Abstract
NF-κB transcription factors activate genes important for immune response, inflammation, and cell survival. P-TEFb and DSIF, which are positive and negative transcription elongation factors, respectively, both regulate NF-κB-induced transcription, but the mechanism underlying their recruitment to NF-κB target genes is unknown. We show here that upon induction of NF-κB, a subset of target genes is regulated differentially by either P-TEFb or DSIF. The regulation of these genes and their occupancy by these elongation factors are dependent on the NF-κB enhancer and the core promoter type. Converting a TATA-less promoter to a TATA promoter switches the regulation of NF-κB from DSIF to P-TEFb. Accumulation or displacement of DSIF and P-TEFb is dictated by the formation of distinct initiation complexes (TFIID dependent or independent) on the two types of core promoter. The underlying mechanism for the dissociation of DSIF from TATA promoters upon NF-κB activation involves the phosphorylation of RNA polymerase II by P-TEFb. The results highlight a regulatory link between the initiation and the elongation phases of the transcription reaction and broaden our comprehension of the NF-κB pathway.
Transcription of protein-coding genes by RNA polymerase II (Pol II) is a multistep process, each step being a target for regulation and critical for the production of mature mRNA (27, 29). A number of factors that control RNA Pol II elongation have been characterized in recent years. Among these are the positive elongation factor P-TEFb, which induces Pol II processivity by facilitating the transition from the early to the late elongation phase (24), and two negative elongation factors, DSIF (DRB sensitivity inducing factor) (31) and NELF (negative elongation factor) (37). In vitro, P-TEFb alleviates transcription inhibition by DSIF (25, 32).
NF-κB is a transcription factor central to the cellular response to a broad range of extracellular signals, including inflammatory cytokines, tumor promoters, and chemotherapeutic agents. In response to these agents, NF-κB induces the expression of cell cycle regulators, pro- and antiapoptotic factors, inflammatory cytokines, chemokines, adhesion molecules, and many other factors (22). In unstimulated cells, NF-κB is retained in the cytoplasm by IκB proteins. NF-κB-activating signals trigger degradation of IκB and nuclear translocation of NF-κB, which result in activation of responsive genes (14). A subset of early response genes that includes IκBα and A20 are themselves negative regulators of the NF-κB pathway and so form a negative feedback loop. Transcriptional control of these genes is likely to influence the strength and the duration of the inflammatory signal.
Induction of NF-κB target genes is remarkably fast, and the mechanism underlying their rapid transcriptional activation was investigated previously. It was found that the promoters of NF-κB-regulated genes are bound by the general transcription machinery prior to NF-κB activation, and subsequent activation by NF-κB increases the rate of the transcription cycles (reinitiation) rather than promoting preinitiation complex formation (2). Further experiments with the A20 NF-κB target gene revealed that both the basal and the NF-κB-induced transcription are repressed at the level of elongation. We identified the inhibitory factor as DSIF, which in this system acts without NELF (1). On the other hand, NF-κB-induced transcription of the interleukin 8 but not the IκBα gene was shown to be regulated by the positive elongation factor P-TEFb (4, 18). Thus, NF-κB target genes are subjected to regulation by both positive and negative transcription elongation factors. However the mechanism underlying the differential control and recruitment of these factors to NF-κB target genes is currently unknown.
Here we investigated the regulation of NF-κB-mediated transcription by DSIF and P-TEFb. Our data revealed that DSIF attenuation of NF-κB is promoter dependent and requires the NF-κB response element to be in the context of a TATA-less core promoter, which, in turn, enhances DSIF occupancy upon NF-κB induction. By contrast, TATA box-containing NF-κB promoters are not targeted for inhibition by DSIF, and in these genes NF-κB diminishes DSIF occupancy. Remarkably, the core promoter also influences regulation and recruitment of the positive elongation factor P-TEFb, but inversely to DSIF. We found that the two core promoter types dictate formation of distinct initiation complexes that are activated by NF-κB, thereby linking the initiation machinery to elongation control. Thus, the core promoter type, via the formation of distinct initiation complexes, affects the extent of NF-κB activation by reducing or facilitating transcription elongation rate.
MATERIALS AND METHODS
Plasmid constructions.
The A20, A20 mNF-κBs, 2κB-A20, 2κB(A20)-α-actin, and DSIF RNA interference (RNAi) 1 were described previously (1, 2). The DSIF RNAi 2 and cdk9 RNAi were constructed according to Brummelkamp et al. (6), using pSuper plasmid and synthetic oligonucleotides targeting the 5′-GTTCATTGCCTACCAGTTC and 5′-CCAAAGCTTCCCCCTATAA sequences, corresponding to positions 784 to 802 of DSIF p160 and positions 358 to 376 of cdk9 mRNAs, respectively. A20-TATA was constructed by replacing a PmlI-XmaI fragment from the A20 promoter with a double-stranded oligonucleotide containing the TATA mutation: 5′-CCTACAACCCGTATAAAACTGAAACGGGGC; reverse, 5′-GCCCCGTTTCAGTTTTATACGGGTTGTAGG. The promoters of IP-10, RANTES, and IκBα were amplified by PCR from genomic DNA and cloned in the promoterless reporter gene pGL2-basic (Promega). The primers used are as follows: IP-10, 5′-CAAGGCACTCATCTGATTTC and 5′-GACAAAGCTTCGGGATGTCTCTCAGCGGTG; RANTES, 5′-CCTATGACCAGGATGAAAGC and 5′-AGCCAAGCTTAGAGGCTGTGCGAGGTCCAC; IκBα, 5′-AAGGCTCACTTGCAGAGGG and 5′-GGACTGCTGTGGGCTCTG. All the constructs were verified by sequencing.
Transient-transfection assays and ChIP.
293T cells (human embryonic kidney fibroblasts) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transfections were performed by using the standard CaPO4 method. To avoid basal NF-κB activity, cells were kept from reaching confluence and replated no more than nine times. For reporter assays, subconfluent cells were transfected in a 24-well plate using 1.1 μg pSuper or DSIF RNAi plasmid, 20 ng of the reporter plasmids, 1 ng Rous sarcoma virus (RSV) promoter-driven Renilla luciferase reporter plasmid, 10 ng CMV-GFP, and 1 ng p65/RelA. At 48 h after transfection, cells were harvested and their luciferase and Renilla luciferase activities were measured. For the chromatin immunoprecipitation (ChIP) assay, subconfluent cells in 100-mm dishes were transfected with 0.2 μg of the reporter plasmids, and 24 h later, cells were left untreated or treated with tumor necrosis factor alpha (TNF-α) for 1 h (20 ng/ml) and fixed with formaldehyde for 10 min. Chromatin extract was then prepared from the cells and used for immunoprecipitation as described previously (2). To correct for differences in transfection efficiency, the amount of input DNA was measured by PCR prior to the analysis of the immunoprecipitated DNAs, and differences in the total amount of transfected DNA in the samples were normalized by adjusting the amount of precipitated DNA taken for the PCR analysis. The forward primer for each reporter is derived from the promoter, and the reverse primer is from the luciferase gene (5′-CCATCCTCTAGAGGATAGAATG). The primers from the 1-kb upstream region of the A20 promoter that was used as a control for specificity are 5′-GGTTAGCTCCTTCGGTCCTC and 5′-CCGTATTGACGCCGGGCAAG. For the endogenous genes, the amount of input DNA was first measured by PCR, and then differences in the input DNA in the samples were normalized by adjusting the amount of precipitated DNA taken for the PCR analysis. The primers used are as follows: for the A20 promoter, 5′-CAGCCCGACCCAGAGAGTCAC and 5′-CTTGGCCCGCCACGAA; for the A20 coding region, the same as for the RT-PCR; for the IκBα promoter, 5′-AAGGCTCACTTGCAGAGGG and 5′-GGACTGCTGTGGGCTCTG; for the IκBα coding region, 5′-TCCTGAGCTCCGAGACTTTC and 5′-GTAGTTGGTAGCCTTCAGG; for the RANTES promoter, 5′-CTTATGATACCGGCCAATGC and 5′-GTGCGAGGTCCACGTGCTGTC; for the RANTES coding region, 5′-CACAGGTGAGAGGCCCTTCG and 5′-CAGCTGAACTTCTTCTCGCCC; for the β-actin promoter, 5′-AAAGGAGGGGAGAGGGGGTAA and 5′-AAAGGCGAGGCTCTGTGCTC; for the BLR1 promoter, 5′-CATTACAAGTTGTGAGCC and 5′-CATCAGTGCTAGTCAAGC. Pol II antibodies were from BAbCO, and p65/RelA, cdk9, and TAF1 antibodies were from Santa Cruz Biotechnology, Inc. DSIF antibodies were described previously (1). The ChIP data were quantified by densitometric analysis using Quantity One one-dimensional analysis software (Bio-Rad).
RNA preparation and quantitative RT-PCR analysis.
Human 293T cells in 100-mm plates were left untreated or treated with TNF-α for 1 h (20 ng/ml). Total RNA was prepared using the TRIzol reagent (Gibco BRL), according to the manufacturer's instructions. RNA preparations were treated with RQ1 DNase I (Promega) to avoid contamination of genomic DNA. First-strand cDNA was synthesized from 1 μg of total RNA using an oligo(dT)15 primer and SuperScript II reverse transcriptase (Invitrogen). The PCR was performed in 20-μl glass capillary tubes using a LightCycler system (Roche Molecular Biochemicals) equipped with a thermal cycler and real-time detector of fluorescence. The total cDNAs were amplified using a LightCycler-FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals), according to the manufacturer's instructions. The oligonucleotides used for real-time PCR are as follows: for human GAPDH, 5′-CTGAGCTGAACGGGAAGCTC and 5′-CACCTGGTGCTCAGTGTAGC; for A20, 5′-GCTGCTGCCTCAGGGAAAGTC and 5′-CTCTTCTGTCCTTTTGGCCTC; for IκBα, 5′-CCTTCCTCAACTTCCAGAACAACC and 5′-GGCTAAGTGTAGGCAGGTGTGGC. The primers for RT-PCR analysis of ts13 hamster cells are as follows: for A20, 5′-CAAAATGCTAAGAAGTTTGG and 5′-CTCTGTTAACAAGTGGAACAG; for IκBα, same as for the human gene; for β-actin, 5′-CCCTGGAGAAGAGCTACGAGCTGCC and 5′-GCTTGCTGATCCACATCTGCTGGAAGG.
RESULTS
Core promoter context determines DSIF attenuation of NF-κB-mediated transcription.
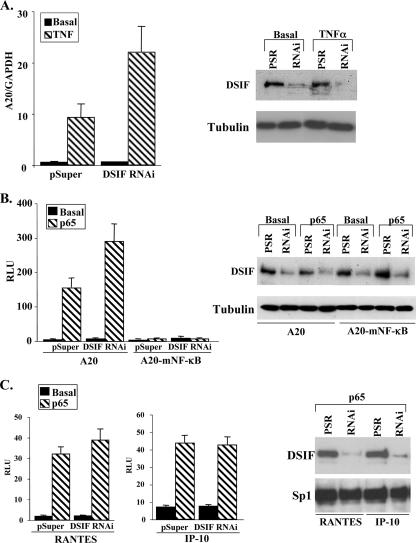
The A20 gene is highly responsive to the cytokine TNF-α through its activation of NF-κB. Treatment of HEK 293T cells with TNF-α for 1 h resulted in induction of A20 mRNA, as determined by quantitative real-time PCR analysis (Fig. 1A). Upon transfection of these cells with DSIF RNAi, there was a significant increase in the TNF-α-induced A20 mRNA levels, suggesting that DSIF attenuates A20-induced transcription.
FIG. 1.
Attenuation of NF-κB-mediated transcription by DSIF. (A) 293T cells were transfected with either pSuper or DSIF p160 RNAi, and 48 h posttransfection, cells were treated with TNF-α for 1 h. Total RNA was extracted and subjected to a quantitative RT-PCR for the A20 and GAPDH mRNAs, using a Light Cycler system. Data are means and standard deviations of three independent experiments. A representative immunoblot verifying down regulation of p160 DSIF is shown on the right. (B) 293T cells were transfected with either the wild-type A20 or the A20-mNF-κB (in which the two NF-κB sites are mutated) promoter, with or without the p65/RelA (p65) expression vector, and with either pSuper (parental vector) or DSIF p160 RNAi as indicated. Cells were harvested 48 h posttransfection, and luciferase activity was measured. RSV promoter-driven Renilla luciferase reporter plasmids served to normalize the transfection efficiency. Shown is the relative luciferase activity (luciferase units divided by the activity of cotransfected RSV promoter-driven Renilla reporter luciferase, in relative light units [RLU]). Data are means and standard deviations of seven independent experiments, each with independent duplicates (left). A representative immunoblot showing DSIF knockdown in transfected cells is shown on the right. (C) 293T cells were cotransfected with p65/RelA expression plasmid together with luciferase reporter genes driven by the RANTES and IP-10 promoters and with either pSuper or DSIF p160 RNAi as indicated. The averages of seven independent transfection experiments are shown in the graphs. A representative immunoblot showing DSIF knockdown in cells transfected with the indicated reporter plasmids together with p65/RelA is shown on the right.
Next we determined the effect of DSIF knockdown on a luciferase reporter gene driven by the wild-type A20 promoter or one with the NF-κB sites mutated in the presence of the cotransfected NF-κB protein p65/RelA. As expected, the A20 promoter was activated by NF-κB, whereas no induction was seen with the mutant A20 promoter (Fig. 1B). Consistent with previous results (1), down regulation of DSIF by RNAi (Fig. 1B, right) enhanced the NF-κB induced transcription of the wild-type A20 promoter but had no effect on the mutated promoter (Fig. 1B, left), suggesting that NF-κB itself is required for inhibition by DSIF.
To examine whether DSIF inhibitory activity is common to other NF-κB target genes, we determined the effect of DSIF knockdown on luciferase activity driven by RANTES and IP-10 promoters in the presence of the cotransfected NF-κB protein p65/RelA (Fig. 1C). Unlike the A20 promoter activity, the NF-κB-induced activity of these promoters was not significantly affected by DSIF depletion. These results suggest that even though the inhibition of NF-κB by DSIF requires NF-κB, the effect is not general but promoter specific.
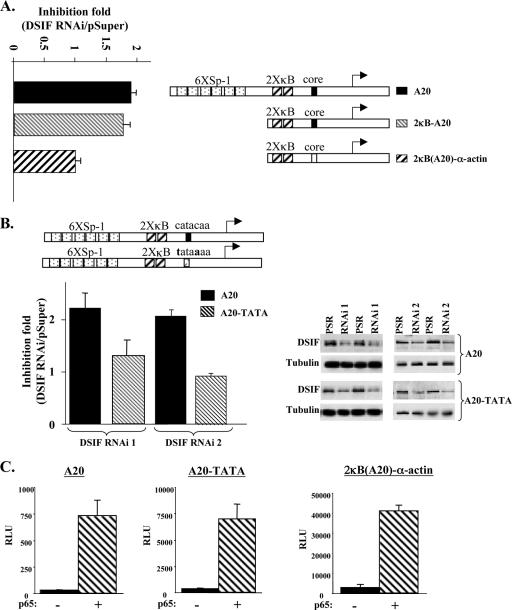
To understand the basis for the promoter-mediated regulation of NF-κB transcription by DSIF, we set out to determine the region in the A20 promoter that in conjunction with NF-κB generates the inhibitory effect of DSIF. The A20 promoter has six Sp1 binding sites followed by two NF-κB binding sites in front of a TATA-less core promoter (Fig. 2A). To determine whether Sp1 plays a role in DSIF activity, the Sp1 sites were deleted, leaving only the two NF-κB binding sites and the core promoter. The results (Fig. 2A) show that the A20 minimal promoter (2κB-A20) retained the DSIF inhibitory effect (presented as the ratio between the activities in the presence and absence of DSIF RNAi), indicating that Sp1 is not required for DSIF activity. Thus, the differential regulation of NF-κB transcription by DSIF could be dependent either on the specific sequence of the NF-κB sites or on the core promoter. To test this, we replaced the A20 core promoter with the core promoter of the α-actin gene and examined the effect of DSIF RNAi on NF-κB activity (Fig. 2A). While 2κB-A20 was sensitive to DSIF knockdown, the construct bearing the same two NF-κB sites in front of the heterologous α-actin core promoter [2κB(A20)-α-actin] was unaffected by DSIF knockdown (Fig. 2A) (the amount of NF-κB used in these experiments is well below that required to achieve maximal activation). This suggests that it is the context of the core promoter that is important for attenuation of NF-κB by DSIF.
FIG. 2.
DSIF attenuation of NF-κB is dependent on a TATA-less core promoter. (A) 293T cells were cotransfected with A20 promoter mutants (schematically shown on the right) with a suboptimal concentration of p65/RelA expression vector and with either pSuper (parental vector) or DSIF p160 RNAi as indicated and analyzed as described for Fig. 1B. The effect of DSIF knockdown on different A20 promoter mutants activated by p65/RelA concentration (i.e., inhibition) is presented as the ratio of the relative luciferase activity in the presence of DSIF RNAi to the activity in the presence of the parental vector pSuper. (B) Effect of DSIF knockdown by two distinct RNAis on the wild-type A20 promoter and a mutant (A20-TATA) which was converted into a canonical TATA box promoter by substitution of two nucleotides (bold letters) at −30 and −26 relative to the transcription start site, in the presence or absence of p65/RelA (suboptimal concentration). DSIF RNAi 1 is the one used for Fig. 1, and DSIF RNAi 2 is directed against a different region of the p160 subunit (see Materials and Methods). The inhibition results are presented as in panel A and are the averages of seven (DSIF RNAi 1) or six (DSIF RNAi 2) independent duplicate transfection experiments. Representative immunoblots showing down regulation of DSIF p160 by the two RNAis are shown on the right. (C) Responsiveness of the A20 mutant reporter genes to p65/RelA NF-κB protein.
Analysis of the core promoters of the genes used in Fig. 1 and Fig. 2A revealed that the DSIF-responsive promoter A20 is TATA-less, whereas the less sensitive promoters IP-10, RANTES, and 2κB-α-actin contain a canonical TATA box (TATAWA) at the appropriate location (−25 to −30 relative to the transcription start site). To confirm that it is indeed the nature of the core promoter that determines the DSIF-inhibitory activity of NF-κB, the TATA-less A20 promoter was converted into a canonical TATA promoter by substituting two nucleotides in the core promoter as shown in Fig. 2B, and the effects of two distinct DSIF RNAis were analyzed in the presence of cotransfected p65/RelA (using a concentration well below that required to achieve maximal activation). The TATA box mutation (A20-TATA) increased the basal activity but not the extent of activation by NF-κB (Fig. 2C). Expression of each DSIF p160 RNAi enhanced the activity of the wild-type A20 but not of the A20-TATA mutant ∼2-fold. The difference in the effect of DSIF on A20 and A20-TATA is statistically significant (P = 0.0025). Thus, the inhibitory effect of DSIF on NF-κB-induced transcription requires an NF-κB site(s) to be in the context of a TATA-less core promoter.
DSIF occupancy is enhanced by NF-κB in a core promoter-dependent manner.
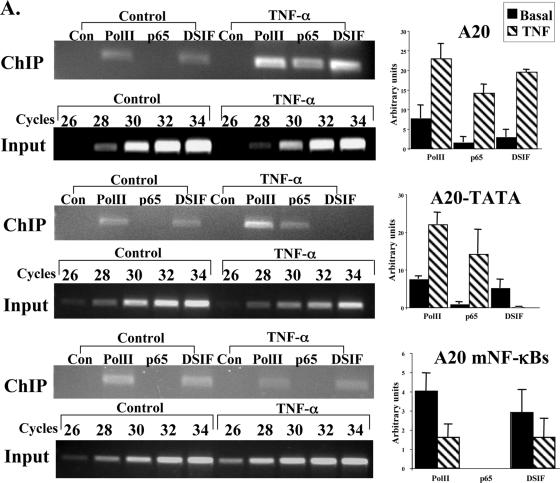
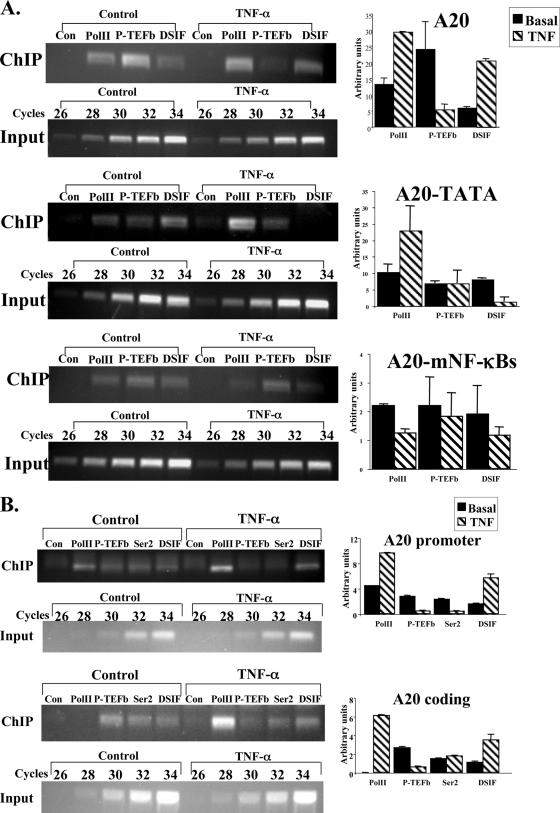
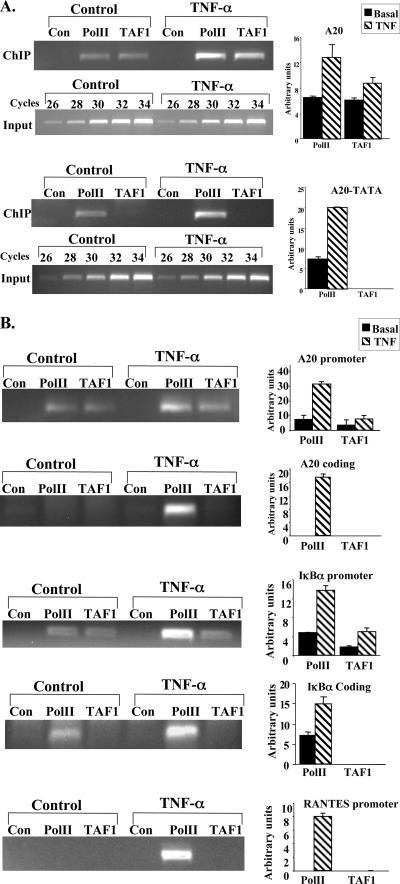
If the attenuation of NF-κB-induced transcription by DSIF is a direct effect, it should involve association of DSIF with the promoter in a manner dependent on NF-κB and the core promoter. We therefore transfected 293T cells with the different A20 reporter genes, treated them with TNF-α for 1 h to induce NF-κB activity, and then subjected them to a ChIP assay using antibodies against RNA Pol II, p65/RelA, and the p160 subunit of DSIF, and an irrelevant antibody as a control. After reverse cross-linking, PCRs were performed with primers corresponding to the 5′ end of the A20 promoter and to the beginning of the luciferase gene. As shown in Fig. 3A, under basal conditions Pol II and DSIF constitutively bind wild-type A20 and the A20-TATA mutant. Upon TNF-α treatment, NF-κB p65/RelA associates with A20 and A20-TATA, and there is a concomitant increase in Pol II occupancy. DSIF occupancy, on the other hand, is enhanced only in A20 and 2κB-A20 but is reduced in the TATA-containing promoter A20-TATA. Enhancement of Pol II and DSIF occupancies is NF-κB dependent, as an A20 promoter bearing mutations in the two NF-κB binding sites which prevented binding of p65/RelA after TNF-α induction (Fig. 3A, bottom) failed to enhance Pol II and DSIF binding. The association of Pol II, p65/RelA, and DSIF with the promoter is specific, as a nonpromoter sequence located 1 kb upstream was not enriched by them (data not shown).
FIG. 3.
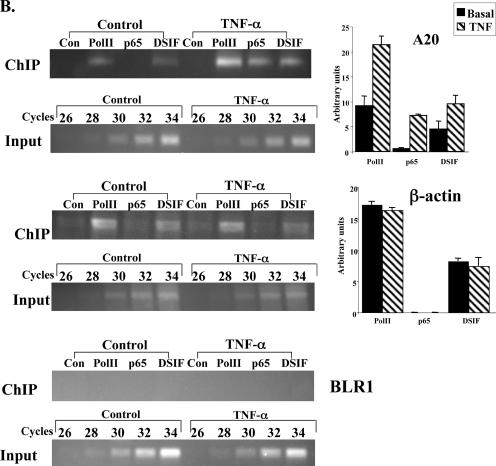
The core promoter controls differential occupancy of DSIF on A20 gene upon NF-κB induction. (A) The wild-type and mutant A20 promoters-driven reporter plasmids, as indicated, were each transfected into 293T cells. Cells were treated 24 h later with TNF-α for 1 h and then subjected to ChIP with the indicated antibodies and an irrelevant antibody as a control, followed by PCR analysis after normalization to the input (see Materials and Methods). The forward primer is specific to each of the promoters, and the reverse primer is derived from the proximal region of the luciferase gene. Representative ChIP results are shown on the left. Input DNA (0.1%) was subjected to an increasing number of PCR cycles in order to find the linear range to serve as a reference for the immunoprecipitation. Quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Jurkat T cells were untreated or treated with TNF-α for 1 h, followed by ChIP assay with the indicated antibodies and an irrelevant antibody. The immunoprecipitated DNAs were subjected to PCR with primers specific to the promoter and beginning of each of the A20, β-actin, and BLR1 endogenous genes. Quantified results, normalized to the input, derived from three independent experiments are shown on the right.
To confirm that the endogenous A20 gene is also directly regulated by DSIF upon NF-κB induction, we performed ChIP assays on the native A20 gene. Figure 3B shows that upon NF-κB induction by TNF-α, DSIF occupancy of the A20 promoter is increased, as found with the transfected promoter. Enhancement of DSIF occupancy is specific to NF-κB, as DSIF occupancy at the noninducible housekeeping gene β-actin is unaffected by NF-κB induction. To confirm the specificity of the interaction of Pol II, p65, and DSIF with the A20 promoter, a similar analysis was performed with the promoter of the BLR1 gene, a B-cell-specific NF-κB target gene (22) that is not expressed in Jurkat T cells. This promoter was not enriched by any of these factors. Together the results suggest that the effect of DSIF on the A20 gene upon NF-κB induction is directly correlated to promoter occupancy in a manner dependent on NF-κB and the core promoter.
The core promoter has opposite effects on DSIF and P-TEFb occupancies upon NF-κB induction.
Previous studies demonstrated that the positive elongation factor P-TEFb alleviates inhibition of transcription elongation by DSIF in vitro (25, 32). To determine the interplay between DSIF and P-TEFb in the regulation of the A20 gene, we compared P-TEFb and DSIF occupancy of the various A20 promoter derivatives before and after stimulation of NF-κB by TNF-α (Fig. 4A). The results revealed that on the TATA-less A20 reporter, P-TEFb occupancy is clearly detectable under basal conditions, but in contrast to DSIF, it is down regulated upon NF-κB induction (Fig. 4A, A20). This effect is NF-κB dependent, since P-TEFb occupancy is unaffected when the NF-κB sites are mutated (Fig. 4A, A20-mNF-κBs). P-TEFb associates specifically with the promoter region, as P-TEFb was not detected in a nonpromoter sequence located 1 kb upstream (data not shown). Notably, converting A20 to a TATA promoter reverses the effect of NF-κB: P-TEFb occupancy is still clearly detected before stimulation as in the wild type, but now its occupancy is unchanged after NF-κB induction as opposed to DSIF levels, which are reduced (Fig. 4A, A20-TATA). Thus, in the NF-κB-induced system, switching the core promoter type inverts the ratio between positive and negative elongation factors. This suggests that the ratio between functionally opposing elongation factors is important for elongation control and can be modulated by core promoter type.
FIG. 4.
P-TEFb occupancy is the inverse of DSIF upon NF-κB induction. (A) ChIP of transfected A20 wild-type and mutant reporter genes, as described for Fig. 3A, using antibodies against Pol II, the cdk9 subunit of P-TEFb, and the p160 subunit of DSIF and an irrelevant antibody as a control. Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) ChIP assay of the endogenous A20 gene from control Jurkat T cells or cells treated for 1 h with TNF-α as described for Fig. 3B, using antibodies to unphosphorylated Pol II, the cdk9 subunit of P-TEFb, serine 2-phosphorylated Pol II (Ser2), and the p160 subunit of DSIF and an irrelevant antibody as a control (Con). The immunoprecipitated DNAs were subjected to PCR with primers specific to the promoters or to internal regions of these genes (coding). Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right.
P-TEFb is known to phosphorylate serine 2 of Pol II CTD heptapeptides and to facilitate its processivity. We therefore assessed the occupancy of unphosphorylated and serine 2-phosphorylated forms of Pol II as well as of P-TEFb and DSIF at the promoter and at downstream coding sequences of the endogenous A20 gene in Jurkat T cells treated with TNF-α for 1 h (Fig. 4B). Consistent with the results from transfected promoters, P-TEFb and serine 2-phosphorylated Pol II, which are detected under basal conditions, are clearly reduced in the A20 promoter upon TNF-α induction (Fig. 4B, A20 promoter), as opposed to unphosphorylated Pol II and DSIF, which are clearly increased. Within the A20 gene, Pol II is present only in its serine 2-phosphorylated form, along with P-TEFb and DSIF under basal conditions. Following TNF-α treatment, substantial amounts of unphosphorylated Pol II are detected, whereas P-TEFb is reduced and DSIF is increased. Surprisingly, the amount of serine 2-phosphorylated Pol II does not decrease in spite of P-TEFb release, possibly because a small fraction of the large quantity of induced Pol II is phosphorylated by the residual P-TEFb. The effect of NF-κB, then, on the occupancies of DSIF and P-TEFb on the promoter continues into the coding region, indicating its relevance to the elongation phase of the A20 gene.
P-TEFb regulation of NF-κB is dependent on a TATA box.
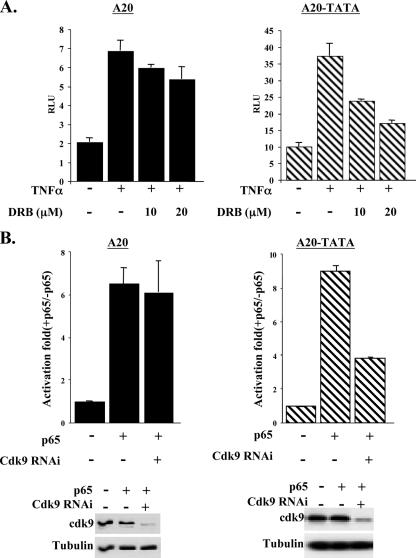
The P-TEFb ChIP experiments (Fig. 4A) suggest that P-TEFb, like DSIF, regulates NF-κB in a core promoter-dependent manner. To examine this, 293T cells were transfected with wild-type A20 and A20-TATA mutant reporter genes and 24 h later were treated with either TNF-α alone or TNF-α together with increasing amounts of the drug DRB, a potent inhibitor of P-TEFb (24). The results (Fig. 5) show that TNF-α stimulated the luciferase activity of both reporters more than threefold, but the induced activity of A20-TATA is more sensitive to inhibition by DRB than that of A20.
FIG. 5.
Differential regulation of NF-κB by P-TEFb is core promoter type dependent. (A) 293T cells were transfected with the indicated reporters and 24 h later were treated either with TNF-α or with TNF-α plus increasing amounts of DRB for 4 h. The luciferase activity was normalized to the values of the cotransfected RSV promoter-driven Renilla reporter luciferase plasmid. The graphs show means and standard deviations of three duplicate experiments. (B) 293T cells were transfected with either wild-type A20 or A20-TATA mutant promoters together with p65/RelA expression vector and with either pSuper (parental vector) or cdk9 RNAi as indicated. Cells were harvested 48 h posttransfection, and luciferase activity was measured. Data are means and standard deviations of six independent duplicated experiments (left). A representative immunoblot showing cdk9 depletion in transfected cells is shown on the bottom.
To examine further the role of P-TEFb in NF-κB-mediated transcription, we used RNAi to reduce the endogenous P-TEFb levels. The wild-type A20 and the A20-TATA mutant reporter genes were transfected into 293T cells together with a plasmid directing the expression of RNAi specific for the cdk9 subunit of P-TEFb. Analysis of cdk9 expression by immunoblotting shows its specific depletion from cells transfected with cdk9 RNAi but not from cells transfected with the parental plasmid (Fig. 5B). Akin to the effect of DRB, down-regulation of cdk9 had no effect on the NF-κB-activated transcription of the wild-type A20 promoter, but it significantly reduced NF-κB induction of the A20-TATA promoter. These findings indicate that P-TEFb is particularly important for TATA-containing NF-κB target genes and are consistent with the ChIP assay data showing that the P-TEFb/DSIF ratio on these promoters is increased upon NF-κB induction. In agreement with this model, the interleukin 8 gene, previously shown to be regulated by P-TEFb upon NF-κB induction (4, 18), has a TATA box promoter, whereas the TATA-less IκBα is not regulated by P-TEFb (18).
Differential occupancy of other NF-κB target genes by P-TEFb and DSIF is correlated with core promoter type.
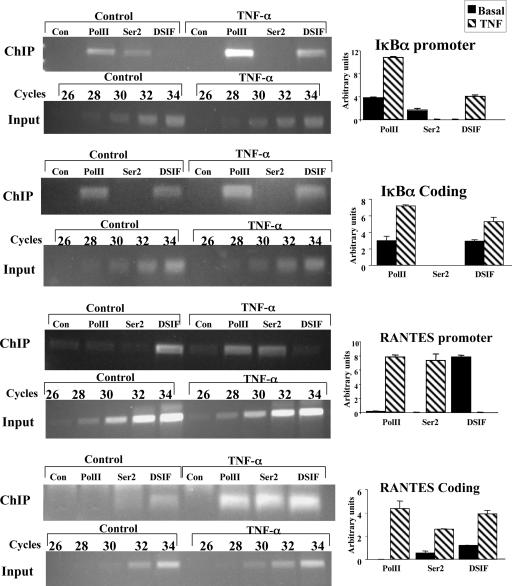
To gain further support for the dependency of DSIF and P-TEFb occupancies on the core promoter type upon NF-κB induction, we examined two additional NF-κB-responsive genes, one which is TATA-less (IκBα) and one which has a canonical TATA element (RANTES). These genes were analyzed by ChIP before and 1 h after treatment with TNF-α by using antibodies against unphosphorylated (Pol II) and serine 2-phosphorylated (Ser2) forms of Pol II, DSIF, or irrelevant control antibodies. PCRs were performed with primers corresponding to the promoter region (promoter) and an internal region of the gene (coding). As shown in Fig. 6, in the TATA-less IκBα promoter and gene, unphosphorylated Pol II and DSIF occupancies are enhanced but the promoter serine 2 Pol II is reduced upon TNF-α induction, as observed with A20. With the TATA-containing RANTES promoter, the effects are reversed: DSIF is released and serine 2-phosphorylated Pol II is enhanced by NF-κB. In the coding region of the RANTES gene, both the unphosphorylated and serine 2-phosphorylated forms of Pol II are increased, but DSIF is not diminished in the coding sequences upon TNF-α induction. The lack of correlation of DSIF occupancy between the promoter and the coding argues that in the RANTES gene DSIF regulation during elongation is not mediated by the promoter. This regulation is likely to involve additional elements of the elongating Pol II.
FIG. 6.
ChIP analysis of other NF-κB target genes. The TATA-less IκBα and the TATA-containing RANTES genes were subjected to ChIP assay before or 1 h after TNF-α treatment as described for Fig. 3B. Antibodies to unphosphorylated Pol II, serine 2-phosphorylated Pol II (Ser2), and the p160 subunit of DSIF and an irrelevant control antibody (Con) were used for immunoprecipitation. The precipitated DNAs were subjected to PCR with primers specific to the promoters or the coding regions of the genes. Representative results of ChIP and input are shown on the left. Quantified results, normalized to the input, derived from two independent experiments are shown on the right.
Together these results strengthen the notion that differential occupancy by DSIF and P-TEFb during the initial stages of transcription, and by and large during elongation, is dependent on core promoter type.
DSIF occupancy is counteracted by serine 2 phosphorylation of Pol II.
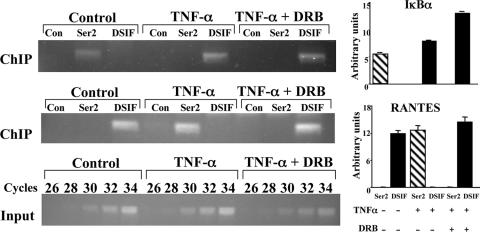
Given that upon NF-κB activation, DSIF and P-TEFb occupancies are mutually exclusive, and that in vitro P-TEFb relieves the inhibitory effect of DSIF (25, 32), it is possible that DSIF accumulation or dissociation upon NF-κB activation is the result of, respectively, P-TEFb release or recruitment. Alternatively, DSIF may be recruited or released by NF-κB itself. However, whereas P-TEFb has been shown to interact directly with the NF-κB protein p65/RelA (4), we failed to detect any direct interaction between p65/RelA and DSIF in vitro or in cell extracts, suggesting that the latter possibility is less likely. To examine the role of P-TEFb in DSIF occupancy, we performed ChIP experiments on TNF-α-induced Jurkat T cells treated with the P-TEFb inhibitor DRB, using antibodies against DSIF and the serine 2-phosphorylated form of Pol II (Fig. 7). Analysis of the TATA promoter RANTES revealed that the TNF-α-induced serine 2 phosphorylation by P-TEFb is diminished by DRB (Fig. 7, compare TNF-α to TNF-α+DRB lanes). This inhibition of P-TEFb activity resulted in failure to displace DSIF from the promoter. In contrast, DRB had no effect on DSIF accumulation in the IκBα promoter. Thus, it is most likely that the reduced DSIF occupancy upon NF-κB activation in TATA-containing promoters is a consequence of P-TEFb recruitment.
FIG. 7.
Serine 2 phosphorylation of Pol II on the TATA promoter releases DSIF. Jurkat cells were untreated or treated with TNF-α without or with DRB (20 μM) for 1 h and then subjected to ChIP assay with antibodies to serine 2-phosphorylated Pol II (Ser2) and the p160 subunit of DSIF and an irrelevant antibody as a control (Con) followed by PCR analysis of the TATA-less IκBα and the TATA-containing RANTES promoters. Representative results from two experiments are shown. Quantified results, normalized to the input, derived from two independent experiments are shown on the right.
Notably, under basal conditions DSIF occupies the RANTES promoter in the absence of unphosphorylated or serine 2-phosphorylated Pol II (Fig. 6 and 7). As DSIF is recruited to genes via its association with Pol II, we checked for the presence of another form of Pol II, the serine 5-phosphorylated form. Indeed, we found that serine 5-phosphorylated Pol II occupies this promoter exclusively and that this form disappears upon stimulation by TNF-α (data not shown).
The core promoter type determines the nature of the initiation complex activated by NF-κB.
It is well established that enhancers and core promoters direct the initiation step of transcription. The experiments described above suggest that at least for NF-κB target genes, the core promoter type is also involved in regulation of transcription elongation. Given that the core promoter is the site at which the transcription initiation complex assembles, we reasoned that the formation of different initiation complexes on the two types of core promoter may dictate recruitment or displacement of distinct elongation factors. Therefore, we wanted to determine the nature of the initiation complex that is formed on the different NF-κB target genes. Our experiments were based on studies with Saccharomyces cerevisiae in which, depending on the core promoter structure, initiation complex assembly is mediated by either TFIID or SAGA (5, 8, 13, 15, 20). To examine the role of the core promoter in determining the type of the transcription initiation complex activated by NF-κB, we analyzed the occupancy of A20 promoter derivatives by TAF1, a TFIID-specific subunit not present in the mammalian SAGA-related complexes STAGA, TFTC, and PCAF (12, 19, 21, 35). 293T cells were transfected with the wild-type A20 or A20-TATA promoter, treated 24 h later with TNF-α for 1 h to induce NF-κB, and then subjected to ChIP assays using antibodies against TAF1 and RNA Pol II and an irrelevant antibody. The results (Fig. 8A) show that TAF1 occupies the TATA-less A20 promoter before and after NF-κB stimulation, whereas it is completely absent from A20-TATA. The presence of Pol II and TAF1 under basal conditions is consistent with our previous findings that the basal transcription machinery is already assembled in rapidly induced NF-κB target genes (2).
FIG. 8.
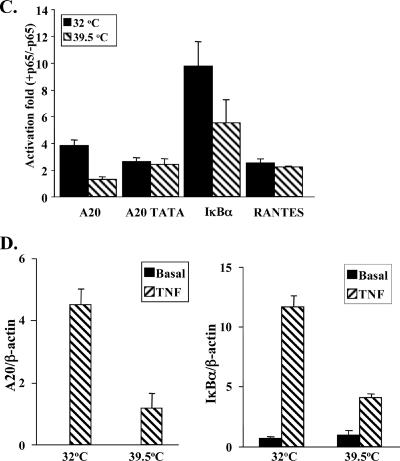
Dependency of the different NF-κB-responsive promoters on TAF1. (A) Wild-type A20 and A20-TATA mutant promoters were each transfected into 293T cells and analyzed by ChIP as described for Fig. 3A, using antibodies against TAF1 and Pol II and an irrelevant antibody as a control (Con). Representative results of ChIP and input are shown on the left, and quantified results, normalized to the input, derived from two independent experiments are shown on the right. (B) Fixed chromatin extract was prepared from Jurkat T cells induced by TNF-α for 1 h. ChIP assays were performed as described for Fig. 3B using antibodies to Pol II and TAF1 and an irrelevant antibody as a control. Representative results are shown on the left, and quantified results, normalized to the input (as in Fig. 4B) derived from two independent experiments on the right. (C) Temperature-sensitive hamster ts13 cells were cotransfected with the indicated reporter plasmids together with the p65/RelA expression plasmid. The cells were incubated at the permissive temperature (32°C) for 6 h, washed, and then separated into two groups. One was grown at 32°C and the second at the nonpermissive temperature (39.5°C) for an additional 48 h, after which the cells were harvested and luciferase activity measured. RSV promoter-driven Renilla reporter luciferase plasmids served to normalize transfection efficiency within each group, and c-fos-luciferase (TAF-independent promoter) served to normalize transfection efficiency between the groups. Results are means of four independent experiments, each with independent duplicates. (D) Hamster ts13 cells were grown at 32°C and then shifted to the nonpermissive temperature (39.5°C) for 6 h, followed by a 1-h treatment with mouse TNF-α. Total RNA was extracted and subjected to a quantitative RT-PCR assay to measure the mRNAs of A20, IκBα, and β-actin genes, using a Light Cycler system. Data are means and standard deviations of two independent experiments.
Next, we determined TAF1 and Pol II occupancy of the endogenous TATA-less A20 and IκBα genes and of the TATA-containing RANTES gene in Jurkat T cells that had been treated with TNF-α for 1 h. TAF1 was found to be specifically associated with the A20 and IκBα promoters but not with the coding regions of these genes before and after TNF-α induction (Fig. 8B). In contrast, TAF1 was undetectable on the RANTES promoter under both basal and TNF-α-stimulated conditions. These findings suggest that the core promoter sequence of NF-κB target genes controls the pathway of the transcription initiation complex assembly, which can be either TFIID dependent (TATA-less) or TFIID independent (TATA).
To gain further support for the idea of a differential requirement for TFIID in NF-κB transcription, we used the temperature-sensitive hamster cell line ts13, in which TAF1 contains a point mutation that renders TFIID inactive at 39.5°C. These cells were cotransfected with NF-κB-dependent reporter plasmids and the NF-κB protein p65/RelA. As shown in Fig. 8C, activation by NF-κB of the TATA-less A20 and IκB promoters decreased at the nonpermissive temperature, 39.5°C, compared to the permissive temperature, 32°C, demonstrating TFIID dependency. Activation of the TATA-containing RANTES promoter, on the other hand, was unaffected by changing from the permissive to the nonpermissive temperature. Moreover, modifying the TATA-less sequence of the A20 promoter to canonical TATA (A20-TATA) abolished its dependency on TFIID.
We also assessed the involvement of TFIID in the transcription of endogenous NF-κB target genes by measuring the mRNA levels of the A20 and IκBα genes in ts13 cells before and after treatment with TNF-α for 1 h (the endogenous RANTES gene, whose promoter was analyzed above, is not expressed in these cells). Quantitative RT-PCR measurements (Fig. 8D) showed a significant decrease in the TNF-α-induced mRNA levels of A20 and IκBα at the nonpermissive temperature. The mRNA of the β-actin gene was unchanged, and we used it to normalized the results.
Considering the correlation between the core promoter structure and differential recruitment of DSIF and P-TEFb, we can conclude that DSIF attenuation of NF-κB is dependent on the initiation complex assembled via TFIID, whereas P-TEFb regulation of NF-κB requires TAF-independent assembly of the initiation complex.
DISCUSSION
Two types of DNA element, enhancers and the core promoter, regulate transcription. Several studies have indicated that specific combinations of these elements play a regulatory role in transcription (7, 9, 10, 15, 20, 28, 30, 33, 34). Such combinations are considered important for the initiation step of transcription. The data presented in this study demonstrate the importance of the arrangement of the enhancer with a core promoter type in gene-specific effects of elongation regulatory factors. Our results thus reveal new links between the initiation and elongation phases of the transcription reaction that are relevant to gene regulation. Thus, it appears that the genetic information encoded by transcription regulatory regions affects more steps of the transcription cycle than initially believed. A role of the promoter in RNA Pol II processivity was reported for the HIV long terminal repeat and c-myc transcription (17, 38), where it was found that a paused but not processive transcription is dependent on an intact TATA box, though the basis for the TATA box requirement was not explored. Our findings extend those previous studies by showing that in the NF-κB pathway the TATA box is required for NF-κB-induced facilitation of elongation by P-TEFb upon NF-κB activation. Given the involvement of P-TEFb in TAT activation of HIV long terminal repeat elongation, it remains to be seen whether those findings are linked to ours.
Specifically, we demonstrated that attenuation of NF-κB-induced transcription by the negative elongation factor DSIF occurs in genes carrying NF-κB binding sites in the context of a TATA-less core promoter. This specific configuration of the promoter is also responsible for enhanced recruitment of DSIF to these genes upon NF-κB induction. Contrary to TATA-less NF-κB target genes, transcription of TATA box-containing genes is not inhibited by DSIF, and DSIF is actually lost from these genes upon NF-κB induction. In addition, the core promoter also influences gene occupancy and regulation by the positive elongation factor P-TEFb in a manner that correlates inversely with that of DSIF. The finding that activation of the A20 and IκBα genes results in a large increase in hypophosphorylated Pol II within the coding region but no increase of Ser-2-phosphorylated Pol II was surprising and suggests that in some genes, transcription elongation occurs by a mechanism that does not involve Ser-2 phosphorylation. This result adds to the growing number of genes whose transcription is reported to be independent of P-TEFb (11, 18).
The mechanism by which the elongation factors DSIF and P-TEFb differentially regulate NF-κB target genes involves both NF-κB and the initiation complex that is formed on the different types of core promoter. When the initiation complex is assembled via TFIID on TATA-less promoters, P-TEFb is released upon NF-κB induction. As a consequence, DSIF occupancy is enhanced along with the unphosphorylated form of Pol II. In contrast, when the initiation complex is formed by a TFIID-independent pathway on TATA-containing promoters, reminiscent of the SAGA complex in yeast (5, 8, 15, 20), NF-κB recruits or retains P-TEFb, which then phosphorylates the Pol II C-terminal domain on serine 2, resulting in DSIF displacement.
How does differential regulation of NF-κB target genes by positive and negative elongation factors affect its biological activity? DSIF has been implicated in the coordination of transcription with mRNA capping (16, 23). Whether this function of DSIF plays a role in activation of NF-κB target genes is yet to be determined. However, the increase in the level of the luciferase protein in DSIF RNAi-treated cells would seem to indicate that the luciferase mRNA has been capped. DSIF attenuates an important subset of NF-κB-induced genes, A20 and IκBα. Although the inhibitory effect of DSIF on these genes is moderate (∼2-fold), it is likely to have a significant biological impact. NF-κB is latent in the cytoplasm and translocates into the nucleus upon activation to induce target genes. Among those are A20 and IκBα, which act to terminate NF-κB activity in a negative feedback loop. Thus, a twofold inhibition of these genes by DSIF may alter the duration and the extent of the NF-κB activity.
In addition to the genes analyzed here, there are other genes induced by NF-κB in different cells. We analyzed the core promoter type and expression pattern of 48 documented NF-κB target genes (22) for which the transcription initiation site is known. A TATA box is present in the core promoter of 32 of these genes (data not shown), the majority of which (26/32) are cell type specific (expressed in some but not all cell types). On the other hand, most of the TATA-less genes (14/16) are ubiquitously expressed (expressed in all cell types). The overrepresentation of TATA-less promoters among ubiquitously expressed target genes and TATA promoters among cell type-specific target genes is in agreement with the genome-wide distribution of these two groups of genes (26). Interestingly, genes that are activated by NF-κB to modulate its own signaling pathway, such as A20, IκBα, cIAP2, TRAF1, TRAF2, NF-κB1 (p105), and c-Rel, are predominantly in the group of the TATA-less target genes. Given the dependency of DSIF and P-TEFb on the core promoter type, it can be predicted that most of the genes that mediate the physiological response of NF-κB, including cytokines, chemokines, and adhesion molecules, are controlled by P-TEFb, whereas genes involved in the signaling pathway of NF-κB are attenuated by DSIF.
DSIF is a positive and negative elongation regulatory factor, and the mechanisms of these distinct functions seem to be different. The positive activity of DSIF is facilitated by P-TEFb, which phosphorylates DSIF (36), and cooccupancy of P-TEFb and DSIF on positively DSIF-regulated hsp70 and c-fos genes has been observed (3, 36). On the other hand, the negative activity of DSIF is actually counteracted by P-TEFb (25, 32), and this is consistent with our findings of their mutually exclusive regulation and occupancy in the NF-κB pathway. Moreover, DSIF inhibition is dependent on NF-κB and a TATA-less promoter, in contrast to its positively regulated genes, c-fos and hsp70, which are driven by a TATA promoter.
What then is the function of DSIF and P-TEFb in NF-κB-mediated transcription? NF-κB is a powerful transcription factor responsible for the activation of many genes in response to a variety of external signals. For any particular biological response, the desired levels of activation of different genes by the same activated NF-κB may not be equal. Reducing or enhancing the processivity of the activated Pol II via positive and negative elongation factors is a mechanism that can tune the level of activation by the same NF-κB up or down in a regulated fashion. The differential effects of positive and negative elongation factors on NF-κB target genes that belong to functionally distinct groups may be particularly important for modulating the strength and the duration of the inflammatory response.
Acknowledgments
We thank Sandra Moshonov for critical reading and editing of the manuscript and Nadav Bar and Rofa Elfakess for their assistance throughout the project.
This work was supported by grants from the Israel Cancer Research Foundation and the Israel Science Foundation.
Footnotes
Published ahead of print on 14 May 2007.
REFERENCES
- 1.Ainbinder, E., L. Amir-Zilberstein, Y. Yamaguchi, H. Handa, and R. Dikstein. 2004. Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-κB. Mol. Cell. Biol. 24:2444-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22:6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 5.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-555. [DOI] [PubMed] [Google Scholar]
- 7.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, J. X., M. Floer, P. Ononaji, G. Bryant, and M. Ptashne. 2002. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 12:1828-1832. [DOI] [PubMed] [Google Scholar]
- 9.Conkright, M. D., E. Guzman, L. Flechner, A. I. Su, J. B. Hogenesch, and M. Montminy. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 11:1101-1108. [DOI] [PubMed] [Google Scholar]
- 10.Das, G., C. S. Hinkley, and W. Herr. 1995. Basal promoter elements as a selective determinant of transcriptional activator function. Nature 374:657-660. [DOI] [PubMed] [Google Scholar]
- 11.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 13.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 14.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 15.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 16.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, X., T. M. Welsh, and B. M. Peterlin. 1993. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J. Virol. 67:1752-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luecke, H. F., and K. R. Yamamoto. 2005. The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes Dev. 19:1116-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 20.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 21.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 22.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 23.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639-19648. [DOI] [PubMed] [Google Scholar]
- 24.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 26.Schug, J., W. P. Schuller, C. Kappen, J. M. Salbaum, M. Bucan, and C. J. Stoeckert, Jr. 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shilatifard, A., R. C. Conaway, and J. W. Conaway. 2003. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72:693-715. [DOI] [PubMed] [Google Scholar]
- 28.Simon, M. C., T. M. Fisch, B. J. Benecke, J. R. Nevins, and N. Heintz. 1988. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell 52:723-729. [DOI] [PubMed] [Google Scholar]
- 29.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, I. C., and R. E. Kingston. 1990. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol. Cell. Biol. 10:165-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, W. D., and J. D. Gralla. 1991. Differential ability of proximal and remote element pairs to cooperate in activating RNA polymerase II transcription. Mol. Cell. Biol. 11:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wefald, F. C., B. H. Devlin, and R. S. Williams. 1990. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature 344:260-262. [DOI] [PubMed] [Google Scholar]
- 35.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 36.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. A. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227-237. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 38.Yankulov, K., J. Blau, T. Purton, S. Roberts, and D. L. Bentley. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749-759. [DOI] [PubMed] [Google Scholar]