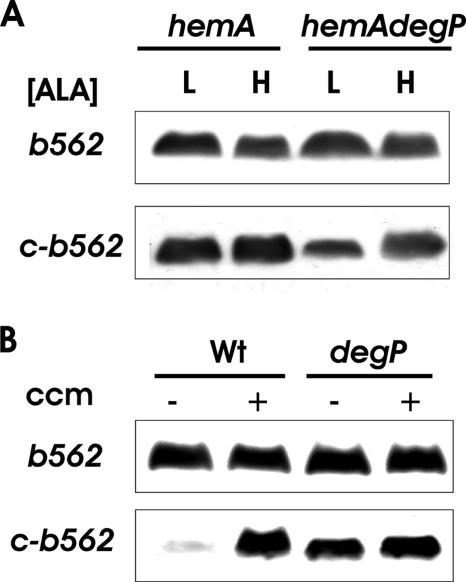
Accumulation of cytochrome c550 polypeptide under various conditions in degP+ and degP backgrounds and in the absence or presence of Ccm. (A) B. japonicum cytochrome c550 was expressed in hemA strain S905 or hemA degP strain S906 grown in medium containing either 60 μM (L) or 600 μM (H) ALA. The cells also contained pEC86, which carries the ccm genes. Cell extracts were analyzed by immunoblotting using antibodies against a His4 tag. Thirty micrograms of protein was loaded per lane. (B) Effect of mutation of the heme-binding site on expression. Cell extracts were prepared from either wild-type strain KS272 (Wt) or degP strain KS474 harboring B. japonicum cytochrome c550 constructs which expressed unmutated (Wt) or 41Cys-to-Ser or 45His-to-Leu mutated cytochrome c550. The cells also contained pEC86, which carries the ccm genes. Proteins were analyzed as described for panel A. (C) Effect of ccm genes on cytochrome c550 expression. B. japonicum cytochrome c550 was expressed in either a degP+ (Wt) or degP E. coli strain in either the absence or presence of the plasmid pEC86 expressing ccm genes. Proteins were analyzed as described for panels A and B.
Cytochromes c contain heme covalently bound to the protein at two cysteinyl thiols in the motif CXXCH, which corresponds to 41CLACH in B. japonicum cytochrome c550 (30). The histidine is also required for binding (2). We showed previously that cytochrome c1 apoprotein did not accumulate in B. japonicum when the heme-binding site was mutated (11). Here we found that substitution of 41Cys to Ser or 45His to Leu within cytochrome c550 abolished protein accumulation in E. coli cells (Fig. 3B). Protein levels were restored in a degP strain, indicating that these mutant derivatives were degraded in the periplasm by DegP in the parent strain.
The Ccm system is required to transport heme to the periplasm and covalently ligate heme to the apoprotein. It was shown previously that cytochrome c550 does not accumulate in Ccm-deficient E. coli cells (5). We wanted to address whether abrogation of cytochrome c accumulation in a Ccm-defective background is the result of DegP-dependent degradation. In the present study, the ccm genes were expressed from a constitutive promoter in trans because they are not expressed in the chromosome in aerobically grown cells of E. coli (3). Thus, we examined cytochrome c550 levels in wild-type and degP backgrounds in the absence of the ccm-containing plasmid. We found that apoprotein expression was strictly dependent on the Ccm system in wild-type cells (Fig. 3C), similar to what was reported previously (5). However, cytochrome c550 peptides accumulated in degP cells independently of Ccm, showing a DegP requirement for degradation and confirming that translocation to the periplasm does not require the ccm gene products (29).
A cytochrome b562 variant with a c-type cytochrome heme-binding motif is dependent on Ccm, but not heme, for stability.
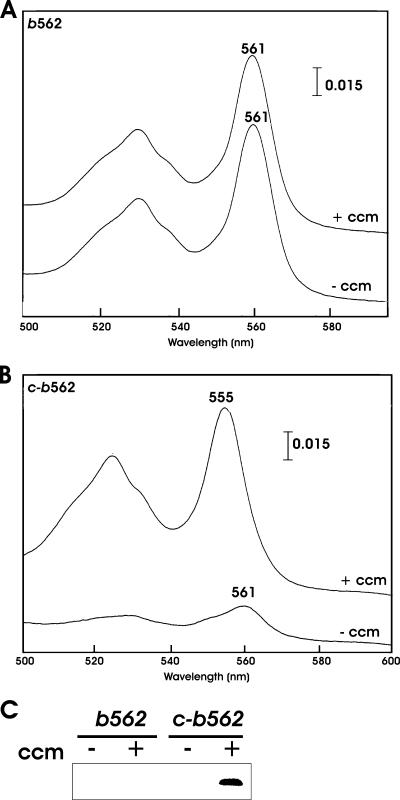
Cytochrome c550 and cytochrome b562 are both periplasmic heme proteins, but the former is unstable in a heme-deficient E. coli strain, whereas the latter is not (Fig. 1). Thus, we wanted to ask which features of the proteins account for the differences in stability. It was shown previously that introduction of a cytochrome c heme-binding motif into cytochrome b562 resulted in a covalently bound heme with spectral features of a c-type cytochrome (1, 7). A variant containing Arg98→Cys and Tyr101→Cys substitutions in cytochrome b562, near the axial ligand 102His, was made, resulting in a CXXCH motif. The wild type and the mutant protein, called c-b562, were expressed in E. coli in the presence of Ccm, and the soluble fraction of cells was analyzed spectrophotometrically (Fig. 4A and B). The wild-type cytochrome b562 had an absorption peak at 561 nm (Fig. 4A). Cytochrome c-b562 showed a shift to 555 nm (Fig. 4B), consistent with covalent binding to the heme vinyl groups found in cytochromes c. To confirm the covalent attachment of heme to the protein, a heme staining assay was carried out using the soluble fraction from cells overexpressing the cytochromes (Fig. 4C) (see Materials and Methods). Because the proteins were resolved under denaturing conditions, only covalently bound heme was detected by SDS-PAGE. Attached heme was detected on cytochrome c-b562 but not on cytochrome b562.
FIG. 4.
Effects of Ccm on E. coli cytochrome b562 and c-b562 absorption spectra and heme staining assay. E. coli cytochrome b562 (A) or a derivative containing heme-liganding cysteines (c-b562) (B) was expressed in the parent strain KS272 in either the presence or absence of the plasmid pEC86 expressing the E. coli ccm genes. Absorption spectra for the soluble cell fraction (12 mg/ml) were determined for the 500- to 600-nm region as described in the text. (C) Cell fractions were separated by SDS-polyacrylamide gel electrophoresis and stained for covalently bound heme as described in Material and Methods. Thirty micrograms of protein was loaded per lane.
To address whether the cytochrome b562 variant containing a cytochrome c heme-binding motif showed altered stability in a heme-deficient strain, the protein was expressed in the hemA strain and grown in either low- or high-ALA medium (Fig. 5A). Accumulation of the mutant protein was high and independent of the heme status, as found for wild-type cytochrome b562. Thus, heme per se does not stabilize the cytochrome c-b562 peptide.
FIG. 5.
Expression of E. coli cytochrome b562 and c-b562 polypeptides under various conditions. (A) Effects of heme and DegP on cytochrome b562 and c-b562 polypeptide accumulation. E. coli cytochrome b562 or c-b562 was expressed in either hemA strain S905 or hemA degP strain S906 grown in medium containing either 60 μM (L) or 600 μM (H) ALA. The cells also contained pEC86, which carries the ccm genes. Cell extracts were analyzed by immunoblotting using antibodies against the His tag. Thirty micrograms of protein was loaded per lane. (B) Effects of Ccm and DegP on cytochrome b562 and c-b562 polypeptide accumulation. E. coli cytochrome b562 or c-b562 was expressed in degP+ (Wt) strain KS272 or degP strain KS474 in either the absence or presence of the plasmid pEC86 containing the E. coli ccm genes. Proteins were analyzed as described for panel A.
Expression of the cytochrome b562 holoprotein was found to be independent of the Ccm system (Fig. 4A), as judged spectrophotometrically, which is in agreement with a previous report (31). Conversely, cytochrome c-b562 holoprotein expression was strongly dependent on Ccm (Fig. 4B). In the absence of Ccm, a low but spectroscopically detectable level of the cytochrome was found, with a major peak at 561 nm, as found for the wild-type protein. This was probably due to the fact that the axial ligands of cytochrome b562 were not removed in the construction of the variant containing the cytochrome c heme-binding motif. In addition, covalently bound heme was not detected in the heme staining assay for cytochrome c-b562 in the absence of Ccm (Fig. 4C).
Examination of cytochrome b562 and c-b562 peptides as a function of the Ccm status by Western blotting revealed that the variant containing the cytochrome c heme-binding motif was expressed strongly in the presence of Ccm but only at low levels in Ccm-deficient cells (Fig. 5B). However, a high protein level was found in a degP strain, independent of Ccm. These findings show that introduction of the CXXCH motif into cytochrome b562 destabilizes the protein in a DegP-dependent manner and that the stable protein requires Ccm. Furthermore, a deficiency in heme or in Ccm results in an apoprotein lacking its prosthetic group but does not yield the same outcome with respect to the stability of the cytochrome c-b562 apoprotein.
A Vhb variant with a cytochrome c heme-binding motif does not bind heme covalently but is dependent on Ccm.
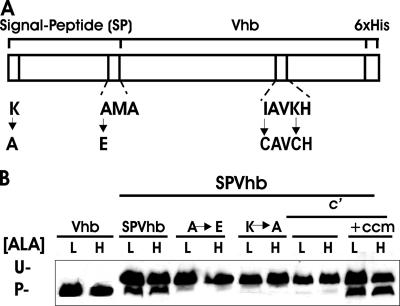
Vhb is expressed as a cytoplasmic protein in E. coli (23). We constructed a variant of Vhb that contains the N-terminal signal peptide from cytochrome c550 (SPVhb) (Fig. 6). This resulted in two proteins observed on a Western blot after SDS-PAGE, with one approximately the same size as wild-type Vhb and the other a larger protein. The large and small bands likely represent unprocessed and processed forms of SPVhb, respectively, based on the observation that mutations in the signal peptide region that prevent targeting (Lys3→Ala) or cleavage (Ala27→Glu) resulted in accumulation of only the larger band. The larger band was found in the membrane fraction of cells (data not shown), indicating that it does not get completely into the periplasm. The SPVhb derivative was modified further by introducing a CXXCH binding site which includes the axial histidine of Vhb (c′SPVhb) (Fig. 6). Surprisingly, spectroscopic analysis of this protein expressed in E. coli did not show spectral features of a c-type cytochrome, indicating that this variant does not contain covalently bound heme (data not shown). Nevertheless, accumulation of the processed protein was dependent on Ccm. These experiments were carried out in a hemA strain grown with low or high levels of ALA. Accumulation of neither the wild-type Vhb protein nor any of the derivatives was heme dependent. These findings suggest that the CXXCH motif is sufficient to destabilize a protein that cannot form a c-type heme ligation. These findings are consistent with the conclusion drawn for cytochrome c-b562 that stability conferred on a CXXCH-containing protein by Ccm does not require heme ligation to the protein.
FIG. 6.
Effects of heme and Ccm on expression of Vhb derivatives. (A) Vhb constructs. SPVhb was generated by fusion of the 30-amino-acid N-terminal signal sequence of B. japonicum c550 to Vhb as described in Materials and Methods. Lys3-to-Ala and Ala27-to-Glu mutations represent the periplasmic targeting and leader peptide cleavage-site mutations of SPVhb, respectively. c′SPVhb was generated by the addition of heme-binding cysteines Cys124 and Cys127 to SPVhb. (B) Wild-type or mutant derivatives of Vhb were expressed in hemA strain S905 grown in medium containing either 60 μM (L) or 600 μM (H) ALA. c′SPVhb, which contains heme-liganding cysteines, was grown in either the absence of presence of the plasmid pEC86 expressing the E. coli ccm genes. Cell extracts were analyzed by immunoblotting using antibodies against the His tag. Thirty micrograms of protein was loaded per lane.
DISCUSSION
In the present study, we found that cytochrome c expression is regulated by heme availability in E. coli at the level of protein stability. In the absence of heme, the peptide is degraded by the protease DegP. Furthermore, we show that protein stability conferred by the Ccm system can be separated from its role in heme ligation to the protein.
DegP is a periplasmic protease, and thus the persistence of cytochrome c550 polypeptide in the degP strain shows that a heme deficiency does not prevent targeting to the periplasm but rather affects stability within that space. In agreement with posttranslational control, mutations in cytochrome c550 that prevented heme binding also nearly abrogated polypeptide accumulation in a DegP-dependent manner (Fig. 3B). Finally, the loss of cytochrome c550 polypeptide in Ccm-deficient cells observed here (Fig. 3C) and reported by others (28) is DegP dependent as well.
Unlike cytochrome c550, polypeptides of cytochrome b562 or Vhb were expressed in cells, independent of the heme status. For some b-type cytochromes, heme may bind to a preformed pocket (19), suggesting at least some stability of the apoprotein. Biophysical studies of cytochrome b562 in vitro showed that removal of heme alters the tertiary structure but not the highly helical secondary structure (10).
In order to determine which feature of cytochrome c550 or its synthesis rendered it unstable compared with b-type cytochromes, we characterized a cytochrome b562 derivative containing the cytochrome c binding motif, CXXCH. This mutant derivative bound heme covalently and contained spectral features of a c-type cytochrome, which agrees with earlier reports (1, 4). Here we showed that the CXXCH motif in cytochrome c-b562 destabilized the polypeptide in the absence of Ccm even though accumulation of the derivative did not require heme. This differs from the case for cytochrome c550, which was unstable in heme-deficient cells, although it also required Ccm for stability (Fig. 3 to 5). Thus, whereas analysis of cytochrome c550 indicates that any condition resulting in apoprotein not bound to heme leads to degradation, the stability of the cytochrome c-b562 polypeptide depends on the method by which the apoprotein is generated. The Ccm system is responsible for transporting heme into the periplasm, chaperoning heme to the polypeptide, keeping the cysteinyl thiols reduced, and catalyzing covalent linkage of the two moieties (15, 20, 25). Our findings rule out any of the heme-dependent steps in the stabilization of cytochrome c-b562 by Ccm. However, construction of the cytochrome c heme-binding motif in cytochrome b562 introduces cysteine residues that have the potential to oxidize in the oxidizing environment of the periplasm. A disulfide bond within the protein may prevent normal folding and lead to degradation. Thus, it is plausible that Ccm stabilizes cytochrome c-b562 by keeping the cysteine thiols reduced, at least until the protein is folded. This could explain why Ccm stabilizes cytochrome c-b562 without involving heme ligation. Additional studies are needed to confirm or refute this idea.
We note a previous study in which the expression of cytochrome c-b562 in Ccm-deficient cells resulted in an accumulation of holoprotein with the heme rotated 180 degrees relative to the correct orientation, which was also the major product formed from uncatalyzed heme ligation in vitro (1). This differs from the current work, in which little protein accumulated at all in Ccm-deficient cells. Although we do not know the basis for the different results, it is possible that the cytochrome c-b562 was expressed at higher levels in the previous work, allowing retention of the protein for nonenzymatic ligation with heme.
Some c-type cytochromes are stable in the absence of heme, as judged by the retention of polypeptide in heme-deficient cells (21, 22), like that found for cytochrome c-b562. It would be interesting to learn whether those peptides require Ccm activity for stability.
Acknowledgments
We thank Mark Sutton for help with E. coli P1 transduction constructions and Linda Thöny-Meyer, Hauke Hennecke, and Jon Beckwith for strains and plasmids.
This work was supported by National Institutes of Health grant R01 GM067966 to M.R.O.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Allen, J. W., P. D. Barker, and S. J. Ferguson. 2003. A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J. Biol. Chem. 278:52075-52083. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. W., N. Leach, and S. J. Ferguson. 2005. The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem. J. 389:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arslan, E., H. Schulz, R. Zufferey, P. Kunzler, and L. Thöny-Meyer. 1998. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251:744-747. [DOI] [PubMed] [Google Scholar]
- 4.Barker, P. D., E. P. Nerou, S. M. Freund, and I. M. Fearnley. 1995. Conversion of cytochrome b562 to c-type cytochromes. Biochemistry 34:15191-15203. [DOI] [PubMed] [Google Scholar]
- 5.Bott, M., L. Thöny-Meyer, H. Loferer, S. Rossbach, R. E. Tully, D. Keister, C. A. Appleby, and H. Hennecke. 1995. Bradyrhizobium japonicum cytochrome c550 is required for nitrate respiration but not for symbiotic nitrogen fixation. J. Bacteriol. 177:2214-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikshit, K. L., and D. A. Webster. 1988. Cloning, characterization and expression of the bacterial globin gene from Vitreoscilla in Escherichia coli. Gene 70:377-386. [DOI] [PubMed] [Google Scholar]
- 7.Faraone-Mennella, J., F. A. Tezcan, H. B. Gray, and J. R. Winkler. 2006. Stability and folding kinetics of structurally characterized cytochrome c-b562. Biochemistry 45:10504-10511. [DOI] [PubMed] [Google Scholar]
- 8.Feissner, R., Y. Xiang, and R. G. Kranz. 2003. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal. Biochem. 315:90-94. [DOI] [PubMed] [Google Scholar]
- 9.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, J. A. Loughman, K. W. Earley, and R. G. Kranz. 2006. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol. Microbiol. 60:563-577. [DOI] [PubMed] [Google Scholar]
- 10.Feng, Y. Q., and S. G. Sligar. 1991. Effect of heme binding on the structure and stability of Escherichia coli apocytochrome b562. Biochemistry 30:10150-10155. [DOI] [PubMed] [Google Scholar]
- 11.Gao, T., and M. R. O'Brian. 2005. Iron-dependent cytochrome c1 expression is mediated by the status of heme in Bradyrhizobium japonicum. J. Bacteriol. 187:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 13.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 14.Kolmar, H., P. R. Waller, and R. T. Sauer. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29:383-396. [DOI] [PubMed] [Google Scholar]
- 16.Kranz, R. G., C. S. Beckett, and B. S. Goldman. 2002. Genomic analyses of bacterial respiratory and cytochrome c assembly systems: Bordetella as a model for the system II cytochrome c biogenesis pathway. Res. Microbiol. 153:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Leong, S. A., D. S. Ditta, and D. R. Helinski. 1982. Heme synthesis in Rhizobium. Identification of a cloned gene coding for δ-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724-8730. [PubMed] [Google Scholar]
- 18.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 19.Moore, C. D., and J. T. Lecomte. 1993. Characterization of an independent structural unit in apocytochrome b5. Biochemistry 32:199-207. [DOI] [PubMed] [Google Scholar]
- 20.O'Brian, M. R., and L. Thöny-Meyer. 2002. Biochemistry, regulation and genomics of haem biosynthesis in prokaryotes. Adv. Microb. Physiol. 46:257-318. [DOI] [PubMed] [Google Scholar]
- 21.Page, M. D., and S. J. Ferguson. 1990. Apo forms of cytochrome c550 and cytochrome cd1 are translocated to the periplasm of Paracoccus denitrificans in the absence of haem incorporation caused either mutation or inhibition of haem synthesis. Mol. Microbiol. 4:1181-1192. [DOI] [PubMed] [Google Scholar]
- 22.Page, M. D., and S. J. Ferguson. 1989. A bacterial c-type cytochrome can be translocated to the periplasm as an apo form; the biosynthesis of cytochrome cd1 (nitrite reductase) from Paracoccus denitrificans. Mol. Microbiol. 3:653-661. [DOI] [PubMed] [Google Scholar]
- 23.Ramandeep, K., W. Hwang, M. Raje, K. J. Kim, B. C. Stark, K. L. Dikshit, and D. A. Webster. 2001. Vitreoscilla hemoglobin. Intracellular localization and binding to membranes. J. Biol. Chem. 276:24781-24789. [DOI] [PubMed] [Google Scholar]
- 24.Richard-Fogal, C. L., E. R. Frawley, R. E. Feissner, and R. G. Kranz. 2007. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J. Bacteriol. 189:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setterdahl, A. T., B. S. Goldman, M. Hirasawa, P. Jacquot, A. J. Smith, R. G. Kranz, and D. B. Knaff. 2000. Oxidation-reduction properties of disulfide-containing proteins of the Rhodobacter capsulatus cytochrome c biogenesis system. Biochemistry 39:10172-10176. [DOI] [PubMed] [Google Scholar]
- 26.Smith, K. M. 1975. Porphyrins and metalloporphyrins. Elsevier Scientific, Amsterdam, The Netherlands.
- 27.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thöny-Meyer, L., F. Fischer, P. Kunzler, D. Ritz, and H. Hennecke. 1995. Escherichia coli genes required for cytochrome c maturation. J. Bacteriol. 177:4321-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thöny-Meyer, L., and P. Kunzler. 1997. Translocation to the periplasm and signal sequence cleavage of preapocytochrome c depend on sec and lep, but not on the ccm gene products. Eur. J. Biochem. 246:794-799. [DOI] [PubMed] [Google Scholar]
- 30.Thöny-Meyer, L., P. Kunzler, and H. Hennecke. 1996. Requirements for maturation of Bradyrhizobium japonicum cytochrome c550 in Escherichia coli. Eur. J. Biochem. 235:754-761. [DOI] [PubMed] [Google Scholar]
- 31.Throne-Holst, M., L. Thöny-Meyer, and L. Hederstedt. 1997. Escherichia coli ccm in-frame deletion mutants can produce periplasmic cytochrome b but not cytochrome c. FEBS Lett. 410:351-355. [DOI] [PubMed] [Google Scholar]
- 32.Tomlinson, E. J., and S. J. Ferguson. 2000. Conversion of a c type cytochrome to a b type that spontaneously forms in vitro from apo protein and heme: implications for c type cytochrome biogenesis and folding. Proc. Natl. Acad. Sci. USA 97:5156-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, J., K. Ishimori, and M. R. O'Brian. 2005. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J. Biol. Chem. 280:7671-7676. [DOI] [PubMed] [Google Scholar]