Abstract
Tumor necrosis factor alpha (TNF-α) is a potent cytokine which regulates inflammation via the induction of adhesion molecules and chemokine expression. Its expression is known to be regulated in a complex manner with transcription, message turnover, message splicing, translation, and protein cleavage from the cell surface all being independently regulated. This study examined both cell lines and primary cells to understand the developmental regulation of epigenetic changes at the TNF-α locus. We demonstrate that epigenetic modifications of the TNF-α locus occur both developmentally and in response to acute stimulation and, importantly, that they actively regulate expression. DNA demethylates early in development, beginning with the hematopoietic stem cell. The TNF-α locus migrates from heterochromatin to euchromatin in a progressive fashion, reaching euchromatin slightly later in differentiation. Finally, histone modifications characteristic of a transcriptionally competent gene occur with myeloid differentiation and progress with differentiation. Additional histone modifications characteristic of active gene expression are acquired with stimulation. In each case, manipulation of these epigenetic variables altered the ability of the cell to express TNF-α. These studies demonstrate the importance of epigenetic regulation in the control of TNF-α expression. These findings may have relevance for inflammatory disorders in which TNF-α is overproduced.
Tumor necrosis factor alpha (TNF-α) was isolated in 1975 as the agent capable of inducing necrosis of tumors and has been exploited to treat malignancy (11); however, its major role appears to be in early inflammatory responses and the host response to intracellular organisms. TNF-α, along with interleukin-6 (IL-6), primarily mediates the acute-phase response. It is capable of inducing the expression of adhesion molecules that regulate migration of neutrophils in an early inflammatory response, and it induces the expression of NO in mice and an unknown mediator in humans that is required for intracellular killing of mycobacteria (2, 15, 45, 57). Given these important roles, it is no surprise that therapeutic inhibition of TNF-α leads to an increase in infections and lymphoma (4, 20).
The regulation of expression of TNF-α is complex. Transcription is regulated in a tissue-specific and stimulus-specific manner (17, 18, 59). While many cells can produce TNF-α, the dominant producers of TNF-α are myeloid cells and T cells (8). Myeloid-lineage cells produce TNF-α upon stimulation of Toll-like receptors, activation via cytokines, and activation via lipid mediators (24, 35, 37, 53). In each case, TNF-α is rapidly upregulated with transcription initiating within minutes and protein production beginning within a few hours. In the response to lipopolysaccharide (LPS), the rapid production of TNF-α protein is dependent on transcriptional effects, regulation of message splicing, regulation of message turnover, and regulation of translation (14, 35, 41, 59). Distal effects include cleavage of the mature trimer from the cell surface by a metalloprotease, TNF-α-converting enzyme (TACE) (1). This complex regulatory strategy is felt to be critical to control a cytokine with significant pathological consequences associated with both overexpression and underexpression.
The transcriptional regulation has been extensively studied, and expression in transient transfection assays is primarily controlled by a small promoter fragment spanning −120 to −1 (19, 47, 58). Transcription factor binding is tissue specific and stimulus specific (46, 60, 66). Nuclear factor of activated T cells (NFAT) plays a major role in T-cell expression, while Egr-1 plays a major role in monocyte expression. CBP/p300 and activating transcription factor 2 (ATF2) have both been demonstrated to play a role in TNF-α transcription, and both are directly capable of acetylating adjacent histones (17, 23, 26). The gene coding for TNF-α is one of a set of genes inducibly expressed after LPS stimulation and belongs to the category of early response genes. TNF-α and other early response genes have rapid transcription after stimulation, which is independent of new protein synthesis. Secondary response genes include other inflammatory mediators such as IL-6 and IL-12, which are regulated distinctly. Although early response genes and secondary response genes typically rely on similar transcription factors (NF-κB, AP-1, and C/EBP), the binding of NF-κB is delayed in the secondary response genes and this appears to reflect chromatin accessibility (49).
Very little is known about the epigenetic regulation of TNF-α. An enhancer in intron 3 appears to specify T-cell transcription and displays DNase I hypersensitivity in T cells but not myeloid cells (5). This Ets-binding enhancer was found to be important in the regulation of TNF-α expression in the murine monocytic cell line RAW264 in a transfection system, leaving open the question of its potential role in myeloid cells (56). Maturation of monocytic cells and high-glucose conditions have both been shown to increase histone acetylation at the TNF-α locus, and this was, in turn, associated with increased TNF-α expression (34, 38). The TNF-α gene in a murine monocyte cell line exhibits significant histone acetylation and is constitutively associated with BRG-1, a component of the SWI/SNF nucleosome remodeling complex. Depletion of BRG-1 did not affect transcription of TNF-α, suggesting that it may be involved in the establishment of the chromatin environment but is dispensable for maintenance (44). Studies of other cytokines have shown that tissue-specific expression may be controlled epigenetically, and these data suggest that histone modifications represent a common regulatory strategy for cytokines (32).
The ability of a promoter to bind relevant transcription factors appears to reflect a summation of events that define competence, including nucleosome position, histone modifications, nuclear localization, and DNA methylation status. The localization of a gene within permissive euchromatin or repressive heterochromatin has not been extensively studied but appears to strongly predict competence for transcription (10, 48). Localization to transcription factories may also be important for the coordinate regulation of genes (43). Allelic exclusion of immunoglobulin genes is mediated in part through sequestration of the inactive allele in heterochromatin (29, 48), demonstrating the importance of correct nuclear localization. DNA demethylation is also a precondition for transcription in general. DNA demethylation is typically required for active transcription and is most often developmentally regulated (3, 62, 63). In addition, genes may be regulated through modification of histones. Histone H3 lysine 4 dimethylation and trimethylation occur at the transcription initiation site, with trimethylation having a high correlation with active transcription and dimethylation signaling a state of readiness or competence (7). H3 lysine 4 methylation facilitates interactions with BRG-1 and SNF2H chromatin remodeling factors via Nurf (30, 51, 64). Greater understanding of the sequence of epigenetic changes at the TNF-α locus may lead to improved therapeutic strategies.
Our previous work demonstrated that maturation of monocyte lineage cells was accompanied by increasing histone acetylation at the TNF-α locus (34). Furthermore, inhibition of histone deacetylases, which leads to globally increased histone acetylation, was associated with an increased ability of a cell line to produce TNF-α. Other studies have suggested that the TNF-α gene may be poised, with acetylated histones, prebound Brg-1, and an open chromatin conformation (44). This article describes the developmentally regulated epigenetic changes leading to transcriptional competence, demonstrates that epigenetic changes directly regulate transcriptional competence, and describes additional changes occurring with transcription. As TNF-α is a cytokine implicated in a wide range of inflammatory disorders, definition of the variables governing transcription has become extremely important.
MATERIALS AND METHODS
Cell culture, transfections, and reagents.
K562, THP-1, and HL60 cells and monocytes/macrophages were cultured in RPMI with 10% fetal bovine serum. The NTERA-2cl.D2 cell line was cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Monocytes were obtained from normal human donors and purified by elutriation at the Penn Center for AIDS Research. They were approximately 90% pure as defined by CD14 expression. Macrophages were differentiated by adherence to plastic for 7 days. Hematopoietic stem cells were obtained from peripheral blood after stimulation (Fred Hutchinson Cancer Center, Seattle, WA) and were purified by Miltenyi Clinimacs to 90% purity and frozen according to standard protocols.
Transfection of cells was performed by electroporation. Two million cells were transfected by electroporation according to the manufacturer's recommendation (Amaxa Biosystems, Gaithersburg, MD) with 1.5 μg of small interfering RNA (siRNA). The cells were rested for 48 h prior to analysis. The siRNA for RbBP5 was synthesized by the A4 protocol by Dharmacon to the target NNUGAACGUUGUGGCAUCUUU. The siRNA for Ash2L was synthesized by the A4 protocol by Dharmacon to the target NNAAAGAUGGCUAUCGGUAUA. Both of these siRNAs have been independently validated for their targets (13).
The histone methyltransferase inhibitor 5′-deoxy-5′ methylthioadenosine (MTA) was resuspended in an acidic solution, and the diluent was used in all of the control wells. The concentrations are indicated in Fig. 5. Azacytidine and the histone deacetylase inhibitor trichostatin A (TSA) were added to cells at final concentrations of 250 nM and 100 nM, respectively. LPS (chromatography purified; catalog no. P-8139 [Sigma Aldrich, St. Louis, MO]) was used at 1 μg/ml unless otherwise indicated. Phorbol myristate acetate (PMA [Sigma Aldrich]) was used at 100 ng/ml unless otherwise indicated. Immune complexes were produced using rabbit anti-bovine serum albumin as previously described and were used at 1 μg/ml (34).
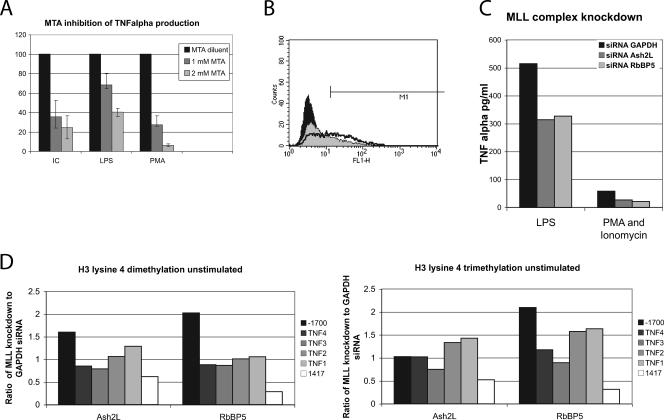
FIG. 5.
(A) The histone methylation inhibitor MTA markedly decreased the ability of the cells to produce TNF-α, as detected by ELISA. The results are expressed as a percentage of the TNF-α produced by each stimulus in the absence of the MTA inhibitor. “IC” represents rabbit anti-bovine serum albumin immune complexes at 1 μg/ml. MTA diminishes the response to all stimuli. (B) The methylation inhibitor MTA reduced the ability of the cells to produce TNF-α by flow cytometry. Only 0.6% of the cells were positive by flow cytometry when unstimulated. After treatment with 1 μg/ml of LPS for 6 h, 47.6% of the cells were positive and had a mean fluorescent intensity of 56.3 U (black line). MTA pretreatment followed by LPS stimulation led to 19.5% of the cells being positive, with a mean fluorescent intensity of 39.9 U (gray fill). The black fill represents unstimulated, untreated cells. (C) ELISA study of siRNA-transfected THP-1 cells. Cells were stimulated for 6 h with 100 ng/ml of LPS 48 h after transfections, and supernatants were tested for TNF-α. siRNAs directed at Ash2L and RbBP5 have been shown to compromise H3 lysine 4 dimethylation and trimethylation. (D) The two MLL knockdown siRNAs and siRNA to GAPDH were transfected in to THP-1 cells in parallel cultures. The data are expressed as the signal in the Ash2L- and RbBP5-transfected cells normalized to GAPDH. H3 lysine 4 dimethylation was increased in the MLL knockdowns in the 3′ region of LTα and clearly decreased in the TNF-α third intron enhancer region.
hESC culture and hEB formation.
The human embryonic stem cell (hESC) lines H9 and H1 (55), cultured in mouse embryonic fibroblast-conditioned medium (MEF-CM) (12, 65), were passed in MEF-CM. MEF-CM was collected as previously described (12, 65) and contained 8 ng/ml of basic fibroblast growth factor (bFGF). Maintenance of hESC lines was performed as previously described (12, 65). In brief, hESC colonies were dissociated with collagenase IV (Gibco, Burlington, Canada) for 5 min and passaged every 6 to 7 days. Occasionally, hESC colonies maintained in MEF-CM were physically dissociated during passaging in order to reduce the numbers of fibroblast-like cells. Formation of human embryoid bodies (hEBs) was performed as previously described (12, 65) in EB medium alone or EB medium plus cytokines and BMP-4 (see Fig. 6). hEBs were collected at days 4, 10, and 15. Single cells from hESCs and hEBs were prepared as previously described (65).
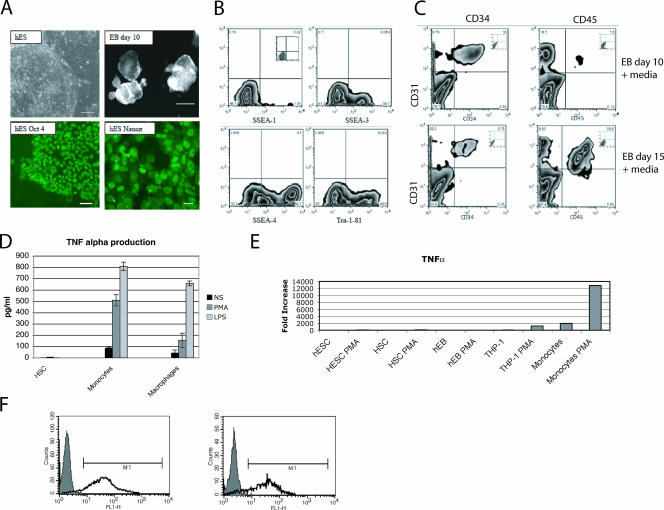
FIG. 6.
Primary cell characterization. (A) Undifferentiated hESCs and hEBs. Undifferentiated hESC culture, phase contrast, bar = 100 μm; Oct 4-stained undifferentiated hESC colony, bar = 50 μm; Nanog-stained undifferentiated hESC colony, bar = 50 μm; hEBs, day 10, bar = 500 μm. (B) Flow cytometry analysis of undifferentiated hESCs. SSEA-1, SSEA-3, SSEA-4, and Tra-1-81 (inset is the isotype control) were used to confirm the features of ESCs. (C) Flow cytometry analysis of hEBs in EB medium plus cytokines plus BMP-2 at day 10 and day 15. Hematopoietic markers: CD31 (y axis), CD34, and CD45. Insets are isotype controls. (D) Hematopoietic stem cells (HSC) do not produce TNF-α by ELISA. Cells were stimulated with PMA and ionomycin, 1 μg/ml of LPS, or diluent (NS) for 6 h. TNF-α was measured by ELISA. (E) Cells from early differentiation states do not produce TNF-α message. Cells were mock treated or treated with 100 ng/ml PMA and 500 ng/ml of ionomycin for 3 h. RNA was isolated and reverse transcribed. The untreated ESCs were used as the calibrator. cDNA quantity is expressed as increase (fold) over unstimulated ESCs. (F) Peripheral blood monocytes are largely competent for the production of TNF-α. Two different preparations of monocytes stimulated with LPS are shown using intracellular flow cytometry.
Flow cytometry, protein analysis, and RNA analysis.
Cells were treated with BD Golgiplug (BD Pharmingen, San Diego, CA), stimulated for 6 h, and stained according to the manufacturer's instructions. For sorting and analysis, a FACSCalibur flow cytometer with CellQuest software was utilized (BD Biosciences, Mountain View, CA). Enzyme-linked immunosorbent assays (ELISAs) for TNF-α were performed as previously described (35). RNA was quantitated by reverse transcription and quantitative PCR using ABI (Foster City, CA) TNF-α and 18S primers on a Taqman SDS 7900HT. The cDNA was normalized to the 18S signal and then expressed as an increase (fold) over the ESC signal (calibrator).
Bisulfite analysis.
DNA was extracted with a PUREGENE Genomic DNA isolation kit (Gentra Systems, Minneapolis, MN). DNA (5 μg) was digested with HindIII (New England Biolabs, Beverly, MA) and purified with a GeneClean II kit (Bio 101, Vista, CA). The purified DNA was denatured and treated with bisulfite as previously described (54). PCR-generated fragments were amplified as two overlapping fragments using nested primers. The following oligonucleotides were used for fragment 1: PCR round 1, 5′-GATTTTAGAGTTTTTTGGAAGTTAAGA-3′ and 5′-ATAATAAACCCTACACCTTCTACT-3′; and round 2, 5′-GTTAAGATTGAAATTAGTATTATGAGT-3′ and 5′-AAAAATTAAAAACACACAAACATCAAA-3′. The following oligonucleotides were used for fragment 2: PCR round 1, 5′-GTTTTTAGGGTTTTATATATAAATTAG-3′ and 5′-TCCATTTCCCCTTAAATAAAAAAAT-3′; and round 2, 5′-GTATTTTTGATGTTTGTGTGTTTTTAA-3′ and 5′-TCACTCAAAATACAACAAACAAAAA-3′. PCR products were subcloned into pGEM-T Easy plasmid (Promega, Madison, WI), and approximately 10 individual colonies were isolated and sequenced per experiment.
FISH analysis.
For fluorescent in situ hybridization (FISH), cells were resuspended in 0.56% KCl for 20 min at 37°C. Fixative was added, and the cells were centrifuged and resuspended in fixative (3:1 methanol-glacial acetic acid) at a concentration of about 1 × 106/ml and maintained at −20°C. The slides were prepared as previously described (40). The centromere probe set corresponding to the autosome centromeres was labeled with fluorescein isothiocyanate (FITC) (Vysis, Downers Grove, IL), the bacterial artificial chromosome (BAC) probe containing the TNF-α locus (RPCI-11-184F16; CHORI, Oakland, CA) was labeled with Spectrum orange (Vysis) by nick translation, and they were cohybridized at 37°C overnight with the cell DNA in hybridization buffer (Vysis) after denaturation. The slides were washed in 0.4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.3% IGEPAL (Sigma, St. Louis, MO) at 74° for 5 min and then 2× SSC-0.1% IGEPAL at room temperature for 3 min. DAPI (4′,6′-diamidino-2-phenylindole) counterstain was added, and the slides were viewed on a Leica TCS SP2 spectral confocal system using z-steps of 3.7 to 10 μM.
ChIP.
TNF 1, 2, 3, 4 primer-probe combinations have been previously published (34). The TNF-1700 primer-probe combination lies within the 3′ untranslated region of the lymphotoxin α (LTα) gene: downstream primer 5′-CCCCTGGGATGCCTAGAATT-3′, upstream primer 5′-CAACAGCTCAAGTCTTCCCTGAT-3′, and 6-carboxyfluorescein (FAM) probe 5′-TTGAAAGCTCCGGTGACT-3′. The 1417 primer-probe combination lies within the third intron enhancer (5): downstream primer 5′-CCTGGCCATGACGTTCTGA-3′, upstream primer 5′-AGGGAAGGGTGGAGGAACAG-3′, and FAM probe 5′-ATCCCACTAAGGCCTG-3′. The first intron primer is located at +720: downstream primer 5′-TCTTTCCCTGAGTGTCTTCTGTGT-3′, upstream primer 5′-GCGGGAAATATGACAGCTAAGG-3′, and FAM probe 5′-AGGAGAGAAGAAGATAGGGTGT-3′. Threshold cycle (CT) values were normalized to 10% of the input of the sample according to the formula 2 (10% input CT − sample CT). Final values reported represent averages of duplicates or triplicates and multiple experiments.
Chromatin immunoprecipitation (ChIP) assays were done according to the protocol obtained from Upstate Biotechnology, with several modifications. Five to 10 million cells were treated with formaldehyde at a final concentration of 1% for 10 min at room temperature. Glycine was added to a final concentration of 0.125 M for 5 min at room temperature. Cells were washed and lysed, and DNA was sonicated to a size of approximately 200 to 400 bp. Chromatin was immunoprecipitated overnight with antibodies for total histone H3 (05-928), acetylated histone H3 (06-599), or acetylated histone H4 (06-866) (Upstate Biotechnology, Waltham, MA) or dimethylated histone H3K4 (ab7766), trimethylated histone H3K9 (ab8898), or trimethylated histone H3K4 (ab8580) (Abcam, Cambridge, MA). A negative control antibody (anti-glutathione S-transferase [GST]; Abcam) was always included but never gave a significant signal and is omitted from some of the figures for simplicity. Immune complexes were collected using a protein A slurry (Invitrogen, Carlsbad, CA), washed, and eluted. The DNA was reverse cross-linked overnight at 65°C and extracted via phenol-chloroform and ethanol. Protein was removed with proteinase K incubated at 65°C with 100 μg of glycogen. Quantitation was performed on a Taqman SDS 7900HT.
RESULTS
DNA is demethylated at the TNF-α locus in cells that rapidly express TNF-α.
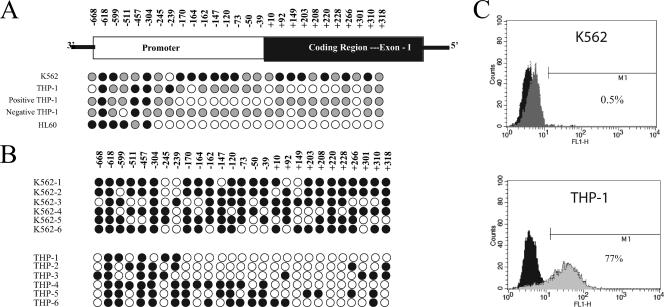
Bisulfite analysis was performed on two cell lines to define the status of DNA methylation at the TNF-α locus. The TNF-α gene does not contain a classical CpG island. A region of the promoter at −150 and a region in the first exon at +450 contain CpG-rich sequences. The TNF-α locus from −668 to +349 was sequenced from multiple clones after bisulfite treatment (Fig. 1). K562 cells are multipotent malignant hematopoietic cells and do not produce TNF-α after LPS or PMA stimulation. These cells have methylated or partially methylated DNA throughout this region (Fig. 1A). Examination of individual K562 clones revealed that all clones exhibited sporadic methylation (Fig. 1B). THP-1 cells fairly uniformly produce TNF-α in response to PMA or LPS, although some cells are refractory (Fig. 1C). These cells were more extensively demethylated than the less mature K562 cells (Fig. 1A). Individual clones were more heterogeneous than the K562 clones, with some clones exhibiting more extensive demethylation and some clones exhibiting more extensive methylation. THP-1 cells were sorted by flow cytometry based on intracellular cytokine staining. The nonexpressing cells were analyzed by bisulfite analyses, as were the expressing cells. The TNF-α-expressing THP-1 cells (positive) were more extensively demethylated than the nonexpressing cells (negative) (Fig. 1A). As an independent verification that DNA methylation status correlates with the potential to produce TNF-α, HL60 cells were examined. HL60 is another myeloid cell line which produces TNF-α. The TNF-α promoter of HL60 cells was slightly more demethylated than the TNF-α-expressing THP-1 cells (Fig. 1A).
FIG. 1.
Bisulfite analysis of DNA methylation in cell lines. Filled black circles represent >80% methylation at a given CpG after sequencing 5 to 10 individual clones. A gray circle represents between 20 and 80% methylation at a given CpG. An open circle represents <20% methylation at a given CpG. (A) Three cell lines were analyzed as representative of cells incapable of producing TNF-α (K562) and competent to produce TNF-α (THP-1 and HL-60). THP-1 cells were stimulated with LPS, sorted by flow cytometry based on expression of TNF-α, and analyzed by bisulfite conversion. These results are indicated as positive (+) and negative (−). (B) Individual clones of two partially methylated cell lines (K562 and THP-1). THP-1 cells show significant heterogeneity in the pattern of methylation between clones, whereas K562 has sporadic bases demethylated on all clones. (C) Flow cytometry demonstrating the cell lines' competence to produce TNF-α. K562 cells (top panel) were stimulated with 100 ng/ml of PMA for 6 h. (K562 does not express TLR4 or CD14 receptors for LPS.) THP-1 cells (lower panel) were stimulated with 1 μg/ml of LPS for 6 h. The black fill represents the unstimulated cells stained with the anti-TNF-α antibody. The gray fill represents the stimulated cells.
The TNF-α locus is partially partitioned in euchromatin.
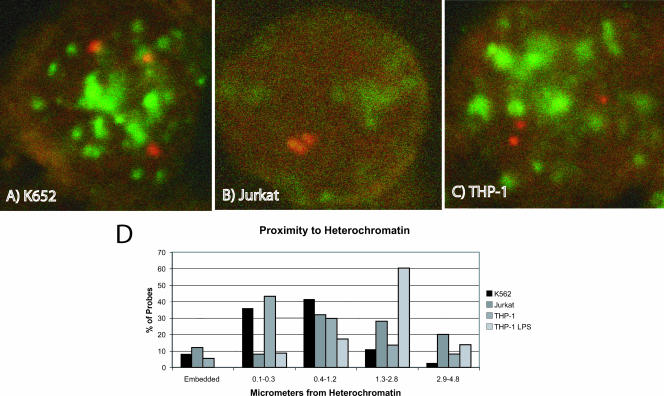
Localization of a gene within euchromatin appears to be a correlate of competence for expression (10, 52). We used FISH analyses to define the position of the TNF-α locus within K562, THP-1, and Jurkat cells (Fig. 2). The latter cell line is a T-cell leukemia line which expresses TNF-α after stimulation with PMA plus ionomycin. A set of centromere probes were used to define heterochromatin, and a BAC clone containing the TNF-α locus was used to define the nuclear localization (Fig. 2A, B, and C). In each cell line, the TNF-α locus was found heterogeneously in both euchromatin and heterochromatin despite the differences in transcriptional competence (Fig. 2D). THP-1 and K562 are triploid for chromosome 6. We defined heterochromatin colocalization as a distance of ≤0.3 μm from a centromeric signal (48).
FIG. 2.
FISH analyses of nuclear localization of the TNF-α locus. The THP-1 and K562 cell lines are triploid for chromosome 6. The confocal panels show representative cells. Centromere probes are shown in green (FITC), and the TNF-α locus is shown in red (Spectrum orange). The graph shows the cumulative findings from 10 to 20 cells. (A) K562 cells. One locus is in heterochromatin in the upper right corner. (B) Jurkat cells. (C) THP-1 cells. All three loci are in euchromatin. (D) Graphical representation of the proximity of the TNF-α loci to the closest centromeric heterochromatin. “THP-1-LPS” refers to THP-1 cells treated with 1 μg/ml of LPS. A distance of ≤0.3 μm is considered localization to heterochromatin.
The TNF-α locus has focused areas of histone modifications in cells that express TNF-α.
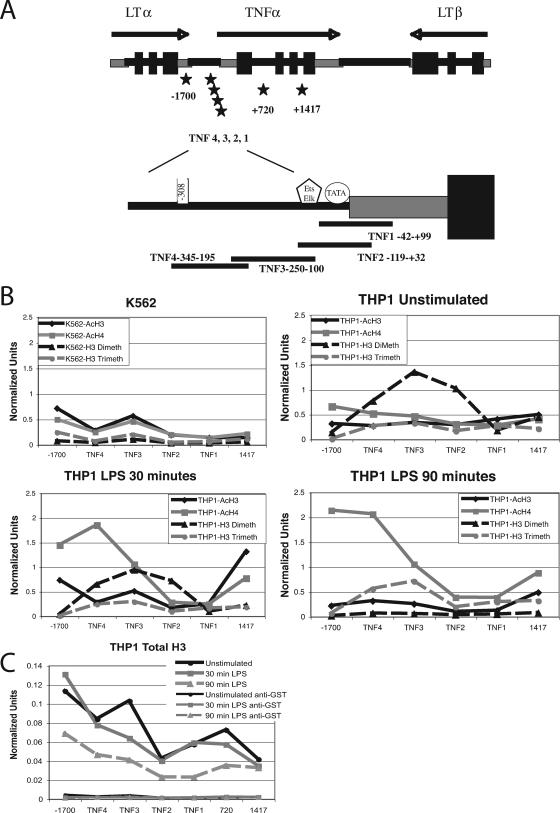
THP-1 cells and K562 cells were examined by ChIP analysis to define whether histone modifications were associated with competence to express TNF-α (Fig. 3). In addition, THP-1 cells were tested 30 min and 90 min after stimulation with 1 μg/ml of LPS to determine whether active stimulation altered histone modifications at this locus. The ChIP assays utilized real-time PCR primers and probes corresponding to four overlapping regions in the TNF-α promoter, one distant upstream region (−1700) within the 3′ untranslated region of the LTα gene, and one distant downstream region (1417) located within the third intron enhancer of TNF-α (Fig. 3A). The Ets-binding enhancer has been reported to function in both T cells and monocytes; however, the data on its role in monocytes are conflicting (5, 56). THP-1 cells, which are competent to express TNF-α, had more H3 dimethylated lysine 4 at the TNF-α locus than K562, which does not produce TNF-α (Fig. 3B). LPS stimulation increased the H4 acetylation of the TNF-α locus at two specific regions, the distal upstream promoter (TNF4) and the enhancer region (+1417). H3 acetylation was increased only in the enhancer region (+1417). Acetylated H3 and H4, along with dimethylated H3 lysine 4, appear to mark both active promoters and enhancers, although there is extensive variation in the specific pattern (25).
FIG. 3.
Histone acetylation and methylation at the TNF-α locus. These results represent an average of duplicates or triplicates of two to three separate experiments. The normalized units represent the signal from the immunoprecipitate normalized to 10% of the original input DNA. (A) Schematic representation of the primers and probes used for real-time PCR quantitation. (B) Graphical representation of ChIP analyses. AcH3, acetylated H3; AcH4, acetylated H4; H3 Dimeth, H3K4 dimethylation; H3 Trimeth, H3K4 trimethylated. (C) Total H3 ChIP.
Histone H3 lysine 4 dimethylation and H3 lysine 4 trimethylation are increased in genes that are transcriptionally competent, with H3 lysine 4 trimethylation correlating with active transcription (7, 22, 50). Exceptions to this pattern have been described (27, 42). In particular, the upstream trimethylation mark is thought to cue phosphorylation of the C-terminal domain of RNA polymerase II, which is required for transcription initiation (22). The dimethylation of lysine 4 decreased late in transcription and was replaced by the trimethyl mark. H4 acetylation persisted at 90 min. We hypothesized that the pattern of histone modifications overall, with lower levels of histone modifications at TNF1 and TNF2, was due to a lack of nucleosomes in this region. Confirming this, a ChIP was performed with an antibody recognizing total H3, which demonstrated that the proximal promoter region has little nucleosome content. The promoter appears to undergo some nucleosome remodeling after LPS stimulation (Fig. 3C). The enhancer region and proximal promoter are most depleted of nucleosomes. An amplimer at +720 in the first intron demonstrates that the nucleosome depletion at the third intron enhancer is specific for the enhancer and not due to generalized loss as a consequence of active transcription. In this figure, the negative control antibody, anti-GST, is shown. It was run as a control in all ChIP assays; however, for simplicity, it is shown in only this figure.
Epigenetic variables control TNF-α gene expression.
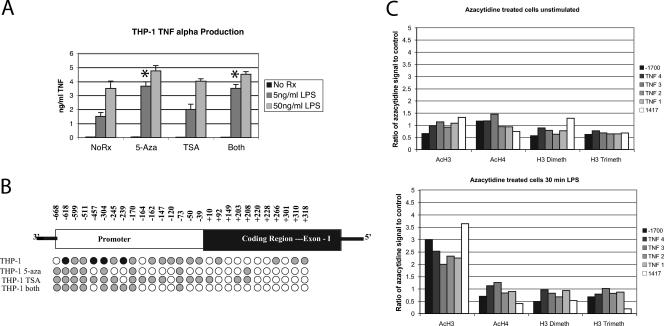
To determine whether DNA demethylation or histone acetylation could enhance TNF-α expression, THP-1 cells were treated with 5-azacytidine for 24 h (Fig. 4A). TSA treatment was also included as a histone deacetylase inhibitor. At 24 h, when most of the cells had divided once, TSA treatment increased the TNF-α produced after low-dose stimulation modestly. 5-Azacytidine augmented the ability of the cells to respond to LPS stimulation. 5-Azacytidine treatment led to demethylation of the TNF-α promoter (Fig. 4B). These data support a direct role for DNA demethylation in the regulation of TNF-α expression. The biological effect was significant, with a doubling of TNF-α protein after low-dose LPS in spite of the fact that DNA demethylation was incomplete.
FIG. 4.
Modification of epigenetic context alters the competence to produce TNF-α. (A) THP-1 cells were treated for 24 h with 100 nM TSA, 250 nM 5-azacytidine (5-Aza), or both and stimulated with 5 or 50 ng/ml of LPS. TNF-α was measured after 6 h by ELISA. 5-Azacytidine increased TNF-α production more significantly than TSA. At 24 h of treatment, the cells remained >90% viable. Longer courses of treatment (Rx) impaired viability. *, P < 0.01. (B) Prolonged 5-azacytidine treatment led to demethylation of the promoter in THP-1 cells, while TSA treatment did not markedly alter the methylation status. (C) 5-Azacytidine treatment of THP-1 cells for 48 h did not substantially alter the pattern of histone modifications. Levels of modification are displayed as a ratio of 5-azacytidine-treated cells to control THP-1 cells.
In certain systems, DNA methylation and histone modifications are mechanistically linked. Methylated DNA can serve as a docking site for methyl CpG binding proteins, which recruit histone deacetylases (9). Conversely, DNA methyltransferases can bind to H3 lysine 9 methyltransferases to methylate DNA de novo (36). To understand whether DNA methylation influences histone modifications or whether histone acetylation could influence DNA methylation, we treated cells with either TSA or 5-azacytidine. TSA treatment did not affect DNA methylation (Fig. 4B). 5-Azacytidine treatment for 48 h did not substantially alter histone modifications (Fig. 4C). H3 acetylation was increased in the 5-azacytidine-treated cells only after LPS stimulation. This was a consistent observation but of uncertain consequence.
Histone H3 lysine 4 methylation is typically associated with active transcription. To determine whether this epigenetic mark was critical for TNF-α expression, histone methylation was inhibited by MTA. MTA was added, and cells were stimulated 2 h later. Viability was assayed by trypan blue and was always greater than 90%. Six hours after stimulation, the cell supernatants were harvested and tested for TNF-α production by ELISA and intracellular flow cytometry. Various concentrations of MTA were tested in THP-1 cells, a cell line with maximal competence to produce TNF-α. Inhibition of histone methylation markedly reduced the cells' ability to produce TNF-α (Fig. 5A). Analysis by flow cytometry to determine whether individual cells became incompetent or whether all cells had a reduced ability to produce TNF-α revealed that 48% of the cells produced TNF-α 6 h after treatment with 1 μg/ml of LPS, with a mean fluorescent intensity of 56.3 U. With MTA pretreatment at a 1 mM concentration, only 20% of the cells produced TNF-α and the mean fluorescence intensity of the positive cells was reduced (39.9 U) compared to that of the cells that did not receive MTA (Fig. 5B). These data are consistent with a continuum effect.
The H3 lysine 4 methylation evaluated on the ChIP assays is typically deposited via one of the mixed-lineage leukemia (MLL) family of histone methyltransferases which share a SET domain (33). In humans, there are four MLL family histone methyltransferases, which interact directly with the basic transcriptional machinery (16). MLL itself is part of a large complex including WDR5, which regulates interactions with the histone substrate Ash2L, which provides a structural role, and RbBP5, which may also play a structural role (13). In addition, MLL members, both as cleaved forms and as intact protein, interact with menin and ASC2 (39). Knockdown of Ash2L or RbBP5 compromises dimethylation and trimethylation of H3 lysine 4 but not monomethylation (13). To further demonstrate the role of H3 lysine 4 methylation, we transfected THP-1 cells with siRNA for RbBP5 and Ash2L. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) siRNA was used as a control. Figure 5C demonstrates the decrease in TNF-α production by ELISA after transfection with either siRNA to Ash2L or RbBP5, consistent with a role for MLL in the methylation of H3 lysine 4 and demonstrating the importance of H3 lysine 4 methylation in the regulation of TNF-α. This effect was independent of stimulation, as one would expect. ChIP assays on transfected cells revealed a clear decrease in H3 lysine 4 methylation at intron 3 in resting cells; however, an effect at the promoter was not observed, perhaps reflecting the low levels of H3 lysine 4 dimethylation and/or a limitation of the transfection efficiency (Fig. 5D).
It is not possible to alter nuclear localization; however, we attempted to address whether nuclear localization was fixed or mutable. THP-1 cells were treated with 1 μg/ml LPS to induce TNF-α expression. Two hours later, FISH was performed to define nuclear localization (Fig. 2D). LPS stimulation drove the TNF-α locus completely into euchromatin, suggesting that nuclear localization is an independent variable governing TNF-α expression. This result is somewhat surprising. The limited data on nuclear localization have suggested that this is largely developmentally regulated. These data demonstrate that nuclear localization is modifiable by acute stimuli.
Evidence from primary cells.
These data suggested that an orderly progression of epigenetic changes lead to transcriptional competence and production of TNF-α. DNA demethylation appeared to correlate with hematopoietic cells acquiring competence to produce TNF-α. Nuclear localization was heterogeneous in all three cell lines. Each of the cell lines examined had a TNF-α locus partitioned in both euchromatin and heterochromatin. Finally, histone acetylation appeared to correspond with both competence to express TNF-α and active transcription. H3 lysine 4 dimethylation correlated with transcriptional competence, while H3 lysine 4 trimethylation was induced with active transcription. We wished to confirm these findings as much as possible in primary cells because it has become clear that malignant transformation can be due to altered epigenetic control of gene expression (28).
hESCs, EBs at various stages of development, hematopoietic stem cells, liver tissue, and peripheral blood monocytes and macrophages were examined. The developmental statuses of the ESCs, EBs, and hematopoietic stem cells were confirmed by flow cytometry and microscopy (Fig. 6). The ESCs and EBs had distinct morphologies by phase contrast (Fig. 6A). The embryonic stem cells were also positive for the POU transcription factor Oct 4 and the homeodomain protein Nanog (Fig. 6A). To further confirm the identity of the ESCs, flow cytometry was performed. ESCs are typically defined as SSEA-1 negative or SSEA-3, SSEA-4, and Tra-1-81 positive (Fig. 6B). EBs containing endoderm, mesoderm, and ectoderm develop CD34-positive hematopoietic precursors by day 10 (Fig. 6C). These studies confirm the identity and purity of the ESCs and the EBs. Hematopoietic stem cells and less mature cells do not produce TNF-α, while peripheral blood monocytes and fully differentiated macrophages do produce TNF-α protein and message after stimulation (Fig. 6D and E). The monocytes used in these studies were purified by elutriation. Although their surface phenotype is somewhat heterogeneous, 90% of the cells produced TNF-α after stimulation, as defined by intracellular cytokine staining (Fig. 6F). The surface phenotype varied somewhat from donor to donor but was typically 100% DR+, 100% HLA class I positive, 60 to 70% CD11b+, 60% CD14hi, 30% CD14lo, 20% CD16+, and 0% mannose receptor positive.
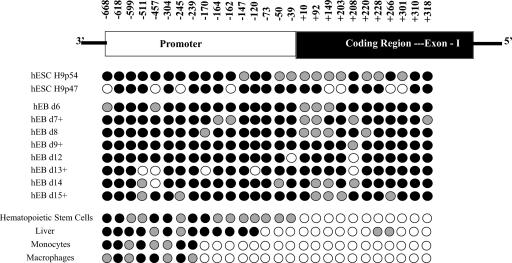
DNA methylation was examined by bisulfite analysis, as before, in these primary cells (Fig. 7). These data demonstrate quite clearly that the TNF-α locus undergoes substantial demethylation during the transition from EB to hematopoietic stem cell. These EBs are capable of differentiating into the three germ cell layers and at day 11 include cells with hematopoietic potential (12, 61). At all stages, EBs contain heterogeneous cell types. At the point of hematopoietic stem cell development, exon 1 appears to be largely demethylated, while the promoter is partially methylated. With further differentiation into peripheral blood monocytes and fully differentiated macrophages, the promoter undergoes additional demethylation. This full demethylation extends to a point in the primary cells that was also demarcated in the cell lines between −170 and −239.
FIG. 7.
DNA methylation status of the TNF-α locus in primary cells. The legend is as for Fig. 1. Cell sources were undifferentiated hESCs, hEBs harvested at various times in culture (+, growth in cytokine-supplemented media), hematopoietic stem cells (HSC) purified from a peripheral blood harvest, human liver, peripheral blood monocytes purified by elutriation, and peripheral blood monocytes differentiated by adherence for 7 days into macrophages.
Nuclear localization in primary cells was more clear cut than in cell lines. The TNF-α locus was found exclusively in heterochromatin in ESCs, EBs, and hematopoietic stem cells. In primary monocytes, the locus was found in euchromatin consistently (Fig. 8). It became clear that in the primary cells, there was a gradual transition from heterochromatin, with the TNF-α locus stepping progressively farther away from the centromeric heterochromatin (Fig. 8B). In undifferentiated ESCs, the TNF-α locus was always embedded in centromeric heterochromatin. In EBs, the TNF-α locus was found both embedded in heterochromatin and adjacent to heterochromatin probes. Hematopoietic stem cells also had their TNF-α locus in heterochromatin, as defined by a proximity of ≤0.3 μm; however, none of the probes were directly embedded within heterochromatin. The functional significance of this is uncertain. Monocytes always had their TNF-α loci in euchromatin.
FIG. 8.
(A) Nuclear localization of the TNF-α locus in primary cells. Centromere probes are shown in green (FITC), and the TNF-α locus is shown in red (Spectrum orange). Each section is between 3 and 10 μm thick. (B) Graphical representation of the proximity of the TNF-α probe to the nearest centromeric heterochromatin. Differentiation is accompanied by gradual movement from heterochromatin to euchromatin. Undiff, undifferentiated.
Hematopoietic stem cells and primary monocytes were examined by ChIP to define H3 and H4 acetylation and methylation status (Fig. 9A). Hematopoietic cells do not produce TNF-α and do not bear any of the histone modifications characteristic of activation examined in this study. Monocytes demonstrated slightly higher histone H4 acetylation and H3 lysine 4 methylation at their baseline than THP-1 cells. (Note that the y axes are different between Fig. 3 and 9.) Both H4 acetylation and H3 lysine 4 trimethylation increased dramatically early in transcription. The pattern was similar to that in the cell lines, but the magnitude was greater (with the axes expanded for the primary cells). The histone modifications are largely back to baseline 90 min after LPS treatment. We also examined hematopoietic stem cells and monocytes for the presence of a modification that is characteristically associated with developmental repression, H3 lysine 9 trimethylation (Fig. 9B). Both hematopoietic cells and monocytes had extremely low levels of this modification at the TNF-α locus. As a positive control, we used NTERA-2cl.D2 cells, a testicular embryonal carcinoma line which can differentiate down a neural path with retinoic acid. These cells had a higher level of H3 lysine 9 trimethylation than either hematopoietic stem cells or monocytes, suggesting that this repressive mark had been removed prior to hematopoietic cell differentiation.
FIG. 9.
Histone modifications in primary cells. (A) ChIP analyses were performed utilizing the same primer-probe combinations as in Fig. 3. The abbreviations are as defined in the legend to Fig. 3. Hematopoietic stem cells (HSC) were examined untreated. Primary monocytes were examined fresh as well as 30 min and 90 min after 1-μg/ml LPS stimulation. These results represent an average of duplicates or triplicates and at least two separate experiments. (B) H3 lysine 9 trimethylation, a repressive mark, was examined in hematopoietic stem cells and peripheral blood monocytes. The NTERA-2cl.D2 cell line was used as a positive control.
DISCUSSION
The regulation of TNF-α expression has long been recognized as complex. The transcription factors known participate in the regulation of TNF-α are fairly broadly distributed, and yet relatively few cells produce substantial amounts of TNF-α. This suggests that another layer of control regulates which cells are competent to produce TNF-α. Early data demonstrated that fusion of a nonexpressing fibroblast cell line with a monocyte line silenced the formerly competent TNF-α genes from the monocyte line, and this was accompanied by increased DNA methylation (31). Recent data have identified histone acetylation and prebound BRG-1 as characteristic of early response genes such as TNF-α. These data together suggested that epigenetic factors might contribute substantially to the control of expression.
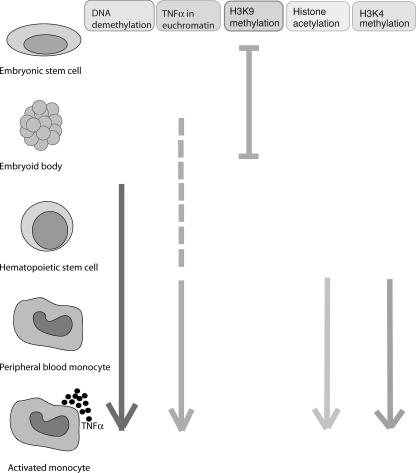
This study was focused on identifying developmentally mediated chromatin changes that establish competence for transcription in myeloid cells. We specifically examined H3 and H4 acetylation as well as H3 lysine 4 methylation. All of these modifications are supported by abundant mechanistic data demonstrating their roles in global genome transcription patterns (6, 7, 25, 50). MLL, a histone methyltransferase, has been demonstrated to bind the TNF-α promoter, and ATF2 and CBP, both histone acetylators, have been implicated in transcriptional regulation of TNF-α, supporting the strategy of evaluating these specific histone modifications (21, 23, 59; M. G. Guenther, personal communication). Our data appear to show an orderly series of events that accompany myeloid differentiation and that these events progressively lead to competence for the production of TNF-α (Fig. 10). Myeloid differentiation proceeds stepwise from ESCs through mesenchymal differentiation to hematopoietic stem cells and then monocytes. Thus, it should not be surprising that differentiation, which is a series of progressive steps, is accompanied by a series of steps that progressively alter the epigenetic landscape of the TNF-α gene, ultimately leading to transcriptional competence.
FIG. 10.
Summary of epigenetic changes at the TNF-α locus during development.
The first event is DNA demethylation of the TNF-α locus, which seems to arise from the coding region and extend 5′ into the promoter. The DNA demethylation begins to occur between the EB stage and differentiation into hematopoietic stem cells. Further demethylation continues as the cells differentiate into monocytes. A cell such as the hepatocyte, which is capable of producing TNF-α only after a priming stimulus, is only partially demethylated. A second step in the acquisition of transcriptional competence is the relocation of the TNF-α locus into euchromatin. This appears to occur just after hematopoietic stem cell development but may begin at the EB stage with a move from being embedded in centromeric heterochromatin domains to being adjacent to centromeric heterochromatin. Maturation into monocytes is accompanied by H4 acetylation and H3 lysine 4 methylation. When the cells were stimulated, histone acetylation and H3 lysine 4 methylation increased.
The primary cells were both similar and different from the cell lines. The major difference lay in the nuclear localization of the TNF-α locus. Another difference was in the pattern of histone modifications. The primary cells came from a variety of donors, and we did not observe differences between donors; however, it is possible that the peripheral blood monocytes had received some stimulation through adherence prior to the assay, accounting for the more advanced appearance of their histone modifications. This concept is supported by the finding of low levels of TNF-α transcripts by reverse transcription-PCR in unstimulated monocytes. A surprising finding was the dramatic changes occurring at the third intron enhancer with stimulation. Histone modifications at enhancers have not been widely studied, although they appear to have a different histone signature from promoters (25). This enhancer was examined in two previous publications with conflicting results (5, 56). Our data support a significant role for the third intron enhancer in primary monocytes.
Manipulation of histone acetylation, histone methylation, and DNA methylation each altered the ability of the cells to produce TNF-α, suggesting that these are not mere correlates of transcriptional competence but are mechanistically important. This study only began to examine the details of the histone modifications. ATF2 and CBP have already been demonstrated to regulate TNF-α, and MLL has now been implicated in H3 lysine 4 methylation, which appears to be important for transcription (13, 59, 60). The pathways responsible for activation of these molecules in a manner which would allow gene-specific regulation are largely unknown and likely differ from cell type to cell type. This study focused on epigenetic changes in myeloid lineage cells because they produce the bulk of TNF-α. Nevertheless, many cells can produce small amounts of TNF-α, and whether production of TNF-α in these cells is also governed by epigenetic controls is uncertain.
This work may be of relevance for understanding inflammatory diseases. Rheumatoid arthritis and Crohn's disease are characterized by TNF-α overexpression. Defining the role of epigenetic regulation of TNF-α may lead to new therapeutic strategies for these chronic inflammatory diseases. This study clearly demonstrates that epigenetic modifications are progressively acquired during monocyte differentiation. Further modifications are acquired during activation of transcription. The critical importance of these epigenetic changes was demonstrated by experimental manipulation and defining changes in TNF-α production. Therapeutic interventions directed at epigenetic variables might lead to control of this important inflammatory cytokine.
Acknowledgments
This work was supported by NIH grants AI051323 and AI44127.
The NTERA-2cl.D1 line was provided by Virginia M.-Y. Lee. The Ash2L and RbBP5 siRNA sequences were provided by Yali Dou. We thank both V. M.-Y. Lee and Y. Dou for assistance.
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Amour, A., P. M. Slocombe, A. Webster, M. Butler, C. G. Knight, B. J. Smith, P. E. Stephens, C. Shelley, M. Hutton, V. Knauper, A. J. Docherty, and G. Murphy. 1998. TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 435:39-44. [DOI] [PubMed] [Google Scholar]
- 2.Appelberg, R., A. Sarmento, and A. G. Castro. 1995. Tumour necrosis factor-alpha (TNF-alpha) in the host resistance to mycobacteria of distinct virulence. Clin. Exp. Immunol. 101:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armenante, F., M. Merola, A. Furia, and M. Palmieri. 1999. Repression of the IL-6 gene is associated with hypermethylation. Biochem. Biophys. Res. Commun. 258:644-647. [DOI] [PubMed] [Google Scholar]
- 4.Askling, J., M. Fored, E. Baecklund, L. Brandt, C. Backlin, A. Ekbom, C. Sundstrom, L. Bertilsson, L. Coster, P. Geborek, L. Jacobsson, S. Lindblad, J. Lysholm, S. Rantapaa-Dahlqvist, T. Saxne, L. Klareskog, and N. Feltelius. 2005. Hematopoietic malignancies in rheumatoid arthritis. Lymphoma risk and characteristics following TNF-antagonists. Ann. Rheum. Dis. 64:1414-1420. (First published 20 April 2005; doi: 10.1136/ard.2004.033241.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthel, R., and A. E. Goldfeld. 2003. T cell-specific expression of the human TNF-alpha gene involves a functional and highly conserved chromatin signature in intron 3. J. Immunol. 171:3612-3619. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 8.Beutler, B., and T. Brown. 1991. A CAT reporter construct allows ultrasensitive estimation of TNF synthesis and suggests that the TNF gene has been silenced in non-macrophage cell lines. J. Clin. Investig. 87:1336-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 10.Brown, K. E., S. Amoils, J. M. Horn, V. J. Buckle, D. R. Higgs, M. Merkenschlager, and A. G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602-606. [DOI] [PubMed] [Google Scholar]
- 11.Carswell, E. A., L. J. Old, R. L. Kassel, S. Green, N. Fiore, and B. Williamson. 1975. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA 72:3666-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chadwick, K., L. Wang, L. Li, P. Menendez, B. Murdoch, A. Rouleau, and M. Bhatia. 2003. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102:906-915. [DOI] [PubMed] [Google Scholar]
- 13.Dou, Y., T. A. Milne, A. J. Ruthenburg, S. Lee, J. W. Lee, G. L. Verdine, C. D. Allis, and R. G. Roeder. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13:713-719. [DOI] [PubMed] [Google Scholar]
- 14.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 15.Dunne, J. L., R. G. Collins, A. L. Beaudet, C. M. Ballantyne, and K. Ley. 2003. Mac-1, but not LFA-1, uses intercellular adhesion molecule-1 to mediate slow leukocyte rolling in TNF-alpha-induced inflammation. J. Immunol. 171:6105-6111. [DOI] [PubMed] [Google Scholar]
- 16.Ernst, P., J. Wang, M. Huang, R. H. Goodman, and S. J. Korsmeyer. 2001. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell. Biol. 21:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falvo, J. V., B. M. Brinkman, A. V. Tsytsykova, E. Y. Tsai, T. P. Yao, A. L. Kung, and A. E. Goldfeld. 2000. A stimulus-specific role for CREB-binding protein (CBP) in T cell receptor-activated tumor necrosis factor alpha gene expression. Proc. Natl. Acad. Sci. USA 97:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falvo, J. V., A. M. Uglialoro, B. M. N. Brinkman, M. Merika, B. S. Parekh, E. Y. Tsai, H. C. King, A. D. Morielli, E. G. Peralta, T. Maniatis, D. Thanos, and A. E. Goldfeld. 2000. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 20:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldfeld, A. E., C. Doyle, and T. Maniatis. 1990. Human tumor necrosis factor a gene regulation by virus and lipopolysaccharide. Proc. Natl. Acad. Sci. USA 87:9769-9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Reino, J. J., L. Carmona, V. R. Valverde, E. M. Mola, and M. D. Montero. 2003. Treatment of rheumatoid arthritis with tumor necrosis factor inhibitors may predispose to significant increase in tuberculosis risk: a multicenter active-surveillance report. Arthritis Rheum. 48:2122-2127. [DOI] [PubMed] [Google Scholar]
- 21.Guenther, M. G., R. G. Jenner, B. Chevalier, T. Nakamura, C. M. Croce, E. Canaani, and R. A. Young. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. USA 102:8603-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 23.Hayakawa, J., S. Mittal, Y. Wang, K. S. Korkmaz, E. Adamson, C. English, M. Ohmichi, M. McClelland, and D. Mercola. 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16:521-535. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, M. P., S. L. Freeman, and R. P. Donnelly. 1995. IFN-gamma priming of monocytes enhances LPS-induced TNF production by augmenting both transcription and MRNA stability. Cytokine 7:427-435. [DOI] [PubMed] [Google Scholar]
- 25.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311-318. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, H., L. Schiltz, R. Chiu, K. Itakura, K. Taira, Y. Nakatani, and K. K. Yokoyama. 2000. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405:195-200. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J., L. Jia, W. D. Tilley, and G. A. Coetzee. 2003. Dynamic methylation of histone H3 at lysine 4 in transcriptional regulation by the androgen receptor. Nucleic Acids Res. 31:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, G. 2005. Epigenetics: surveillance team against cancer. Nature 434:150. [DOI] [PubMed] [Google Scholar]
- 29.Kosak, S. T., J. A. Skok, K. L. Medina, R. Riblet, M. M. Le Beau, A. G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296:158-162. [DOI] [PubMed] [Google Scholar]
- 30.Kouskouti, A., and I. Talianidis. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24:347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruys, V., P. Thompson, and B. Beutler. 1993. Extinction of the tumor necrosis factor locus, and of genes encoding the lipopolysaccharide signaling pathway. J. Exp. Med. 177:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, G. R., S. T. Kim, C. G. Spilianakis, P. E. Fields, and R. A. Flavell. 2006. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24:369-379. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., and D. G. Skalnik. 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280:41725-41731. [DOI] [PubMed] [Google Scholar]
- 34.Lee, J. Y., N. A. Kim, A. Sanford, and K. E. Sullivan. 2003. Histone acetylation and chromatin conformation are regulated separately at the TNF alpha promoter in monocytes and macrophages. J. Leukoc. Biol. 73:862-871. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. Y., and K. E. Sullivan. 2001. Gamma interferon and lipopolysaccharide interact at the level of transcription to induce tumor necrosis factor alpha expression. Infect. Immun. 69:2847-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 37.Lin, H. I., S. J. Chu, D. Wang, and N. H. Feng. 2004. Pharmacological modulation of TNF production in macrophages. J. Microbiol. Immunol. Infect. 37:8-15. [PubMed] [Google Scholar]
- 38.Miao, F., I. G. Gonzalo, L. Lanting, and R. Natarajan. 2004. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J. Biol. Chem. 279:18091-18097. [DOI] [PubMed] [Google Scholar]
- 39.Milne, T. A., C. M. Hughes, R. Lloyd, Z. Yang, O. Rozenblatt-Rosen, Y. Dou, R. W. Schnepp, C. Krankel, V. A. Livolsi, D. Gibbs, X. Hua, R. G. Roeder, M. Meyerson, and J. L. Hess. 2005. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. USA 102:749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nath, J., and K. L. Johnson. 1998. Fluorescence in situ hybridization (FISH): DNA probe production and hybridization criteria. Biotech. Histochem. 73:6-22. [DOI] [PubMed] [Google Scholar]
- 41.Osman, F., N. Jarrous, Y. Ben-Asouli, and R. Kaempfer. 1999. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13:3280-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins, E. J., B. L. Kee, and D. A. Ramsden. 2004. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic Acids Res. 32:1942-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ragoczy, T., M. A. Bender, A. Telling, R. Byron, and M. Groudine. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez-Carrozzi, V. R., A. A. Nazarian, C. C. Li, S. L. Gore, R. Sridharan, A. N. Imbalzano, and S. T. Smale. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20:282-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiling, N., A. Blumenthal, H. D. Flad, M. Ernst, and S. Ehlers. 2001. Mycobacteria-induced TNF-alpha and IL-10 formation by human macrophages is differentially regulated at the level of mitogen-activated protein kinase activity. J. Immunol. 167:3339-3345. [DOI] [PubMed] [Google Scholar]
- 46.Rhoades, K. L., S. Cai, S. H. Golub, and J. S. Economou. 1995. Granulocyte-macrophage colony-stimulating factor and interleukin-4 differentially regulate the human tumor necrosis factor-α promoter region. Cell. Immunol. 161:125-131. [DOI] [PubMed] [Google Scholar]
- 47.Rhoades, K. L., S. H. Golub, and J. S. Economou. 1992. The regulation of the human tumor necrosis factor a promoter region in macrophage, T cell, and B cell lines. J. Biol. Chem. 267:22102-22107. [PubMed] [Google Scholar]
- 48.Roldan, E., M. Fuxa, W. Chong, D. Martinez, M. Novatchkova, M. Busslinger, and J. A. Skok. 2005. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 6:31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saccani, S., S. Pantano, and G. Natoli. 2001. Two waves of nuclear factor kappaB recruitment to target promoters. J. Exp. Med. 193:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 51.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 52.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, K., and S. Akira. 2004. TLR signaling pathways. Semin. Immunol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 54.Tato, C. M., G. A. Martins, F. A. High, C. B. DiCioccio, S. L. Reiner, and C. A. Hunter. 2004. Cutting edge: innate production of IFN-gamma by NK cells is independent of epigenetic modification of the IFN-gamma promoter. J. Immunol. 173:1514-1517. [DOI] [PubMed] [Google Scholar]
- 55.Thomson, J. A., J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, V. S. Marshall, and J. M. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145-1147. [DOI] [PubMed] [Google Scholar]
- 56.Tomaras, G. D., D. A. Foster, C. M. Burrer, and S. M. Taffet. 1999. ETS transcription factors regulate an enhancer activity in the third intron of TNF-alpha. J. Leukoc. Biol. 66:183-193. [DOI] [PubMed] [Google Scholar]
- 57.Tomioka, H., T. Shimizu, W. W. Maw, and K. Ogasawara. 2000. Roles of tumour necrosis factor-alpha (TNF-alpha), transforming growth factor-beta (TGF-beta), and IL-10 in the modulation of intercellular adhesion molecule-1 (ICAM-1) expression by macrophages during mycobacterial infection. Clin. Exp. Immunol. 122:335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trede, N. S., A. V. Tsytsykova, T. Chatila, A. E. Goldfeld, and R. S. Geha. 1995. Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B. J. Immunol. 155:902-908. [PubMed] [Google Scholar]
- 59.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfeld. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, L., L. Li, F. Shojaei, K. Levac, C. Cerdan, P. Menendez, T. Martin, A. Rouleau, and M. Bhatia. 2004. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity 21:31-41. [DOI] [PubMed] [Google Scholar]
- 62.Wang, Y., Q. Yu, A. H. Cho, G. Rondeau, J. Welsh, E. Adamson, D. Mercola, and M. McClelland. 2005. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia 7:748-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, G. P., P. M. Watt, B. J. Holt, and P. G. Holt. 2002. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO− T cells. J. Immunol. 168:2820-2827. [DOI] [PubMed] [Google Scholar]
- 64.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 65.Xu, C., M. S. Inokuma, J. Denham, K. Golds, P. Kundu, J. D. Gold, and M. K. Carpenter. 2001. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19:971-974. [DOI] [PubMed] [Google Scholar]
- 66.Yao, J., N. Mackman, T. S. Edgington, and S.-T. Fan. 1997. Lipopolysaccharide induction of the tumor necrosis factor-a promoter in human monocytic cells. J. Biol. Chem. 272:17795-17801. [DOI] [PubMed] [Google Scholar]