Abstract
c-myc is frequently amplified in breast cancer; however, the mechanism of myc-induced mammary epithelial cell transformation has not been defined. We show that c-Myc induces a profound morphological transformation in human mammary epithelial cells and anchorage-independent growth. c-Myc suppresses the Wnt inhibitors DKK1 and SFRP1, and derepression of DKK1 or SFRP1 reduces Myc-dependent transforming activity. Myc-dependent repression of DKK1 and SFRP1 is accompanied by Wnt target gene activation and endogenous T-cell factor activity. Myc-induced mouse mammary tumors have repressed SFRP1 and increased expression of Wnt target genes. DKK1 and SFRP1 inhibit the transformed phenotype of breast cancer cell lines, and DKK1 inhibits tumor formation. We propose a positive feedback loop for activation of the c-myc and Wnt pathways in breast cancer.
Breast cancer is the most common malignancy of women worldwide, affecting nearly 10% of all women in Western countries at some point in their lifetime. Despite considerable research, the genetics of breast cancer remains unresolved. A small fraction of breast cancers arise from familial inheritance of highly penetrant tumor susceptibility genes, whereas the majority of cancers appear to arise through the somatic accumulation of a variety of different lesions to oncogenes and tumor suppressors (27). Among the genes first found to be amplified in breast cancer is c-myc, one of the earliest oncogenes identified and one which can contribute to many forms of cancer (40). Amplification of c-myc in breast cancer was first described in 1986, and reports vary as to how consistent these lesions are in tumors (22, 28). A comprehensive review suggests that at least 15% of breast cancers present with significant amplification of c-myc, and it is likely that a much larger fraction of tumors exhibit overexpression of the c-myc gene or protein through other mechanisms (13).
The link between c-myc overexpression and breast cancer was confirmed by animal model studies. Constitutive c-Myc expression under control of either the mouse mammary tumor virus (MMTV) long terminal repeat promoter or the whey acidic protein promoter in the mammary gland is oncogenic in transgenic mice (12, 44-46). Tumors are focal and form with variable latency, which presumably reflects the need for additional genetic lesions to promote progression of partially transformed cells. No consistent pattern of secondary lesions in c-myc-induced mammary tumors has been identified.
The cellular response to c-Myc overexpression is dependent on the cellular context. c-Myc can increase proliferation rate, cell mass accumulation, and oncogenic transformation but can also induce apoptosis and G2 arrest (15). c-Myc-driven oncogenic transformation is particularly significant because it models the phenotypic changes that c-Myc causes during tumorigenesis, including loss of cell contact inhibition, loss of the requirement for cell matrix attachment, and increased proliferation rate.
c-Myc is a pleiotropic but weak transcription factor (9). Numerous microarray studies demonstrate that expression of Myc activates or represses about 5 to 10% of all genes by 1.5-fold or more (reviewed in reference 11). None of these genes can remotely recapitulate the powerful cell transforming activity of c-Myc; thus, the cooperative action of at least several, possibly many, c-Myc target genes must operate to transform cells. Despite the fact that 80% of cancers are epithelial in origin and that overexpression of c-Myc plays a prominent role in many of these tumors, the only Myc-regulated gene demonstrated to be necessary for epithelial cell transformation is CDK4 (35), and no specific genes have been shown to be necessary for transformation of mammary epithelial cells by Myc.
Another established oncogenic pathway in mammary epithelial cells is constitutive Wnt signaling, which also functions in other epithelial cell lineages. Regulated Wnt signaling is critical to normal development, whereas unrestrained Wnt signaling is found in many tumors and experimentally activating Wnt is oncogenic (7, 17, 38). The family of extracellular Wnt proteins ligates the transmembrane coreceptors Frizzled and LRP5/6, thus inducing the “canonical” pathway, which increases the active nuclear pool of β-catenin by a coordinated set of mechanisms. Wnt ligand binding also induces the “noncanonical” pathway, which signals via Ca2+, Jun N-terminal protein kinase, and other mediators (31). Wnts were first identified as being capable of initiating tumors in a promoter insertion screen for constitutively hyperactivated genes in the mouse mammary gland (39). Subsequently, hyperactivating mutations in β-catenin and inactivating mutations in APC and Axin, which result in increased β-catenin levels, have been commonly found in colon carcinomas and other tumors (17). However, although β-catenin has been found to be stabilized in about 50% of breast carcinomas, mutations in the Wnt signaling pathway are relatively rare (7). Recently it was demonstrated that an autocrine Wnt signaling loop is operational in breast cancer lines and is necessary for full oncogenic activity (3).
In this report, we show that c-Myc is a potent transforming gene for human mammary epithelial cells, supporting the consistent observations that amplification of c-myc is a frequent contributory factor in breast cancer. Furthermore, we have identified the Wnt inhibitors DKK1 and SFRP1 as being Myc-repressed genes that account for a significant component of the oncogenic activity of Myc through activation of the Wnt pathway. These findings provide a molecular basis for the prominent role of c-myc in human breast cancer.
MATERIALS AND METHODS
Cell culture, transfection, and retrovirus infection.
PhoeNX retrovirus producer cells, mouse embryonic fibroblasts, breast cancer lines MDA-435 and T47D, and normal HBL100L breast epithelial cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Immortalized mammary epithelial cells (IMECs) and MDA-231 cells were cultured in 1:1 DMEM-F12 medium supplemented with factors (14). Retrovirus infection was performed with PhoeNX cells. Briefly, PheoNX cells were transfected with 4 μg of DNA by using Fugene (Roche) according to the manufacturer's instructions. Virus was harvested from the culture medium 2 days later and used to infect recipient IMECs. Cells were selected 2 days later and maintained in 0.5 mg/ml G418 (Sigma) to select for IRES-NEO vectors (IRES-NEO, DKK1-FLAG-IRES-NEO, and FLAG-SFRP1-IRES-NEO). Hygromycin (150 μg/ml) (Calbiochem) was used to select for LXSH vectors (LXSH, LXSH c-MycWT, LXSH c-MycΔMBII, and LXSH MycER). 4-Hydroxytamoxifen (OHT) (100 nM) (Sigma) was used to activate MycER. Cycloheximide (20 μg/ml) was added 30 min before the addition of tamoxifen. Small interfering RNA (siRNA) was purchased from Dharmacon and used according to the manufacturer's instructions. Cells were transfected for 2 days prior to being harvested.
Vectors.
DKK1-FLAG was subcloned from pCS-2-hDKK1-FLAG, a gift from S. Sokol, into BMN-IRES-Neo. FLAG-SFRP1-IRES-NEO and BMN (IRES-GFP) FLAG-SFRP1 were subcloned from a plasmid provided by J. Rubin. LXSH c-Myc, LXSH c-MycΔMBII, and LXSH c-MycER were also used.
Cell counts.
Cells were plated in regular growth medium at 0.5 × 105 per six-well plate well. Cells were maintained as described above. Cells were harvested each day in 500 μl trypsin and counted with a hemocytometer. At least 100 cells were counted. The experiments were repeated on three independent occasions, and error bars show standard deviations.
Soft agar assay.
Assays were performed in six-well plates, in duplicate. The lower layer consisted of 2 ml medium and 0.6% Noble agar (USB). The upper layer consisted of 2 ml medium, 0.3% Noble agar, and 2 × 104 cells. Agar at 50°C was mixed with medium at 37°C, plated, and left to set for 10 min. Plates were fed every other day with 250 μl regular growth medium. After 2 weeks, undivided cells and colonies over 20 μM were scored with a reticle. One hundred cells were counted from duplicate wells, and the experiment was performed on two independent occasions. Graphs show the mean number of colonies, with standard deviations.
TCF reporter assay.
For T-cell factor (TCF) reporter assays, low-passage cells (105) were plated onto six-well plates (for IMECs coated in 10% Matrigel [BD Biosciences]). Two days following plating, each well was transfected with 0.02 μg TOP or FOP FLASH plasmid (26) and 0.002 μg pRL-SV40. For a positive control, 1 μg and 0.25 μg pCDNA 3.1 β-catenin were also transfected. Two days following transfection, cells were lysed, and luciferase and Renilla activity was measured with Dual-Glo luciferase reagents (Promega). Luciferase readings were normalized against Renilla readings, and TOP FLASH/FOP FLASH ratios for each cell line were calculated. Experiments were performed on at least two independent occasions, and error bars indicate standard deviations.
RT-PCR.
RNA was extracted from log-phase cells with the RNeasy kit (QIAGEN) or TRIzol (Invitrogen), according to the manufacturer's instructions. Reverse transcription (RT)-PCR was carried out with the Platinum Quantitative RT-PCR system (Invitrogen), according to the manufacturer's instructions. The annealing temperature was usually 55°C. RT-PCRs were monitored for each primer pair to be within the linear range. Primers used for human cells were as follows (the number of cycles is indicated in parentheses): DKK1 (20), TTCCAACGCTATCAAGAACCTGC and CAAGGTGGTTCTTCTGGAATACC; SFRP1 (20), AAGCCACCTCAGTGCGTGG and CAGATTTCAACTCGTTGTCACAGG; RAR-γ (RARG) (20), TGCCATGGCCACCAATAAGG and CGTGTACACCATGTTCTTCTGGATGCTTCG; WISP1 (22), TGTGCGCTCAGCAGCTTGG and CGTCGTCCTCACATACC; GAPDH (17), CTCAAGACACCATGGGGAAGGTGA and ATGATCTTGAGGCTGTTGTCATA. Primers used for mouse cells were as follows: DKK1, TTCCAACGCGATCAAGAACCTGC and CAGTGTGGTTCTTCTGGGATATC; SFRP1, CCACAACGTGGGCTACAAGAAGATGG and TTCATCCTCAGTGCAAACTCGCTTGC. Fragments were resolved by 5% Tris-borate-EDTA polyacrylamide gel electrophoresis (PAGE), visualized by phosphorimager, and quantitated with ImageQuant software.
Immunoblotting.
For conditioned medium immunoblots, 1 × 105 cells were plated in six-well-plate wells with 2 ml regular medium. Two days later, medium was filtered through an 0.45-μm filter, and either 10 μl medium was used for immunoblotting (DKK1) or medium was concentrated 5× on Centricon columns and 30 μl was used for immunoblotting (SFRP1, β-tubulin). The protein concentration of the medium prior to concentration was also determined by the Lowry method. For other immunoblots, cells were washed with phosphate-buffered saline (PBS) and lysed in a modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0]; 130 mM NaCl; 1% NP-40; 0.5% deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 20 mM NaF; 1 μg/ml of leupeptin, pepstatin, and aprotinin; 1 mM dithiothreitol; and phosphatase inhibitor cocktail I and II [Sigma]). The protein concentration was determined with the Modified Lowry Protein Assay kit (Pierce). Protein equivalents were subject to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted. Antibodies were as follows: anti-c-myc polyclonal (a gift from Steve Hann), anti-FLAG antibody (Sigma), anti-β-tubulin antibody (Santa Cruz), anti-E-cadherin antibody (BD Biosciences), anti-DKK1 antibody, and anti-SFRP1 antibody (R&D Systems).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed with the Chip Assay kit (Upstate), according to the manufacturer's instructions. Antibodies used were polyclonal anti-c-Myc antibody (N262) and polyclonal anti-β-tubulin antibody (both from Santa Cruz Biotech). Primers used for human cells were as follows: DKK1 promoter (−227 to +259), GCTCGCTGGTAGCCTTCACC and CCGAGTTCAAGGTGGCGCTC; DKK1 coding (+1818 to +2290), TACACATTGGTGCTTCTGTCCATCTGG and CAAACAGAACCTTCTTGTCCTACGAAGG; SFRP1 promoter (−152 to +318), GTTCTAGTAAACCGAACCCGCTCGC and CTGCGCCCGATGCCCATG; SFRP1 coding (+44056 to +44416), GGCCCATCAAGAAGAAGGACCTGAAG and GAATGCTGCAAGAACAAGCCGACTG. Primers used for mouse cells were as follows: DKK1 promoter, AGACTCCACCAGCGGAGGTCC and GGCGCCACACTGACAGCAGAGC; DKK1 gene body, TGTTAATGTCTCAAAGAAGTCTCC and AACTTCTCTGCTCTGGGTGCTAGC; SFRP1 promoter, TTGACCTGGGTCTAGTTCTAGTAAACC and GCTGCTGCACCTACTTGCGACG; SFRP1 gene body, GGAAAACGGTGACAAGAAGATTGTCC and ACAGACTGGAAGGTGGGACACTCG. Sequences of other primers used to scan the Myc binding sites are available upon request. PCRs were labeled with 32P-labeled primers, and PCR products were resolved by gel electrophoresis and quantitated with a phosphorimager. PCR products were normalized to input, and then the ratio of Myc IP signal/β-tubulin IP signal was calculated.
Transgenic mice.
Mammary tumors were induced in transgenic mice by activation of the c-MYC, Neu, Wnt1, or Ras pathway by administering 2.0 mg/ml doxycycline-5% sucrose in drinking water to 6-week-old female MMTV-rtTA/TetO-c-MYC (12), MMTV-rtTA/TetO-NeuNT (36), or MMTV-rtTA/TetO-Wnt1 (19) mice or by administering 0.012 mg/ml doxycycline to 6-week-old female MMTV-rtTA/TetO-v-Hras (B. A. Keister et al., unpublished data) mice. Doxycycline was replaced weekly. Animals were sacrificed when tumors reached 10 mm in diameter; then, tumor tissue was snap-frozen and RNA was prepared by homogenization in guanidine thiocyanate, followed by ultracentrifugation through cesium chloride. RNA was DNase 1 treated on an RNeasy minicolumn (QIAGEN) for 15 min, according to the manufacturer's instructions.
Tumor work.
Cells (106) cells in 100 μl PBS and one Matrigel were injected subcutaneously into both flanks of female nude mice. Mice were monitored for tumors at least weekly. Tumors were first detected after 4 weeks. Tumors larger than 7 mm in diameter were recorded. After 8 weeks, tumors from the mice were extracted and weighed.
RESULTS
c-Myc transforms IMECs.
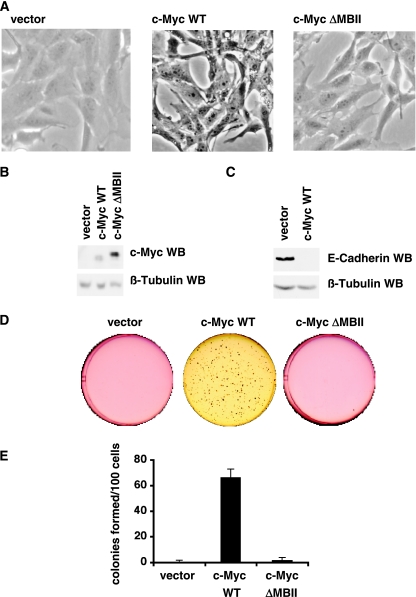
To study the role of c-Myc in mammary epithelial cells, we infected telomere reverse transcriptase (TERT)-IMECs (14) with retrovirus vectors expressing c-MycWT, c-MycΔMBII, and a control (LXSH) and selected pools of infected cells (Fig. 1A). c-MycΔMBII is a transformation- and transactivation-deficient mutant that deletes an evolutionarily conserved segment of the N terminus (9). Western blot analysis of cell extracts confirmed that c-MycWT and c-MycΔMBII were overexpressed in these cell lines (Fig. 1B).
FIG. 1.
c-Myc transforms TERT-IMECs. TERT-IMECs were infected with c-MycWT, c-MycΔMBII, or vector control, and pools were selected with hygromycin. (A) Phase-contrast micrographs of log-phase IMECs. (B) Western blot analyses were performed with equivalent amounts of cell extracts using anti-c-Myc antibody (top) and anti-β-tubulin antibody (bottom). (C) Western blot analyses were performed as for panel B with anti-E-cadherin antibody (top) and an anti-β-tubulin antibody (bottom). (D) Cell lines were subjected to a 14-day soft agar assay in six-well plates, and representative wells were scanned. (E) Colonies larger than 10 μm were counted for two wells from two independent experiments. The graph shows the mean number of colonies per 100 plated cells, and error bars indicate standard deviations.
Immediately following drug selection, a change in morphology was observed in IMECs expressing c-MycWT but not c-MycΔMBII or vector control. Vector IMECs have a typical cuboidal epithelial cell morphology, but c-MycWT IMECs exhibit a fibroblastoid phenotype; i.e., they are less cuboidal, more spindly, and more refractile (Fig. 1A). At confluence, c-MycWT IMECs exhibit less contact inhibition than vector IMECs, but they do not pile up to form foci. This change in morphology suggested that a c-Myc-dependent epithelial mesenchymal transition (EMT) had occurred. We performed a Western blot analysis against E-cadherin, a marker of EMT which is repressed following EMT (47). Consistent with the EMT phenotype, we found that E-cadherin expression is strongly repressed in cells expressing c-MycWT compared to vector control cells (Fig. 1C). In a separate study, we present data that c-Myc repression of E-cadherin is posttranscriptional (8).
Elevated expression of c-Myc increases the cell proliferation rate in many cell systems. To measure whether exogenous expression of c-Myc increases the cell proliferation rate in IMECs, we assessed the growth rate of each cell type over 5 days (see Fig. 4D). Vector IMECs were found to proliferate with a doubling time of 23 h, whereas c-Myc IMECs proliferate significantly faster, with a doubling time of 16 h (see further discussion below).
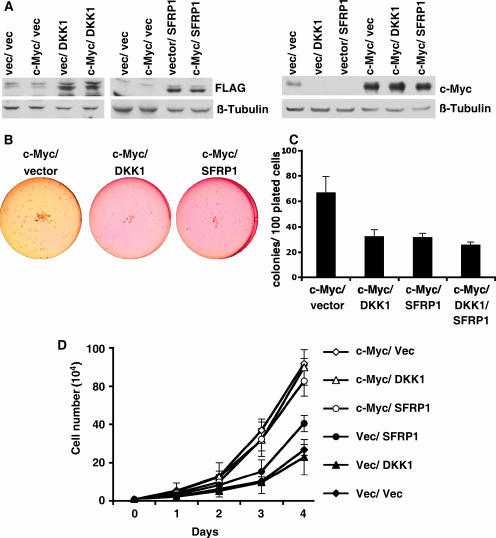
FIG. 4.
c-Myc repression of DKK1 and SFRP1 is necessary for c-Myc-induced transformation. c-MycWT IMECs and vector control IMECs were infected with DKK1-FLAG, FLAG-SFRP1, or empty vector (IRES-NEO), as indicated, and pools were selected. (A) Western blot analyses were performed with equivalent amounts of cell extracts using anti-FLAG antibody to detect DKK1 and SFRP1 and using anti-Myc antibody. Anti-β-tubulin served as a loading control. Cells were subjected to a 14-day soft agar assay in six-well plates. (B) Scan of representative wells. (C) Colonies larger than 10 μm were counted for two wells from two independent experiments. The graph shows the mean number of colonies per 100 plated cells, and error bars indicate standard deviations. (D) Cell proliferation rate of cell lines measured by counting equivalently plated cells on five consecutive days with a hemocytometer. The graph shows the mean cell number for each day for three independent experiments, and error bars show standard deviations. Each of the c-Myc-expressing cell lines proliferated significantly faster than all the vector control cells lines (vector, DKK1, and SFRP1), with P values of >0.03.
Since c-Myc is an oncogene found frequently overexpressed in breast cancer, we investigated whether overexpression of c-Myc could transform IMECs. Transformation assays score whether cells can grow independently of a subset of the signals required by normal cells, thus modeling the changes that occur during tumor formation. We used a soft agar transformation assay that measures whether cells can undergo anchorage-independent growth, which is one of the most consistent indicators of oncogenic transformation (1). After 14 days, vector IMECs and c-MycΔMBII IMECs failed to grow in soft agar, whereas c-MycWT IMECs grew into robust colonies (Fig. 1D). Quantitation of repeated experiments showed that an average of 67% of the c-MycWT IMECs formed colonies in soft agar (Fig. 1E). Therefore, expression of c-Myc alone is sufficient to transform TERT-IMECs and to permit anchorage-independent growth. However, the cells do not form tumors in nude mice (data not shown), indicating that additional mutations would be required for fully oncogenic transformation.
The Wnt inhibitors DKK1 and SFRP1 are repressed by c-Myc.
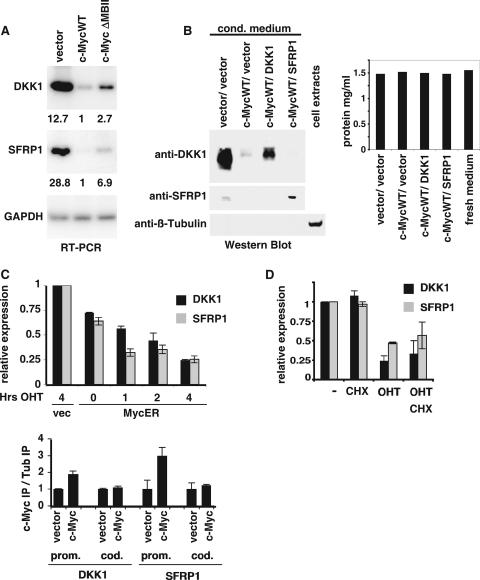
c-Myc is a transcription factor, and we investigated the target genes responsible for the c-Myc-induced transformation of mammary epithelial cells. We conducted a global analysis of the transcriptional response to c-Myc using microarrays, which will be described in detail elsewhere. Two of the genes strongly repressed by overexpression of c-Myc were the inhibitors of the Wnt pathway, DKK1 and SFRP1. We confirmed that c-Myc repressed DKK1 and SFRP1 by performing RT-PCR on RNA from several independently isolated cell lines (Fig. 2A). Quantitation of the reactions confirmed that expression of MycWT strongly repressed the expression of both of these genes. c-Myc expression repressed DKK1 expression by over 12-fold and SFRP1 expression by over 28-fold. We also found that N-Myc represses DKK1 and SFRP1 expression in primary human foreskin fibroblasts and granular neuronal precursors (data not shown).
FIG. 2.
c-Myc represses DKK1 and SFRP1. (A) RNA from IMECs expressing vector control, c-MycWT, or c-MycΔMBII (left) were used as a template for RT-PCR using primers specific for DKK1, SFRP1, and GAPDH. The RT-PCR signal was quantitated and normalized to c-MycWT values, and the relative values are shown below the image. (B) Immunoblotting was performed with equivalent volumes of conditioned medium from IMECs expressing the indicated constructs. Primary antibodies were anti-DKK1, anti-SFRP1, and anti-β-tubulin, as indicated. For the anti-β-tubulin blot, whole-cell extracts were used as a positive control. The protein concentration in the medium was determined by the Lowry method. (C) IMECs expressing vector control or c-MycER were induced with 100 nM OHT for 0, 1, 2, and 4 h, as indicated. RNA was extracted and used as a template for RT-PCR using primers specific for DKK1 (dark gray) and SFRP1 (light gray). (D) Human BJ fibroblasts expressing vector control or MycER were treated for 4 h with OHT and/or cycloheximide (CHX), as indicated. RNA was extracted and used as a template for RT-PCR using primers specific for DKK1, SFRP1, and GAPDH. Expression of DKK1 and SFRP1 normalized to GAPDH and relative to untreated cells is indicated. Throughout the figure, error bars indicate standard deviations. (E) ChIP assay performed on vector IMECs and c-Myc IMECs (left) using anti-c-Myc and anti-β-tubulin antibodies. PCR was performed using primers specific for the DKK1 and SFRP1 promoter (prom.) and coding (cod.) regions. PCR signals were normalized to input, and a ratio of signal from anti-Myc IP to anti-β-tubulin IP was calculated.
DKK1 and SFRP1 are both secreted Wnt inhibitors. Therefore, we investigated DKK1 and SFRP1 protein expression by performing Western blot analyses of conditioned medium (Fig. 2B). In correlation with the mRNA expression levels, Myc was found to repress both DKK1 and SFRP1 protein levels. Overexpressed DKK1 and SFRP1 were used as positive controls, and a β-tubulin control was used to confirm that the conditioned medium did not contain cell contamination.
Many models have been proposed to explain how Myc induces transcriptional repression, but the precise mechanism remains elusive. It is beyond the scope of this study to resolve the mechanism of Myc repression, but Myc can be judged to repress a gene relatively directly if it represses transcription rapidly in an inducible system and in the presence of translation inhibitors. Since IMECs are estrogen receptor negative (14), we were able to take advantage of the Myc-estrogen receptor fusion, c-MycER, which is relatively inactive until the addition of OHT (30). Activation of c-MycER for 1 h was sufficient to repress expression of both SFRP1 and DKK1 but not the GAPDH control (Fig. 2C). Consistent with what has been observed for many genes, activated MycER repression of DKK1 and SFRP1 is weaker than steady-state Myc-induced repression. It may take some time for the existing pools of DKK1 and SFRP1 to be depleted once transcription is repressed. In IMECs, it was not possible to activate MycER in the presence of cycloheximide to establish that repression is independent of translation, because cycloheximide was prohibitively toxic. In human fibroblasts, however, activated MycER repressed DKK1 and SFRP1 expression (Fig. 2D). Repression was judged to be direct since it also occurred in the presence of cycloheximide, which blocks translation. SFRP1 and DKK1 were repressed by c-MycV394D, indicating that repression does not involve Miz1 (21) (see Fig. S1 in the supplemental material).
Many Myc-repressed genes have been found to have Myc bound to their promoters, although the functional significance of this remains unclear. We carried out a ChIP assay using anti-c-Myc or control (β-tubulin) polyclonal antibodies to determine if Myc was bound to the DKK1 and SFRP1 promoters. PCR was performed with a series of overlapping primers that spanned 1 kb to either side of the transcriptional start site. For both DKK1 and SFRP1, Myc binding was found to be two- to threefold higher around the transcriptional start site in Myc IMECs than in vector IMECs (Fig. 2E). As a negative control, Myc binding was not found in the coding regions of either gene. c-Myc could also be found bound to the DKK1 and SFRP1 promoters in mouse embryo fibroblasts and in human breast cancer cell lines (see Fig. S2 in the supplemental material). In summary, c-Myc binds to DKK1 and SFRP1 promoters and rapidly represses these genes in a translation-independent manner, but further studies are required to establish the precise mechanism.
c-Myc increases expression of TCF and Wnt-responsive genes.
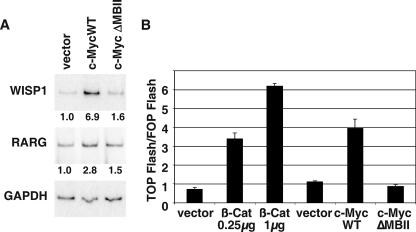
Because we had found that c-Myc expression represses two Wnt pathway inhibitors, we wanted to determine whether this had the functional consequence of activating Wnt signaling. Transcriptional activation of c-Myc and cyclin D1 are often used as a Wnt readout, but they could not be used in this study because c-Myc represses its own expression through an autoregulation loop (41). Myc is also reported to have variable effects on cyclin D1 expression (10, 42), and we found that cyclin D1 is repressed by c-Myc in IMECs (data not shown). Therefore we used two other well-established Wnt-responsive genes, WISP1 and RARG, as a readout for an active Wnt signal (33, 53). c-MycWT but not c-MycΔMBII expression was found to increase expression of both WISP1 and RARG in IMECs (Fig. 3A), and c-MycWT can weakly induce many other reported Wnt target genes (see Fig. S3 in the supplemental material).
FIG. 3.
c-Myc activates Wnt signaling. (A) RNA extracted from IMECs expressing vector, c-MycWT, or c-MycΔMBII was used as a substrate for RT-PCR performed using primers specific for WISP1, RARG, and GAPDH. (B) TOP FLASH TCF reporter assay was performed with IMECs expressing vector control, c-Myc, or β-catenin, as described in Materials and Methods.
An alternative method for measuring cellular Wnt activity is by using the TOP/FOP FLASH reporter assay, which measures TCF transcriptional activity (26). In the TOPFLASH plasmid, luciferase is driven by TCF sites, whereas the FOPFLASH plasmid has mutated TCF sites; therefore, the relative TCF activity can be taken as a ratio of TOPFLASH to FOPFLASH. In this assay, overexpression of c-Myc in IMECs stimulated the TCF reporter activity by twofold (Fig. 3B). IMECs expressing β-catenin were used as a positive control. We conclude that expression of c-Myc in IMECs activates the Wnt pathway. The extent of activation is similar to that found for cellular genes in response to the TCF transcription factor that mediates downstream Wnt signaling (50). We stress that DKK1 and SFRP1 probably regulate other genes and pathways besides those described here, e.g., in the noncanonical Wnt signaling pathway (5, 7, 51).
Repression of DKK1 and SFRP1 is necessary for c-Myc-dependent transformation of human mammary epithelial cells.
Elevated Wnt signaling in the mammary gland is oncogenic in mouse models (39). Therefore, we wanted to investigate whether the c-Myc-mediated repression of the Wnt inhibitors DKK1 and SFRP1 could be necessary for full c-Myc-induced cell transformation. To restore expression experimentally, the vector IMEC and c-Myc IMEC lines were infected with DKK1-FLAG, FLAG-SFRP1, or vector control (IRES-NEO), and stable pools were drug selected (Fig. 4A). Exogenous expression of DKK1 and SFRP1 did not increase or decrease exogenous c-Myc expression; however, endogenous c-Myc expression was reduced by expression of either DKK1 or SFRP1 (Fig. 4A). This is consistent with endogenous c-Myc being a Wnt target gene at the level of both transcription and mRNA stability (20, 37). To assess the extent of transformation, soft agar assays were performed with the different IMEC lines (Fig. 4B and C). As before, c-Myc and vector IMECs grew in soft agar with an average efficiency of 65%. However, restored expression of either DKK1 or SFRP1 in the c-Myc IMECs (c-Myc/DKK1/IMEC and c-Myc/SFRP1/IMEC) led to a reduced efficiency of anchorage-independent growth to below 35% (Fig. 4C). Therefore, c-Myc repression of DKK1 and SFRP1 is necessary for a significant fraction of c-Myc-dependent transformation of IMECs. Expression of DKK1 and SFRP1 simultaneously in c-MycWT IMECs did not reduce transformation more than expression of either inhibitor alone (Fig. 4C), indicating that these inhibitors utilize the same genetic pathway in mammary epithelial cell transformation. In Fig. 1, c-Myc expression is demonstrated to repress E-cadherin expression. E-cadherin is a tumor suppressor, and we report elsewhere that Myc-induced epithelial cell transformation is also dependent on repression of E-cadherin (8).
Because c-Myc accelerates cell proliferation in addition to inducing anchorage-independent growth, we investigated whether c-Myc-mediated repression of DKK1 and SFRP1 was also necessary for the accelerated proliferation rate. We found that c-Myc IMECs expressing exogenous DKK1, SFRP1, or vector control all proliferated with a doubling time of about 16 h, indicating that repression of DKK1 or SFRP1 is not necessary for c-Myc-driven cell proliferation (Fig. 4D). We can conclude that DKK1 and SFRP1 do not repress transformation simply because they repress the proliferation rate. The accelerated growth rate induced by c-Myc must depend on one or more target genes other than DKK1 and SFRP1.
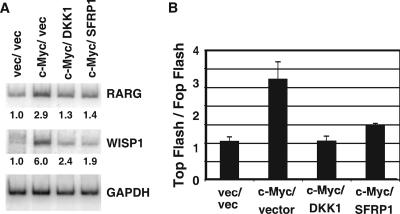
We have shown that c-Myc expression increases Wnt signaling in IMECs (Fig. 3) and hypothesized that c-Myc-mediated repression of DKK1 and SFRP1 was necessary for this Wnt activation. To explore this, we looked at the same Wnt readouts in the c-Myc/DKK1/IMEC and c-Myc/SFRP1/IMEC cells. As expected, expression of DKK1 or SFRP1 inhibited expression of the c-Myc-induced Wnt target genes WISP1 and RARG1 (Fig. 5A). Expression of DKK1 and SFRP1 also inhibited c-Myc-induced TCF reporter gene activity (Fig. 5B). Therefore, we conclude that c-Myc-induced Wnt activity and cell transformation are dependent on c-Myc repression of DKK1 and SFRP1.
FIG. 5.
c-Myc repression of DKK1 or SFRP1 is necessary for c-Myc induction of Wnt/TCF targets. (A) RNA extracted from IMECs expressing both control vectors (LXSH and IRES-NEO), c-Myc/IRES-NEO, c-Myc/DKK1, and c-Myc/SFRP1 was used as a template for RT-PCR using primers specific for WISP1, RARG, and GAPDH. Bands were quantitated and normalized to those of vector control IMECs, indicated under the graphs. (B) The same cell lines were used for the TOP FLASH TCF reporter assay, as described in Materials and Methods.
SFRP-1 is strongly repressed in c-Myc-induced tumors.
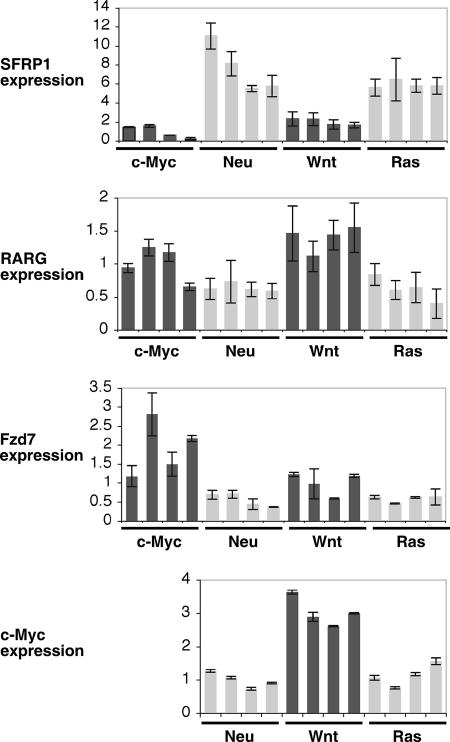
To investigate the in vivo significance of our findings, we investigated DKK1 and SFRP1 expression in a panel of murine mammary tumor models. We analyzed four mouse tumors arising following tetracycline induction of c-Myc, Neu, Wnt, or Ras in the MMTV-rtTA/TetO system (12, 19, 36). SFRP1 was strongly repressed in the c-Myc-induced tumors (Fig. 6). In comparison to the average Myc tumor expression level, SFRP1 was expressed on average 6.9 times higher in Neu tumors, 3.6 times higher in Wnt tumors, and 9.1 times higher in Ras tumors. The SFRP1 expression level in the Myc tumors was statistically lower than in the other groups of tumors, giving P values of 8.2 × 10−4, 1.8 ×10−2, and 8.5 × 10−3 for Neu, Wnt, and Ras tumors, respectively. DKK1 expression was not regulated in the same tumors, and expression was barely detectable.
FIG. 6.
SFRP1 expression is significantly lower in mouse mammary tumors induced by Myc than in those induced by Ras, Wnt, or Neu. RNA was extracted from four independent tumors induced in each of the following mice: MMTV-rtTA/TetO-MYC, MMTV-rtTA/TetO-Neu, MMTV-rtTA/TetO-Wnt, MMTV-rtTA/TetO-Ras. RT-PCR was performed using primers specific for SFRP1 and the Wnt target genes RARG, Fzd7, and c-Myc, as indicated. Error bars indicate standard deviations.
To look for evidence of elevated Wnt signaling in Myc tumors, we investigated the expression level of two other Wnt target genes, RARG and Fzd7, in the tumor panel (33, 52). Wnt- and Myc-induced tumors had the highest average expression of RARG and Fzd7 (Fig. 6). The average expression of RARG in Myc tumors was 1.6-fold higher than that in Neu and Ras tumors (P values of 0.02 and 0.03, respectively), and the average RARG expression in Wnt tumors was 2.2-fold higher than in Neu and Ras tumors. The average expression of Fzd7 in Myc tumors was approximately threefold higher than that in Neu and Ras tumors (P values of 0.01 for both comparisons). Average Fzd7 expression in Wnt tumors was 1.8-fold higher than in Neu and Ras tumors. WISP1 was not expressed at a significant level in these tumors. Thus, Myc induced expression of Wnt target genes to a level comparable to that of a direct Wnt oncogene.
c-Myc is a known Wnt target gene, and we found that endogenous c-Myc expression was elevated in the Wnt tumors. Average c-Myc expression in Wnt tumors was 2.5 to 3 times higher than that in Neu and Ras tumors (Fig. 6), comparable to the other Wnt targets discussed above. Elevated c-Myc expression in Wnt tumors may be responsible for the reduced SFRP1 expression in these tumors compared to Neu and Ras tumors. Although we were able to detect and distinguish c-Myc mRNA transcribed from the human transgene and the endogenous mouse gene, it was not possible to compare their expression in a meaningful manner. The human c-Myc transgene has only protein-coding exons 2 and 3 and has no nontranscribed exon 1 or endogenous untranslated regions and therefore is likely to be translated at a far higher rate than the endogenous mouse c-Myc mRNA (12).
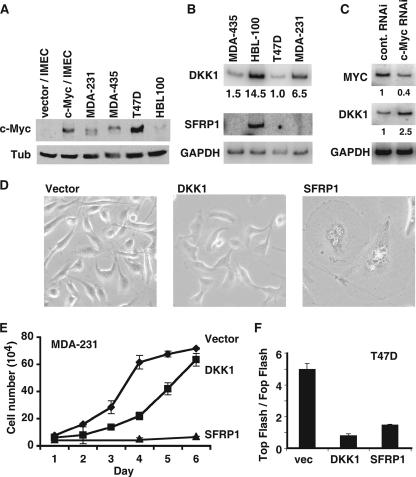
Wnt inhibitors are repressed in breast cancer cell lines.
Previous publications have shown that SFRP1 is repressed in breast cancer (25, 49). We wanted to extend these findings and also measure the expression level of DKK1 in breast cancer cell lines. We measured the expression level of c-Myc in the breast cancer lines MDA-435, MDA-231, and T47D and in one normal mammary epithelial line, HBL100, and compared these to vector IMECs and c-Myc IMECs. c-Myc protein was more highly expressed in all three tumor lines than in the normal epithelial line, HBL100 (Fig. 7A). c-Myc expression in c-Myc IMECs was found to be most equivalent to c-Myc levels in MDA435 but less than in T47D. This confirms that c-Myc in c-Myc IMECs is not overexpressed to nonphysiological levels. RT-PCR was performed with primers specific for DKK1 and SFRP1, and the results were striking (Fig. 7B). SFRP1 was expressed in the HBL100 line but was undetectable in the three tumor lines. DKK1 was detectable in HBL100 but also substantially repressed in all three breast cancer cell lines compared to the normal breast epithelial cell line. Using the ChIP assay, we found that Myc binding was significantly enriched at the DKK1 and SFRP1 promoters in the breast cancer cell lines compared to the normal cell line, HBL100 (see Fig. S2 in the supplemental material).
FIG. 7.
DKK1 and SFRP1 are repressed in breast cancer cell lines. (A)Western blot analyses were performed with equivalent amounts of cell extracts from three breast cancer cell lines, MDA-435, T47D, and MDA-231, and one normal breast epithelial cell line, HBL100, using anti-c-Myc antibody (top) and anti-β-tubulin antibody (Tub) (bottom). (B) RNA extracted from the same cell lines was used as a template for RT-PCR using primers specific for DKK1, SFRP1, and GAPDH. The RT-PCR signal was normalized to T47D for DKK1, and the relative values are shown below the blot. For SFRP1, the RT-PCR signal was detectable only in HBL100 cells, and so no relative quantitation was possible. (C) T47D cells were transfected with control or Myc-directed siRNA for 2 days. RNA was extracted, and RT-PCR was performed with primers for MYC, DKK1, and GAPDH. (D) Cell proliferation rate of the MDA-231 lines was measured by counting equivalently plated cells on six consecutive days with a hemocytometer. The graph shows the mean cell number for each day for two experiments, and error bars show standard deviations. (E) Phase-contrast micrographs of MDA-231 cells infected with vector control, DKK1, or SFRP1. (F) The TOP FLASH TCF reporter assay was performed on T47D cell lines transduced with vector, DKK1, or SFRP1, as indicated in Materials and Methods.
Since Myc expression was highest in T47D cells, we used this cell line to analyze whether DKK1 and SFRP1 repression was Myc dependent. We reduced Myc expression by transfection of siRNA direct against Myc or a scrambled control (Fig. 7C). We found that DKK1 expression was increased following Myc repression, demonstrating that DKK1 repression is Myc dependent. We remained unable to detect SFRP1 expression in this cell line. SFRP1 may have become irreversibly silenced in the breast cancer cell lines due to promoter methylation (see Discussion).
Repression of DKK1 and SFRP1 is necessary for the transformed phenotype and oncogenic potential of breast cancer cell lines.
In order to determine the functional significance of DKK1 and SFRP1 repression in breast cancer cell lines, we exogenously expressed DKK1 and SFRP1 in MDA-231 and T47D cells. Both DKK1 and SFRP1 expression resulted in a reduction in the proliferation rate of the MDA-231 cells (Fig. 7D). MDA-231 vector cells proliferate with a mean doubling time of 24 h, whereas MDA-231 DKK cells have a mean doubling time of 32 h. Expression of SFRP1 in MDA-231 cells led to such a dramatic inhibition of proliferation that there was less than a 50% increase in cell numbers over 6 days. These proliferation defects were reflected in the morphology of the MDA-231 cells (Fig. 7E). Expression of DKK1 and SFRP1 resulted in larger, less refractile cells, with this change in morphology being particularly pronounced on SFRP1 expression. The effect of DKK1 and SFRP1 on T47D cells followed the same trend as for the MDA-231 cells, but the effects were less pronounced (not shown). We used the TOP/FOP flash assay to demonstrate that DKK1 and SFRP1 inhibited TCF reporter activity in T47D cells (Fig. 7F). MDA-231 cells are not able to be transfected; therefore, it was not possible to use this assay.
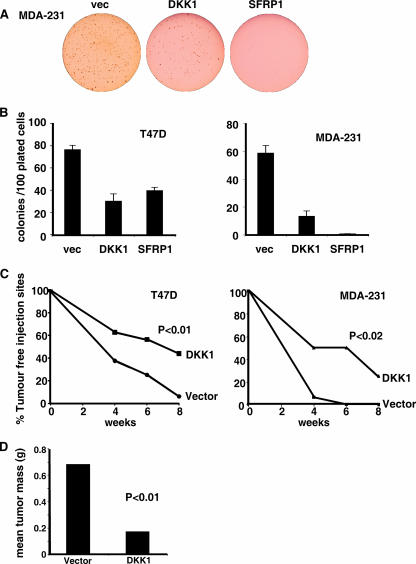
To measure the contribution of DKK1 and SFRP1 repression to the transformed phenotype of the breast cancer lines, we performed a soft agar assay to measure anchorage-independent cell growth. We found that both MDA-231 and T47D cells were able to grow into robust colonies in soft agar (Fig. 8A and B). DKK1 and SFRP1 expression inhibited soft agar colony growth in both cell lines. The results were most striking for MDA-231 cells, in which DKK expression inhibited soft agar colony growth by ∼75% and SFRP1 inhibited colony growth entirely. Therefore, DKK1 and SFRP1 repression is essential for the transformed phenotype of these breast cancer cell lines.
FIG. 8.
SFRP1 and DKK1 suppress transformation of human breast cancer cells. (A) MDA-231 cells expressing vector (vec), DKK1, and SFRP1. Cell lines were subject to a 14-day soft agar assay in six-well plates, and representative wells from the MDA-231 experiment are shown. (B) MDA-231 and T47D cells transduced with the indicated vector were plated in soft agar, and colonies larger than 10 μm were counted for two wells from two independent experiments. The graph shows the mean number of colonies per 100 plated cells, and error bars indicate standard deviations. (C) Nude mice were injected subcutaneously with MDA-231 vector or MDA-231 DKK cells, T47D vector cells, or T47D DKK cells. Injection sites were monitored, and the percentage of tumor-free injection sites is presented. (D) After 8 weeks, MDA-231 tumors were removed and weighed. Mean tumor masses are shown. (P values are shown in the figure.)
To measure the impact of DKK on tumor formation, we injected T47D vector, T47D DKK1, MDA-231 vector, and MDA-231 DKK1 cells subcutaneously into nude mice (Fig. 8C). DKK1 expression significantly increased the time taken for tumors to arise for both breast cancer cell lines. After 8 weeks, tumors had formed in 96% of the T47D vector injection sites but only in 56% of the T47D DKK injection sites. After 4 weeks, tumors had formed in 93% of the MDA-231 vector injection sites but in only 50% of the MDA-231 DKK injection sites. After 8 weeks, the mean mass of the MDA-231 vector tumors was 0.63 g, and the mean mass of the MDA-231 DKK1 tumors was 0.20 g. The T47D DKK tumors were also smaller than the T47D vector tumors, but the difference did not pass statistical tests.
DISCUSSION
c-myc activates the Wnt pathway.
Elevated expression of c-Myc is a common feature of breast cancer, but how c-Myc induces breast tumors is largely uncharacterized. Here we report that elevated expression of c-Myc induces a dramatic change in phenotype in human mammary epithelial cells; i.e., the cells exhibit hallmarks of EMT and become transformed to anchorage-independent growth by c-Myc alone. Furthermore, c-Myc represses transcription of the Wnt pathway inhibitors DKK1 and SFRP1, activating the Wnt pathway and promoting anchorage-independent growth. This c-Myc-driven increase in Wnt signaling is necessary for c-Myc to fully transform mammary epithelial cells. We show that SFRP1 expression in vivo is significantly lower in Myc-induced mouse mammary tumors than in Wnt-1-, Neu-1-, and Ras-induced tumors. We also demonstrate that DKK1 and SFRP1 are necessary for the transformed phenotype of human breast cancer cell lines and that DKK1 inhibits tumor formation in nude mice.
Previously, c-Myc had been shown to be a transcriptional target of the TCF/β-catenin transactivation complex (20). Here we show the converse, that c-Myc activates the Wnt pathway. We propose that in breast cancer, c-Myc and Wnt/TCF can operate in a positive regulatory loop. In normal mammary epithelial cells, Wnt/TCF activity is repressed by Wnt inhibitors such as DKK1 and SFRP1, and c-myc gene expression is held in check. Oncogenic lesions that amplify the c-myc locus increase c-Myc expression, which in turn represses DKK1 and SFRP1 and increases Wnt/TCF signaling. Active β-catenin/TCF would further activate c-myc gene expression, thus constituting a positive feedback loop.
Wnt signaling in breast cancer.
Mutations in the Wnt pathway, such as in APC and Axin, are common in certain tumors and predispose to colon cancer, leading to increased β-catenin levels (17). However, analogous mutations in the Wnt pathway are not commonly found in mammary epithelial cells; e.g., mutations in Axin and APC have not been found to predispose to breast cancer. This is surprising in light of the fact that constitutive expression of Wnt in mouse mammary epithelial cells induces tumor formation, demonstrating that hyperactivation of this pathway is a powerful oncogenic signal (39, 48). In addition, β-catenin is stabilized in a large proportion of breast carcinomas, suggesting that Wnt signaling may be activated (3, 29, 43).
DKK1 and SFRP1 are extracellular Wnt inhibitors with rather distinct structures and functions (23). SFRP1 is a member of the secreted Frizzled-related proteins, which are homologous to the extracellular cysteine-rich domain of the Wnt receptor Frizzled, but it lacks the transmembrane and intracellular domains (34). Therefore, SFRP1 can inhibit Wnt signaling by sequestering Wnt and by heterodimerizing with Frizzled to form a nonfunctional receptor complex (2). DKK1 is a member of the Dickkopf family, which was initially identified due to a head-inducing activity in Xenopus (18). DKK1 exerts its Wnt-inhibitory activity by targeting the Wnt coreceptor LRP5/6 for degradation (32). It is important to note that there is emerging evidence for both canonical (β-catenin/TCF) and noncanonical signaling through the Wnt pathway (5, 7, 51), both of which would be affected by suppression of SFRP1 and DKK1.
Since the discovery of DKK1 and SFRP1, both genes have been found to be repressed in a number of tumors, including colorectal tumors, papillary bladder cancer, malignant pleural mesothelioma, non-small-cell lung cancer, prostate cancer, esophageal adenocarcinoma, and ovarian cancer (reviewed in reference 4). SFRP1 repression is proposed to be an early event in breast carcinogenesis, with its expression being inversely correlated with tumor stage and associated with a poor prognosis (25). In addition, we report here that SFRP1 and DKK1 are repressed in a panel of breast cancer cell lines. We demonstrate that repression of DKK1 and SFRP1 is necessary for the transformed phenotype of two breast cancer cell lines and that DKK1 expression inhibits tumor formation in nude mice. We propose that in breast cancer and other epithelial tumors, c-Myc-induced repression of DKK1 and SFRP1 provides a mechanism for activation of Wnt signaling in the absence of any other overt stimulation of the pathway. Furthermore, repression of DKK1 and SFRP1 by other mechanisms, such as promoter methylation (4), could give a similar activation of the Wnt pathway and subsequent upregulation of c-myc expression. The potent suppressive activity of SFRP1 and DKK1 in the MDA-231 and T47D breast cancer cell lines (Fig. 8) argues strongly that Wnt signaling is required for the continued growth of these lines in culture.
We show that in mouse mammary tumor models, SFRP1 expression is significantly lower in Myc-induced tumors than in those induced by Neu, Wnt, or Ras. Therefore, in vivo, Myc expression inversely correlates with SFRP1 expression. In addition, we present evidence that the Wnt pathway is more active in Myc tumors than in Neu or Ras tumors. RARG and Fzd7, Wnt target genes, are expressed at significantly higher levels in Myc tumors than in Neu or Ras tumors.
A c-myc-dependent human mammary epithelium transformation model.
It is interesting to compare the present study to a previous analysis of oncogene and tumor suppressor activity in the transformation of human mammary epithelial cells (16). In our study, only two genes were introduced, hTERT (14) and c-myc, in contrast to human TERT (hTERT), simian virus 40 large T, and H-rasG12V in the previous study (16). Despite the potent combination in the latter, the transformed cells consistently acquired mutations on chromosome 8 that affected the c-myc gene, either through amplification or some other chromosomal disruption (16). These data suggest that there is a very strong selection for mutations in c-myc in transformed mammary epithelial cells, consistent with the profound phenotypic changes induced by c-myc in our study. The human mammary epithelial cells used for hTERT immortalization are reported to exhibit suppression of p16INK4a, which may synergize with hTERT to promote immortalization prior to oncogene activation (24). p53 mutations are found in 20 to 40% of breast cancers (6), a significant fraction but lower than found in many other cancers. IMECs are wild type for p53 (J. DiRenzo, personal communication), and so the transformation we observe with the c-myc oncogene is p53 independent. It will be interesting to use the Myc IMECs as recipients to assess the contribution of further disruption in oncogene and/or tumor suppressor pathways in mammary epithelial cell transformation. Furthermore, since DKK1/SFRP1 suppression accounts for only part of the c-myc-induced anchorage-independent growth (Fig. 4), other c-Myc target genes can be tested for specific contributions to a human breast cancer model.
Supplementary Material
Acknowledgments
We thank Jim DiRenzo for many useful discussions and Mike Whitfield for assistance with microarrays. We also thank S. Sokol and J. Rubin for the kind gift of reagents. We thank members of the Cole laboratory for useful discussions.
This work was supported by grants CA80320 (M.D.C.) and CA93719 (L.A.C.) from the National Cancer Institute. This work was also supported by a grant (W81XWH-05-1-0405) from the U.S. Army Breast Cancer Research Program (L.A.C.).
Footnotes
Published ahead of print on 7 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Assoian, R. K. 1997. Anchorage-dependent cell cycle progression. J. Cell Biol. 136:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bafico, A., A. Gazit, T. Pramila, P. W. Finch, A. Yaniv, and S. A. Aaronson. 1999. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 274:16180-16187. [DOI] [PubMed] [Google Scholar]
- 3.Bafico, A., G. Liu, L. Goldin, V. Harris, and S. A. Aaronson. 2004. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell 6:497-506. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., and J. E. Ohm. 2006. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 6:107-116. [DOI] [PubMed] [Google Scholar]
- 5.Bejsovec, A. 2005. Wnt pathway activation: new relations and locations. Cell 120:11-14. [DOI] [PubMed] [Google Scholar]
- 6.Borresen-Dale, A. L. 2003. TP53 and breast cancer. Hum. Mutat. 21:292-300. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, K. R., and A. M. Brown. 2004. Wnt proteins in mammary development and cancer. J. Mammary Gland Biol. Neoplasia 9:119-131. [DOI] [PubMed] [Google Scholar]
- 8.Cowling, V. H., and M. D. Cole. 2007. E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene 26:3582-3586. [DOI] [PubMed] [Google Scholar]
- 9.Cowling, V. H., and M. D. Cole. 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16:242-252. [DOI] [PubMed] [Google Scholar]
- 10.Daksis, J. I., R. Y. Lu, L. M. Facchini, W. W. Marhin, and L. J. Penn. 1994. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene 9:3635-3645. [PubMed] [Google Scholar]
- 11.Dang, C. V., K. A. O'Donnell, K. I. Zeller, T. Nguyen, R. C. Osthus, and F. Li. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16:253-264. [DOI] [PubMed] [Google Scholar]
- 12.D'Cruz, C. M., E. J. Gunther, R. B. Boxer, J. L. Hartman, L. Sintasath, S. E. Moody, J. D. Cox, S. I. Ha, G. K. Belka, A. Golant, R. D. Cardiff, and L. A. Chodosh. 2001. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat. Med. 7:235-239. [DOI] [PubMed] [Google Scholar]
- 13.Deming, S. L., S. J. Nass, R. B. Dickson, and B. J. Trock. 2000. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br. J. Cancer 83:1688-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRenzo, J., S. Signoretti, N. Nakamura, R. Rivera-Gonzalez, W. Sellers, M. Loda, and M. Brown. 2002. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer Res. 62:89-98. [PubMed] [Google Scholar]
- 15.Eisenman, R. N. (ed.). 2006. The Myc oncogene, vol. 302. Springer-Verlag, Berlin, Germany.
- 16.Elenbaas, B., L. Spirio, F. Koerner, M. D. Fleming, D. B. Zimonjic, J. L. Donaher, N. C. Popescu, W. C. Hahn, and R. A. Weinberg. 2001. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15:50-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles, R. H., J. H. van Es, and H. Clevers. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Glinka, A., W. Wu, H. Delius, A. P. Monaghan, C. Blumenstock, and C. Niehrs. 1998. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391:357-362. [DOI] [PubMed] [Google Scholar]
- 19.Gunther, E. J., S. E. Moody, G. K. Belka, K. T. Hahn, N. Innocent, K. D. Dugan, R. D. Cardiff, and L. A. Chodosh. 2003. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 17:488-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 21.Herold, S., M. Wanzel, V. Beuger, C. Frohme, D. Beul, T. Hillukkala, J. Syvaoja, H. P. Saluz, F. Haenel, and M. Eilers. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10:509-521. [DOI] [PubMed] [Google Scholar]
- 22.Jamerson, M. H., M. D. Johnson, and R. B. Dickson. 2004. Of mice and Myc: c-Myc and mammary tumorigenesis. J. Mammary Gland. Biol. Neoplasia 9:27-37. [DOI] [PubMed] [Google Scholar]
- 23.Kawano, Y., and R. Kypta. 2003. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 116:2627-2634. [DOI] [PubMed] [Google Scholar]
- 24.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 25.Klopocki, E., G. Kristiansen, P. J. Wild, I. Klaman, E. Castanos-Velez, G. Singer, R. Stohr, R. Simon, G. Sauter, H. Leibiger, L. Essers, B. Weber, K. Hermann, A. Rosenthal, A. Hartmann, and E. Dahl. 2004. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int. J. Oncol. 25:641-649. [PubMed] [Google Scholar]
- 26.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 27.Lacroix, M., R. A. Toillon, and G. Leclercq. 2004. Stable “portrait” of breast tumors during progression: data from biology, pathology and genetics. Endocr.-Relat. Cancer 11:497-522. [DOI] [PubMed] [Google Scholar]
- 28.Liao, D. J., and R. B. Dickson. 2000. c-Myc in breast cancer. Endocr.-Relat. Cancer 7:143-164. [DOI] [PubMed] [Google Scholar]
- 29.Lin, S. Y., W. Xia, J. C. Wang, K. Y. Kwong, B. Spohn, Y. Wen, R. G. Pestell, and M. C. Hung. 2000. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 97:4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logan, C. Y., and R. Nusse. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781-810. [DOI] [PubMed] [Google Scholar]
- 32.Mao, B., W. Wu, G. Davidson, J. Marhold, M. Li, B. M. Mechler, H. Delius, D. Hoppe, P. Stannek, C. Walter, A. Glinka, and C. Niehrs. 2002. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 417:664-667. [DOI] [PubMed] [Google Scholar]
- 33.McGrew, L. L., K. Takemaru, R. Bates, and R. T. Moon. 1999. Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech. Dev. 87:21-32. [DOI] [PubMed] [Google Scholar]
- 34.Melkonyan, H. S., W. C. Chang, J. P. Shapiro, M. Mahadevappa, P. A. Fitzpatrick, M. C. Kiefer, L. D. Tomei, and S. R. Umansky. 1997. SARPs: a family of secreted apoptosis-related proteins. Proc. Natl. Acad. Sci. USA 94:13636-13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miliani de Marval, P. L., E. Macias, R. Rounbehler, P. Sicinski, H. Kiyokawa, D. G. Johnson, C. J. Conti, and M. L. Rodriguez-Puebla. 2004. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol. Cell. Biol. 24:7538-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moody, S. E., C. J. Sarkisian, K. T. Hahn, E. J. Gunther, S. Pickup, K. D. Dugan, N. Innocent, R. D. Cardiff, M. D. Schnall, and L. A. Chodosh. 2002. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2:451-461. [DOI] [PubMed] [Google Scholar]
- 37.Noubissi, F. K., I. Elcheva, N. Bhatia, A. Shakoori, A. Ougolkov, J. Liu, T. Minamoto, J. Ross, S. Y. Fuchs, and V. S. Spiegelman. 2006. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature 441:898-901. [DOI] [PubMed] [Google Scholar]
- 38.Nusse, R. 2005. Wnt signaling in disease and in development. Cell Res. 15:28-32. [DOI] [PubMed] [Google Scholar]
- 39.Nusse, R., and H. E. Varmus. 1982. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109. [DOI] [PubMed] [Google Scholar]
- 40.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 41.Penn, L. J., M. W. Brooks, E. M. Laufer, and H. Land. 1990. Negative autoregulation of c-myc transcription. EMBO J. 9:1113-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipp, A., A. Schneider, I. Vasrik, K. Finke, Y. Xiong, D. Beach, K. Alitalo, and M. Eilers. 1994. Repression of cyclin D1: a novel function of MYC. Mol. Cell. Biol. 14:4032-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryo, A., M. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3:793-801. [DOI] [PubMed] [Google Scholar]
- 44.Sandgren, E. P., J. A. Schroeder, T. H. Qui, R. D. Palmiter, R. L. Brinster, and D. C. Lee. 1995. Inhibition of mammary gland involution is associated with transforming growth factor alpha but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res. 55:3915-3927. [PubMed] [Google Scholar]
- 45.Schoenenberger, C. A., A. C. Andres, B. Groner, M. van der Valk, M. LeMeur, and P. Gerlinger. 1988. Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumours with constitutive milk protein gene transcription. EMBO J 7:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, T. A., P. K. Pattengale, and P. Leder. 1984. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38:627-637. [DOI] [PubMed] [Google Scholar]
- 47.Thiery, J. P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740-746. [DOI] [PubMed] [Google Scholar]
- 48.Tsukamoto, A. S., R. Grosschedl, R. C. Guzman, T. Parslow, and H. E. Varmus. 1988. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55:619-625. [DOI] [PubMed] [Google Scholar]
- 49.Ugolini, F., E. Charafe-Jauffret, V. J. Bardou, J. Geneix, J. Adelaide, F. Labat-Moleur, F. Penault-Llorca, M. Longy, J. Jacquemier, D. Birnbaum, and M. J. Pebusque. 2001. WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene 20:5810-5817. [DOI] [PubMed] [Google Scholar]
- 50.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 51.Veeman, M. T., J. D. Axelrod, and R. T. Moon. 2003. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell 5:367-377. [DOI] [PubMed] [Google Scholar]
- 52.Willert, J., M. Epping, J. R. Pollack, P. O. Brown, and R. Nusse. 2002. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, L., R. B. Corcoran, J. W. Welsh, D. Pennica, and A. J. Levine. 2000. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 14:585-595. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.