Abstract
To explore the global mechanisms of estrogen-regulated transcription, we used chromatin immunoprecipitation coupled with DNA microarrays to determine the localization of RNA polymerase II (Pol II), estrogen receptor alpha (ERα), steroid receptor coactivator proteins (SRC), and acetylated histones H3/H4 (AcH) at estrogen-regulated promoters in MCF-7 cells with or without estradiol (E2) treatment. In addition, we correlated factor occupancy with gene expression and the presence of transcription factor binding elements. Using this integrative approach, we defined a set of 58 direct E2 target genes based on E2-regulated Pol II occupancy and classified their promoters based on factor binding, histone modification, and transcriptional output. Many of these direct E2 target genes exhibit interesting modes of regulation and biological activities, some of which may be relevant to the onset and proliferation of breast cancers. Our studies indicate that about one-third of these direct E2 target genes contain promoter-proximal ERα-binding sites, which is considerably more than previous estimates. Some of these genes represent possible novel targets for regulation through the ERα/AP-1 tethering pathway. Our studies have also revealed several previously uncharacterized global features of E2-regulated gene expression, including strong positive correlations between Pol II occupancy and AcH levels, as well as between the E2-dependent recruitment of ERα and SRC at the promoters of E2-stimulated genes. Furthermore, our studies have revealed new mechanistic insights into E2-regulated gene expression, including the absence of SRC binding at E2-repressed genes and the presence of constitutively bound, promoter-proximally paused Pol IIs at some E2-regulated promoters. These mechanistic insights are likely to be relevant for understanding gene regulation by a wide variety of nuclear receptors.
Signal-regulated transcriptional responses are an important means by which cells respond to physiological and environmental cues. These responses typically involve the direct or indirect activation or inhibition of site-specific DNA-binding transcription factors, which mediate their effects by binding to cognate regulatory sites across the genome. Steroid hormone receptors, such as estrogen receptor alpha (ERα), represent a class of signal-activated DNA-binding transcription factors that respond to small-molecule ligands, such as estrogens (35). Estrogens and other steroid hormones act through their receptors to control patterns of gene expression that specify distinct physiological outcomes, including aspects of reproduction, development, and metabolism (13, 40). Estrogens also play key roles in many disease states, including breast and uterine cancers as well as osteoporosis (14). Understanding how such cellular signaling events lead to specific gene regulatory responses and physiological outcomes is a fundamental question in biology.
The binding of signal-regulated transcription factors to regulatory sites (i.e., enhancers and silencers) across the genome ultimately regulates the recruitment and/or activity of RNA polymerase II (Pol II) at target promoters, thus regulating the transcription of those promoters (see Fig. 1A for an example). In cases of transcriptional activation, the recruitment of a variety of coactivator proteins by DNA-bound transcription factors opens the chromatin structure and stabilizes Pol II binding at the promoter, leading to increased gene transcription (39). In cases of transcriptional repression, the recruitment of corepressor proteins directs the formation of repressive chromatin structures or transcription complexes that inhibit the recruitment and activation of Pol II at the promoter (19). The multistep process of (i) signal-regulated transcription factor binding to enhancers/silencers, (ii) coactivator/corepressor recruitment and activity, and (iii) Pol II recruitment/dissociation and activation/inhibition provides many opportunities for exquisite regulatory control that directs global transcriptional responses (19, 39). Although the underlying mechanisms of signal-regulated transcription factor binding at cognate genomic sites have been well characterized, the specific mechanisms connecting transcription factor activity at enhancers to Pol II recruitment and activation at target promoters across the genome are less well understood.
FIG. 1.
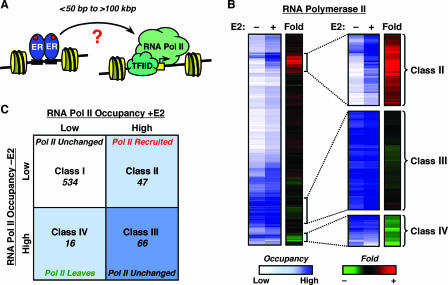
Regulation of RNA Pol II occupancy by E2 at target promoters in MCF-7 cells. (A) Schematic representation of short-range and long-range regulation of RNA Pol II by liganded ERα. Changes in the recruitment or activity of RNA Pol II complexes at the promoter represent the functional outcomes of liganded ERα binding to its cognate enhancers across the genome. TFIID, transcription factor IID. (B) RNA Pol II genomic location analysis by ChIP-chip in MCF-7 cells. Data from all the filtered elements on the custom estrogen-regulated promoter array are represented by heat maps. The relative occupancy of each genomic fragment by RNA Pol II with or without E2 treatment is shown in the blue/white scale, whereas the change (n-fold) of RNA Pol II occupancy upon E2 treatment is shown in the red/green scale. The expanded images on the right highlight some of the interesting classes of genes shown in panel C. (C) Classification of RNA Pol II binding sites. Each quadrant in the graphical representation indicates the number of filtered elements from the array exhibiting one of four different patterns of RNA Pol II occupancy (e.g., for class II, low occupancy, −E2; high occupancy, +E2).
A number of recent studies have examined the binding of signal-regulated transcription factors, including E2Fs, HNFs, Sp1, c-Myc, p53, and ERα, to regulatory sites across the genome using the powerful combination of chromatin immunoprecipitation (ChIP) coupled with DNA microarrays (a “ChIP-chip” approach) (50). These studies have provided a global view of transcription factor binding that was lacking from previous gene-by-gene analyses. Studies with estrogen-bound ERα have shown that, for many estrogen-regulated genes, the receptor binding sites are located at great distances (>100 kilobase pairs) from the target promoters, while for other genes the receptor binding sites are located within or near the proximal promoter regions (9-11, 23, 29, 31) (Fig. 1A). These sites include both direct ERα-binding sites (i.e., estrogen response elements [EREs]) and tethering sites (e.g., AP-1, Sp1, and NF-κB sites). In contrast to the genomic localization of ERα, much less is known about how ERα binding events at promoter-proximal or promoter-distal enhancers lead to estrogen-dependent changes in Pol II occupancy at target promoters and the regulation of gene expression.
In the studies described herein, we used ChIP-chip with a custom genomic microarray to explore how estrogen signaling affects factor recruitment and histone modification at estrogen-regulated promoters across the genome in MCF-7 human breast cancer cells. Specifically, we compared the occupancy of Pol II to the binding of ERα and steroid receptor coactivator (SRC) proteins, acetylation of nucleosomal histones, and transcriptional output from target promoters. The results from our ChIP-chip experiments extend beyond previous ERα genomic localization experiments by (i) examining the ligand-dependent localization of a non-DNA-binding nuclear receptor cofactor (i.e., SRC), (ii) examining the genomic location of four different types of factors simultaneously (i.e., ERα [a DNA-binding factor], SRC [a non-DNA-binding coregulator], histone acetylation [a histone mark], and Pol II), (iii) determining the patterns of localization in both the presence and absence of estradiol (E2), and (iv) correlating patterns of genomic localization with functional output, namely, Pol II recruitment and transcription from target promoters. Our results have allowed us to classify the estrogen-regulated promoters in MCF-7 cells based on factor binding, histone modification, and transcriptional output. Furthermore, examination of both estrogen-treated and untreated conditions has revealed novel mechanistic aspects of ligand-regulated gene expression by ERα. Collectively, our results provide new insights into the transcriptional networks controlling estrogen-regulated gene expression.
MATERIALS AND METHODS
Additional information about the materials and methods is available at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm.
ChIP assays.
ChIPs for ERα, Pol II, the Pol II serine 2 (Ser2)-phosphorylated C-terminal domain, SRC, c-Fos, NELFA (a negative elongation factor [NELF] subunit), and acetylated histones H3 and H4 (AcH) were performed using MCF-7 cells grown in estrogen-free medium essentially as described previously (1, 45). The resulting ChIP DNA material was used to probe custom promoter arrays as described below or in gene-specific quantitative real-time PCR (qPCR) analyses.
ChIP-chip.
A custom spotted estrogen-regulated promoter array was designed and produced using approaches described previously (41, 43). ChIP DNA was blunted using T4 DNA polymerase, ligated to linkers, and amplified using ligation-mediated PCR, as described previously (43). The amplified DNA was labeled with Cy3 or Cy5 fluorophores by use of a Bioprime random primer labeling kit (Invitrogen) and then used to hybridize the promoter arrays as described previously (41, 43); details on the modifications are available at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm. Washed arrays were scanned using a GenePix 4000B scanner (Axon), and the data were collected using the GenePix Pro 6.0 software (Axon).
Statistical methods for ChIP-chip and ChIP-expression analyses.
Statistical analyses were performed using the statistical software R (GNU project; Free Software Foundation); all the scripts that were used are available on request. After the spots flagged for bad quality were filtered out, the signal ratio of immunoprecipitated DNA over input DNA was log2 transformed and normalized based on a set of control genes; descriptions are available at the URL noted above. For each factor studied, the change (n-fold) between untreated and E2-treated cells was calculated and log2 transformed. Analysis of variance was performed across all the replicates, and a nominal P value threshold of <0.05 was used to select promoters for further analyses. In addition to the P value threshold, a log2 change (n-fold) threshold of ±0.37 was used to select target promoters. Promoters that passed both the P value and change (n-fold) thresholds were classified as target sites where the factor occupancy changed upon E2 treatment. Based on standard ChIP-qPCR experiments, our estimated false-positive error rate using the thresholds described above was <9% (number of genes tested = 30). To identify target promoters where ERα or RNA Pol II was localized in both untreated and E2-treated cells, we performed a median percentile rank analysis as described previously (7). Hierarchical clustering of the data was performed using the Cluster software and visualized using the Treeview software (16). For the correlation analyses, Spearman's rank correlation coefficients were calculated using data that passed the P value threshold as described above. The data for all the factors and genomic regions studied are available at the URL noted above.
DNA sequence analyses.
Each DNA sequence on the array was scanned for the presence of EREs and AP-1 binding elements using position weight matrices obtained from the TRANSFAC database (accession numbers M00174 and M00517, respectively) using approaches described in detail at the URL noted above.
Transient transfection reporter gene assays.
MCF-7 cells grown in estrogen-free medium were transfected with the following combination of plasmid DNAs: (i) 1 μg of a luciferase reporter construct containing either the native UGT2B15 promoter (spanning the region from −449 bp to +114 bp) or the native UGT2B15 promoter with a deletion of the AP-1 binding element and (ii) 600 ng of pCMVβ, a constitutive β-galactosidase expression vector used to normalize for transfection efficiency. The transfected cells were treated with or without 10 nM E2 for 16 h and then assayed for luciferase activity and β-galactosidase activity.
RNA extraction and reverse transcription-qPCR.
Total RNA was isolated from MCF-7 cells grown in estrogen-free medium by using RNeasy columns (QIAGEN). cDNA prepared from each sample was analyzed by qPCR in a real-time PCR thermocycler under standard conditions.
Expression microarray analyses.
Total RNA was isolated from MCF-7 cells grown in estrogen-free medium by using TRIzol reagent (Invitrogen) followed by RNeasy columns (QIAGEN). Seven micrograms of total RNA was subjected to one-cycle target labeling assay (Affymetrix) to generate biotinylated cRNA targets for hybridization to Affymetrix U133A 2.0 microarray under standard conditions. The raw data from three independent replicates were processed by Affymetrix GCOS software to obtain detection calls and signal values and then normalized by scaling. Only probe sets having “present” calls on at least two of the three arrays were included for further analysis; those signals were log2 transformed and median centered. The t test was applied to the normalized data matrix to identify differential genes between the E2-treated and untreated control samples.
RESULTS
Regulation of RNA Pol II occupancy by E2 at target promoters.
To understand the specific mechanisms connecting transcription factor activity at enhancers to Pol II recruitment and activation at target promoters, we examined E2-regulated transcriptional responses (i.e., factor binding, histone modification, and transcriptional output) in ERα-positive MCF-7 human breast cancer cells. As an initial functional readout, we examined the localization of Pol II at estrogen-regulated promoters in response to E2 treatment using ChIP-chip with a custom promoter microarray. Since previous studies have demonstrated that Pol II is a good marker for transcriptionally active promoters (3, 21), our expectation was that E2-bound ERα would alter the occupancy of Pol II at the promoters of estrogen-regulated genes whether or not the ERα-binding regulatory site was located in the proximal promoter or at a great distance from the promoter (Fig. 1A). A key aspect of this work not addressed in most of the previous ERα ChIP-chip studies is the examination of both the −E2 and +E2 conditions during the genomic discovery phase of the work (as opposed to the gene-specific confirmation phase). This allowed us to study on a global scale a wider variety of modes for E2-regulated transcription, including (i) E2-dependent repression and (ii) E2-independent constitutive binding of Pol II at E2-regulated promoters.
For our ChIP-chip studies, we produced a custom spotted genomic microarray containing ∼900 human genomic regions (∼1 kb each), including ∼600 estrogen-regulated promoters, ∼250 control promoters, and ∼50 nonpromoter regions. Each promoter was represented by a DNA fragment spanning ca. −800 to +200 bp relative to the transcription start site (TSS), and some of the promoters were tiled in ∼1-kb fragments over a 12-kb region surrounding the TSS. The estrogen-regulated promoters were selected based on (i) previous expression microarray studies, (ii) bioinformatic analyses, and (iii) previous gene-by-gene studies. The promoters included those from estrogen up-regulated and down-regulated genes, as well as genes whose regulation by estrogens is cell type specific. We also included 12 previously characterized nonpromoter ERα-binding regions on the array (9). Overall, our approach provided coverage of more than 50 percent of the E2-regulated transcriptome in MCF-7 cells under the conditions tested (see below).
MCF-7 cells were treated with ethanol or E2 for a short time (i.e., 45 min) so that target promoters most likely directly regulated by E2, not through secondary effects, could be identified. Genomic DNA fragments bound by Pol II were isolated by ChIP, the immunoprecipitated DNA was hybridized to the genomic microarray, and the data were analyzed as shown in Fig. 1B. A number of controls to ensure experimental fidelity in the ChIP-chip experiments were also performed (e.g., mock ChIP hybridization, dye swapping; data not shown). Our data analysis protocol allowed us to determine the relative levels of Pol II in the absence and presence of E2 treatment (“Occupancy” in Fig. 1B), as well as the change (n-fold) in Pol II occupancy upon E2 treatment (“Fold” in Fig. 1B). We then used these analyses to classify the promoters based on the patterns of Pol II localization.
We identified 47 promoters where the occupancy of Pol II increased upon E2 treatment (classified as class II promoters) and 16 promoters where the occupancy of Pol II decreased upon E2 treatment (class IV promoters) (Fig. 1B and C). We also identified 66 promoters that were occupied by Pol II both in the presence and absence of E2 treatment (class III promoters), many but not all of which are well-characterized housekeeping genes that are not regulated by E2. Finally, we identified a group of 534 promoters that were not occupied by Pol II in either the presence or the absence of E2 treatment (class I). The vast majority of the class I promoters represent a group of genes that are not directly regulated by E2 in MCF-7 cells under the experimental conditions that we used (see below). Our results using Pol II as an endpoint of E2 signaling are consistent with previous gene expression studies showing that, in MCF-7 cells, more than 100 genes are regulated more than twofold after a 4-hour treatment with E2, and most of these genes are up-regulated (18).
Classification of direct E2 target genes.
Our initial Pol II ChIP-chip experiments conducted in the absence and presence of E2 allowed us to define a set of 58 direct E2 target genes based on E2-dependent changes in Pol II promoter occupancy (and mRNA expression; see below), as opposed to inferences from bioinformatics and statistical analyses or correlations with RNA expression alone (a more detailed description of how the direct E2 target genes were defined is available at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm. The use of both a short E2 treatment (i.e., 45 min.) and a −E2 condition, which was lacking in previous ChIP-chip analyses (9, 10, 23, 29, 31) (see Table S1 at the URL noted above), is required for this type of classification. While direct regulation by E2 of a number of the target genes on our array has been demonstrated previously (e.g., the TFF1 gene, also known as pS2), our studies using the Pol II-based criteria noted above have confirmed an additional set of more than 40 direct E2 target genes (see Fig. 3A; see also Fig. S2 and Table S1 at the URL noted above). These include UGT2B15, NBPF15, PRUNE, SLC27A2, HSPB8, and FACVL1, which are involved in processes such as xenobiotic metabolism, fatty acid transport, and protein chaperoning. The identification of direct (as opposed to secondary) E2 target genes is an important step in understanding early E2-dependent gene regulatory responses.
FIG. 3.
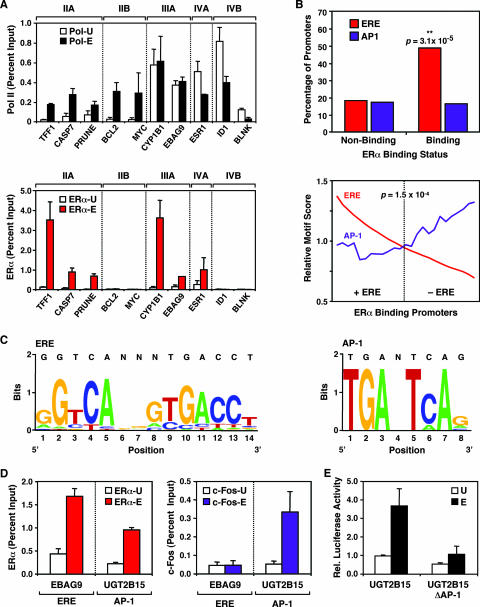
Gene-by-gene validation of RNA Pol II and ERα promoter occupancy coupled with ERα-binding sequence analyses. (A) Validation by ChIP-qPCR of the ChIP-chip results for selected promoters from the estrogen-regulated promoter array. Results for RNA Pol II (upper graph) and ERα (lower graph) are shown. The empty and filled bars represent ChIP enrichment for untreated (-U) and E2-treated (-E) cells, respectively. The class IIA, IIB, IIIA, IVA, and IVB promoters are as described for Fig. 2C. Each bar represents the mean + standard error of the mean (SEM) for at least three separate determinations. (B) Sequence analysis of ERα-binding and nonbinding promoters. Motif-finding algorithms were used to search for ERE- and AP-1-like sequences in the ERα-binding and nonbinding promoters from the promoter microarray, as described in Materials and Methods. (Top) Percentage of promoters in each category (i.e., ERα binding or ERα nonbinding) containing ERE- or AP-1-like sequences; (bottom) analysis of the coexistence of ERE- and AP-1-like sequences in the set of ERα-binding promoters. Relative ERE and AP-1 motif scores are plotted for each sequence. The dotted line represents the significance threshold (P = 1.5 × 10−4) for determining ERE-like sequences. A moving-average analysis was performed by calculating the average motif score of a sliding window of 13 regions from high to low ERE scores. (C) Sequence logos for the ERE-like (left) and AP-1-like (right) elements identified in the ERα-binding promoters are shown. (D) ChIP-qPCR assay of ERα recruitment (left graph) and c-Fos recruitment (right graph) to the EBAG9 promoter, an “ERE-only” (i.e., ERE-positive/AP-1 site-negative) promoter, and the UGT2B15 promoter, an “AP-1-only” (i.e., ERE-negative/AP-1 site-positive) promoter, in response to E2. The empty and filled bars represent ChIP enrichment for untreated and E2-treated cells, respectively. Each bar represents the mean + SEM for at least three separate determinations. Schematics of the EBAG9 and UGT2B15 promoters can be found in Fig. S3 at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm. (E) Luciferase reporter gene assays examining the role of a promoter-proximal AP-1 site in the E2-dependent regulation of the UGT2B15 promoter. UGT2B15-luciferase reporter constructs, with or without the promoter-proximal AP-1 site (UGT2B15 and UGT2B15 ΔAP-1, respectively), were transfected into MCF-7 cells and subsequently treated with E2 as described in Materials and Methods. Each bar represents the mean + SEM for at least three separate determinations.
Correlation between ERα binding at enhancer sites and RNA Pol II recruitment to target promoters.
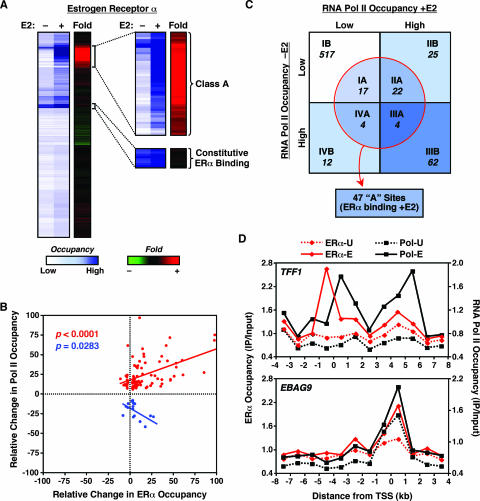
Previous gene-by-gene studies using model estrogen-regulated genes have shown that E2-dependent changes in Pol II promoter occupancy correlate with the binding of ERα to estrogen-responsive enhancers located in the promoters or distal regulatory regions of the genes (15, 37, 45). In this regard, we examined which Pol II-bound promoters were also bound by ERα. In addition, we examined ERα binding to a small subset of previously characterized distal enhancers. We performed ChIP-chip with our custom promoter array to determine the relative levels of ERα in the absence or presence of E2 treatment (“Occupancy” in Fig. 2A) as well as the change (n-fold) in ERα occupancy upon E2 treatment (“Fold” in Fig. 2A). We identified 47 genomic regions where ERα was recruited upon E2 treatment (classified as class A genomic sites, as opposed to class B genomic sites, where no ERα recruitment was observed) (Fig. 2A). Of the 47 class A sites, 37 were in gene promoters (i.e., within 1 kb from the TSS), while the remaining 10 were located at greater distances (i.e., 5 to 400 kb) from the nearest known gene (Table 1). Interestingly, we also identified eight promoters that were occupied by ERα in both the presence and the absence of E2 (Fig. 2A and data not shown).
FIG. 2.
Cooccupancy of RNA Pol II and ERα at target promoters in MCF-7 cells. (A) ERα genomic location analysis by ChIP-chip in MCF-7 cells. Data from all of the elements on the custom estrogen-regulated promoter array are represented by heat maps, as described for RNA Pol II in Fig. 1B. The expanded images on the right highlight the “class A” regions shown in panel C and constitutively ERα-bound promoters. (B) Correlation between ERα and RNA Pol II occupancy at estrogen-regulated promoters. The relative change in factor occupancy is defined as the log2 change (n-fold) in occupancy upon E2 treatment as a percentage of the maximum log2 change (n-fold) observed for each factor. The red data points show the data from a collection of promoters for which estrogen-dependent ERα recruitment correlates positively with RNA Pol II recruitment (correlation coefficient = 0.43 [P < 0.0001]). The blue data points show the data from a collection of promoters for which estrogen-dependent ERα recruitment correlates negatively with RNA Pol II recruitment (correlation coefficient = −0.53 [P = 0.0283]). All elements from the promoter array showing significant E2-dependent enrichment (P < 0.05) were included in the analysis. (C) Classification of ERα-binding sites based on RNA Pol II occupancy. The class A regions (within the circle) include 47 sites where ERα is recruited upon E2 treatment, 37 of which are gene promoters. The class B regions are those for which no ERα recruitment was observed. The distribution of the class A and B regions is superimposed on the representation of the RNA Pol II classes I, II, III, and IV from Fig. 1C. (D) ChIP-chip tiling for the TFF1 and EBAG9 genes. Occupancy, expressed as ChIP enrichment ratios (IP/input), for ERα and RNA Pol II throughout the indicated genomic region is shown in the absence (-U) or presence (-E) of E2. “0” represents the annotated TSS; regions upstream of the TSS are indicated by negative numbers.
TABLE 1.
Factor binding to nonpromoter regions and E2-dependent regulation of the nearest neighboring genes
| Cluster no.a | Binding sitea | Chromosomal location
|
Enhancer bindingb
|
Nearest genec | Distance (kb)c | Promoter bindingb
|
Gene expressiond | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERα | Pol II | SRC | AcH | ER | Pol | SRC | AcH | |||||||
| 3 | Blk3 | 21 | 15171656 | + | − | − | − | NRIP1 | 83.2 | − | + | − | + | + |
| 4 | Blk4 | 15467150 | + | + | − | − | NRIP1 | 226 | − | + | − | + | + | |
| Blk5 | 15493593 | + | + | − | − | |||||||||
| Blk7 | 15503950 | + | + | + | + | |||||||||
| 13 | Blk21 | 40606306 | − | − | − | − | PCP4 | 386 | − | − | − | − | − | |
| Blk22 | 40609671 | + | + | − | + | |||||||||
| 16 | Blk32 | 42680784 | + | − | − | − | TMPRSS3 | 4.80 | − | + | + | + | + | |
| Blk33 | 42690200 | + | − | − | − | |||||||||
| 23 | Blk42 | 22 | 19822950 | + | + | + | + | SLC7A4 | 172 | − | − | − | + | ND |
| Blk44 | 19943560 | + | + | + | + | |||||||||
| 24 | Blk45 | 27534171 | − | − | − | − | XBP1 | 15.5 | − | + | − | + | + | |
| Blk46 | 27539034 | + | + | − | − | |||||||||
The binding sites are those defined by Carroll et al. (9) and are listed using their nomenclature. These binding sites can be grouped in six distinct clusters based on their chromosome location.
The binding of ERα, Pol II, SRC, and AcH at the designated enhancer and promoter regions in MCF-7 cells were determined by ChIP-chip and ChIP-qPCR, respectively. + indicates a ≥1.5-fold increase following E2 treatment; − indicates a <1.5-fold increase following E2 treatment.
The nearest gene to each cluster and the distance between the gene and the center of the cluster are listed.
The change in gene expression following E2 treatment was determined by expression microarray analysis. + indicates a ≥1.5-fold increase following E2 treatment; − indicates a <1.5-fold increase following E2 treatment. ND indicates that no transcript was detectable.
To examine the relationship between Pol II and ERα binding at target gene promoters, the results for ERα binding were compared with the results obtained for Pol II. Spearman rank correlation analysis revealed two distinct types of correlation between Pol II and ERα localization at target gene promoters (Fig. 2B). The first is a significant positive correlation between ERα and Pol II occupancy at a set of cooccupied promoters upon E2 treatment (Fig. 2B; correlation coefficient = 0.43, P < 0.0001). This result is consistent with previous studies showing that the recruitment of ERα to target promoters promotes the recruitment of the Pol II machinery to those promoters (15, 37, 45). The second is a significant negative correlation between ERα and Pol II occupancy at a smaller set of promoters upon E2 treatment (Fig. 2B; correlation coefficient = −0.53 [P = 0.0283]). This finding is consistent with studies showing that for certain estrogen-regulated genes, ERα recruitment represses gene expression (36, 49), although the associated mechanisms are not yet fully understood.
To analyze further which Pol II-bound promoters were bound by ERα upon E2 treatment, we examined how the class A (i.e., ligand-dependent) ERα-binding sites were distributed among the class I, II, III, and IV Pol II binding sites (Fig. 2C). ERα was recruited to about half of the promoters to which Pol II was recruited upon E2 treatment (class IIA; 22 out of 47), whereas no ERα binding was detected at the rest of these promoters (class IIB). Moreover, ERα was recruited to 4 promoters from which Pol II was released upon E2 treatment (class IVA), whereas no ERα binding was detected at the other 12 promoters (class IVB). These results reflect the ability of ERα to regulate its target genes by binding either at proximal promoters (classes IIA and IVA) or at enhancers located outside of the promoter regions (classes IIB and IVB) (10, 29, 31, 37). Interestingly, ERα was also recruited in an E2-dependent manner to four promoters that were occupied by Pol II both in the presence and absence of E2 (class IIIA). Such an observation is consistent with the presence of a constitutively bound “paused” Pol II that is not productively engaged in transcription until the recruitment and activation of ERα by E2 (44; see Fig. 6C and 7 below). Finally, ERα was recruited in an E2-dependent manner to 17 promoters where no Pol II binding was observed (class IA). For these genes, many of which show E2-dependent regulation of expression (see Fig. 6C below), the detection of Pol II may fall below the limit of sensitivity of our ChIP-chip assay. Alternatively, Pol II may not be detected at these promoters due to rapid movement into the body of the gene. The recruitment of ERα and Pol II to promoters was generally consistent with the expression of the associated genes (see Fig. 6B and C; also see Fig. S5C at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Collectively, our studies indicate that the pattern of ERα and Pol II binding at E2-regulated promoters, which is more complicated than previously realized, can be used to group the promoters into mechanistically distinct classes.
FIG. 6.
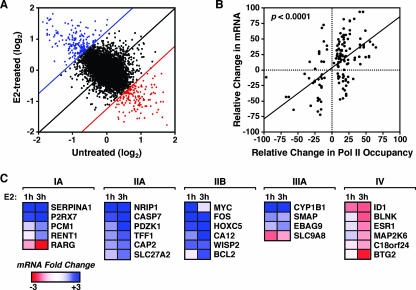
E2-dependent gene expression correlates with RNA Pol II recruitment to target promoters. (A) RNA expression analysis of MCF-7 cells in the presence or absence of a 3-h treatment with E2 by use of Affymetrix U133A 2.0 microarrays. Scatterplot presentation of normalized signal for each gene on the microarray in E2-treated cells (y axis) plotted versus the signal for each gene from untreated cells (x axis). The blue and red lines represent the twofold increase or decrease cutoffs for expression, respectively. Genes showing E2-dependent increases in expression are in blue, whereas genes showing E2-dependent decreases in expression are in red. Genes regulated less than twofold by E2 are in black. (B) Correlation between E2-dependent RNA Pol II recruitment and mRNA expression. The relative change in mRNA or Pol II occupancy is defined as the log2 change (n-fold) upon E2 treatment as a percentage of the maximum log2 change (n-fold) observed for each factor. The correlation coefficient is 0.45 (P < 0.0001). All promoters showing significant changes (P < 0.05) for RNA expression and RNA Pol II ChIP recruitment were included in the analysis. (C) Gene-by-gene confirmation of expression microarray results. MCF-7 cells were treated with or without E2 for 1 h or 3 h, and then RNA was isolated and analyzed by qPCR for genes in classes IA, IIA, IIB, IIIA, and IV (from Fig. 2C). The mRNA expression scale is shown.
FIG. 7.
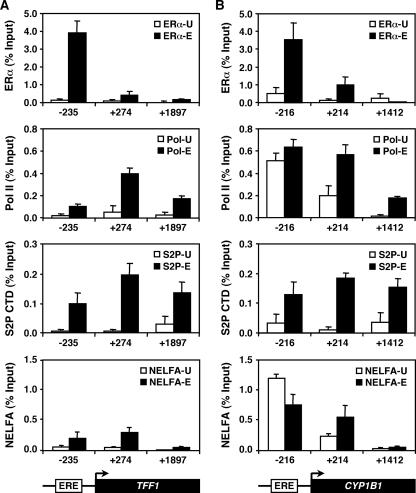
E2-dependent regulation of constitutively bound Pol II at target promoters. ChIP-qPCR was used to determine the occupancy by ERα, Pol II, Ser2-phosphorylated Pol II, and NELFA at the TFF1 (A) and CYP1B1 (B) promoters. The empty and filled/hatched bars represent ChIP enrichment for untreated (-U) and E2-treated (-E) cells, respectively. “−” and “+” indicate the regions upstream and downstream of the TSS, respectively. Each bar represents the mean + SEM for at least three separate determinations.
We also examined the relationship between ERα binding to distal enhancers and Pol II recruitment to the nearest neighboring promoters. Of the 12 previously characterized nonpromoter ERα-binding regions included on the ChIP-chip array, 10 showed detectable binding of ERα in response to E2 and were classified as class A (i.e., ligand-dependent) ERα-binding sites (Table 1). These 10 nonpromoter class A sites correspond to six different enhancer clusters, each presumably controlling the expression of a neighboring gene (Table 1). The E2-dependent recruitment of ERα to the enhancer clusters correlated with Pol II recruitment to the nearest neighboring promoter, as well as expression of the associated gene, in four out of the six cases examined (Table 1). However, in two out of the six cases examined, ERα recruitment to the distal enhancer did not appear to regulate the nearest neighboring promoter. In many cases, Pol II was also detected at the distal enhancers, as described previously (9, 10). Interestingly, however, no ERα recruitment was detected at the promoters of any of the nearest neighboring genes. These results are consistent with long-range activation by liganded ERα from distal enhancers involving, perhaps, a looping mechanism (9).
To substantiate our ChIP-chip results, as well as generate a more detailed view of Pol II and ERα occupancy at target promoters, we performed ChIP-chip tiling across a 12-kb region surrounding the TSSs of four previously identified E2 target genes (i.e., the TFF1, EBAG9, c-Myc, and CASP7 genes) (Fig. 2D; also see Fig. S1A and D at the URL noted above). This approach allowed us to distinguish more clearly the sites of binding and nonbinding across the promoter regions. For example, with the TFF1 gene, a clear peak of ERα recruitment was present at ∼0.5 kb upstream of the TSS in the vicinity of a well-characterized ERE (1, 37), whereas the peak of the Pol II recruitment was immediately downstream of the TSS (Fig. 2D, top panel). In addition, a peak of Pol II was observed ∼5 kb downstream of the TSS, coinciding with the end of the TFF1 gene. This observation is consistent with previous studies suggesting that Pol II temporarily pauses during transcriptional termination (20). The other three promoters also showed good correlations between previously characterized EREs and the peaks of ERα signal (e.g., the EBAG9 gene has a single ERE at +43 bp relative to the TSS) as well as between known TSSs and the peaks of Pol II signal (e.g., the c-Myc gene has three TSSs within its complex promoter region [5]). These results provide an indication of the sensitivity and fidelity of our ChIP-chip approach as well as information about the detailed relationship between ERα binding and Pol II recruitment in response to E2.
Validation of Pol II and ERα ChIP-chip results by qPCR.
To validate our ChIP-chip experiments and calculate the false-positive error rates for our results, we performed standard ChIP-qPCR for Pol II and ERα on a set of randomly selected target promoters identified in our ChIP-chip studies (Fig. 3A; also see Fig. S2 at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Approximately 93 percent (25 out of 27) of the Pol II binding sites identified by ChIP-chip also showed Pol II binding by ChIP-qPCR. Similarly, about 91 percent (21 out of 23) of the ERα-binding sites identified by ChIP-chip showed ERα binding by ChIP-qPCR. Thus, our ChIP-chip experiments had false-positive error rates of less than 10 percent, which are similar to if not lower than those reported for other ChIP-chip analyses.
Sequence analysis of ERα-binding promoters.
After characterizing the binding of ERα to a set of genomic sequences by ChIP-chip, we assessed whether the ERα-binding promoters contain ERE-like sequences by using a statistical motif-searching algorithm. Every DNA region on the microarray was queried for the presence or absence of an ERE using a previously determined ERE positional weight matrix (42), and the results were expressed relative to ERα binding status (Fig. 3B). The likelihood of an ERE-like sequence occurring in one of the ERα-binding promoters was significantly higher (P = 3.1 × 10−5) than that found for promoter regions where no ERα binding was observed (Fig. 3B, top). Among the ERα-binding promoters, about half contained an ERE-like sequence (18 out of 37 [P < 1.5 × 10−4]) (Fig. 3B, bottom, and C, left). The majority of the ERα-binding promoters lacking an ERE-like sequence contained one or more ERE half-sites (data not shown). In total, about 17 percent (10 out of 58) of the direct E2 target promoters on our array (as defined above) bind ERα and contain an ERE-like sequence.
In contrast, a similar approach using an AP-1 binding site position weight matrix (24) revealed no difference in the occurrence of AP-1-like sequences between the ERα-binding and nonbinding promoters (Fig. 3B, top). However, for the ERα-binding promoters, AP-1-like sequences were enriched in the promoters that did not contain an ERE-like sequence (P = 0.038; Fig. 3B, bottom, and C, right). That is, for the set of ERα-binding promoters, the presence of an AP-1 site was more likely in the absence of an ERE sequence. These results implicate ERα action at promoters through AP-1 (i.e., the “tethering” pathway [28]) as a possible mechanism for E2-dependent regulation of ERE-less promoters.
To connect our genomic binding data and bioinformatic analyses to the biology of estrogen signaling through EREs and AP-1 sites, we examined by gene-specific ChIP-qPCR the recruitment of ERα and c-Fos (a component of AP-1) to two ERα-binding promoters: an “ERE-only” promoter (i.e., the EBAG9 gene; ERE-positive/AP-1 site-negative) (see also Fig. S3 at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm) and an “AP-1-only” promoter (i.e., the UGT2B15 gene; ERE-negative/AP-1 site-positive) (see Fig. S3 at the URL noted above). As expected, both promoters showed recruitment of ERα, but only the UGT2B15 promoter showed recruitment of c-Fos upon E2 treatment (Fig. 3D). These results are consistent with the hypothesis that ERα is recruited to the ERE-negative/AP-1 site-positive UGT2B15 promoter through tethering with components of the AP-1 complex. In transient transfection reporter gene assays in MCF-7 cells with a UGT2B15-luciferase construct, E2 treatment caused a fourfold increase in luciferase activity (Fig. 3E). Furthermore, deletion of the AP-1 site in the UGT2B15 promoter dramatically reduced reporter gene activity in the presence of E2 (Fig. 3E). These results confirm the predictions made based on the bioinformatic analyses and suggest that UGT2B15 is a novel ERα/AP-1-regulated gene. Interestingly, UGT2B15 encodes a UDP-glucuronosyltransferase enzyme involved in the glucuronidation of steroids and xenobiotics and may play a role in reducing estrogen and androgen concentrations, thereby reducing their signaling in breast cancer cells (22).
Relationships between Pol II, ERα, acetylated histones, and SRC at E2-regulated promoters.
In addition to Pol II and ERα, E2-dependent transcriptional regulation involves the recruitment of a variety of histone-modifying enzymes to target promoters (27). One major group of histone-modifying enzymes is the histone acetyltransferases (HATs; e.g., p300, CBP, and PCAF [p300/CBP-associated factor]), which establish specific patterns of lysine acetylation on the amino-terminal tails of nucleosomal histones (19, 27, 39). Generally, histone acetylation at promoters correlates with a more open chromatin structure and increased transcriptional output (12). In many cases, the recruitment of HATs by liganded ERα is thought to involve intermediary coactivators, such as the SRC proteins (SRCs 1, 2, and 3, collectively referred to herein as “SRC”), which bind simultaneously to both the liganded receptor and HATs, thus directing the HATs to ERα-bound genomic sites (25, 32).
To examine potential relationships between Pol II, ERα, acetylated histones, and SRC, we performed additional ChIP-chip analyses for SRC (using a panspecific anti-SRC antibody) and acetylated H3 and H4 (using anti-acetylated H3 K9/14 and anti-acetylated H4 K5/8/12/16 antibodies, respectively) with our custom promoter array. Then, using Spearman rank correlation analysis, we searched for possible correlations between pairwise combinations of Pol II, ERα, AcH, and SRC. The strongest and most significant correlations that we observed were between Pol II and AcH (correlation coefficient = 0.72983 [P < 0.0001]) and between ERα and SRC (correlation coefficient = 0.77549 [P < 0.0001]) (Table 2). These results are described in more detail below.
TABLE 2.
Correlation analysis of factor binding and RNA expression upon E2 treatment
| Gene | Correlation coefficient (P value) for indicated pairwise combinationa:
|
||||
|---|---|---|---|---|---|
| ERα | Pol II | SRC | AcH | RNA | |
| ERα | 1 | 0.40922 (<0.0001) | 0.77549 (<0.0001) | 0.34251 (0.0305) | 0.2055 (0.0487) |
| Pol II | 1 | 0.23582 (0.0494) | 0.72983 (<0.0001) | 0.4546 (<0.0001) | |
| SRC | 1 | 0.42857 (0.0257) | −0.0244 (0.8805) | ||
| AcH | 1 | 0.4458 (0.0001) | |||
Spearman's rank correlation coefficients and P values are listed for all possible pairwise combinations of factor binding/histone modification upon E2 treatment. The strongest correlations are shown in bold.
Pol II occupancy correlates with histone acetylation at the promoters of E2-regulated genes.
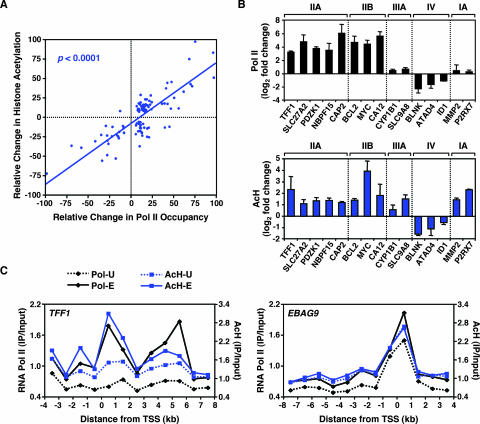
In our ChIP-chip analyses, we identified a significant correlation between changes in Pol II occupancy and AcH levels at promoters upon E2 treatment (Table 2). The recruitment of Pol II correlated with increased levels of AcH, and the release of Pol II correlated with decreased levels of AcH (Fig. 4A). These results were confirmed for more than 25 promoters by ChIP-qPCR (Fig. 4B; also see Fig. S4 at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). In addition, the correspondence between Pol II occupancy and AcH levels at promoters was clearly evident in ChIP-chip tiling assays with the TFF1, EBAG9, c-Myc, and CASP7 genes (Fig. 4C; also see Fig. S1B and E at the URL noted above). For all four genes, the AcH levels were elevated in the proximal promoter region and increased upon E2 treatment in a pattern very similar to the pattern of Pol II occupancy. The correspondence between Pol II occupancy and AcH levels was also evident for promoters apparently regulated by a distal ERα-binding enhancer (Table 1). Collectively, our results indicate that the levels of AcH at the promoters of E2-regulated genes correlate with Pol II occupancy and that histone acetylation is likely to play a role in regulating the expression of E2 target genes across the genome. Furthermore, as described below, our results indicate that Pol II and AcH are good markers for active promoters.
FIG. 4.
RNA Pol II occupancy correlates with histone acetylation at the promoters of E2-regulated genes. (A) Correlation between E2-dependent RNA Pol II recruitment and histone H3 and H4 acetylation. The relative change in histone acetylation or Pol II occupancy is defined as the log2 change (n-fold) upon E2 treatment as a percentage of the maximum log2 change (n-fold) observed for each factor. The correlation coefficient is 0.73 (P < 0.0001). All array elements with significant ChIP enrichment ratios (P < 0.05) were included in the analysis. (B) Validation by ChIP-qPCR of the ChIP-chip results for selected promoters from the estrogen-regulated promoter array. Results for RNA Pol II (upper graph; black) and AcH (lower graph; blue) are shown. The class IIA, IIB, IIIA, IV, and IA promoters are as shown in Fig. 2C. Each bar represents the mean + SEM for at least three separate determinations. (C) ChIP-chip tiling for the TFF1 and EBAG9 genes. Occupancy, expressed as ChIP enrichment ratios (IP/input), for RNA Pol II and AcH throughout the indicated genomic region is shown in the absence (-U) or presence (-E) of E2. “0” represents the TSS; regions upstream of the TSS are indicated by negative numbers.
ERα binding correlates with SRC recruitment at the promoters of E2-stimulated genes.
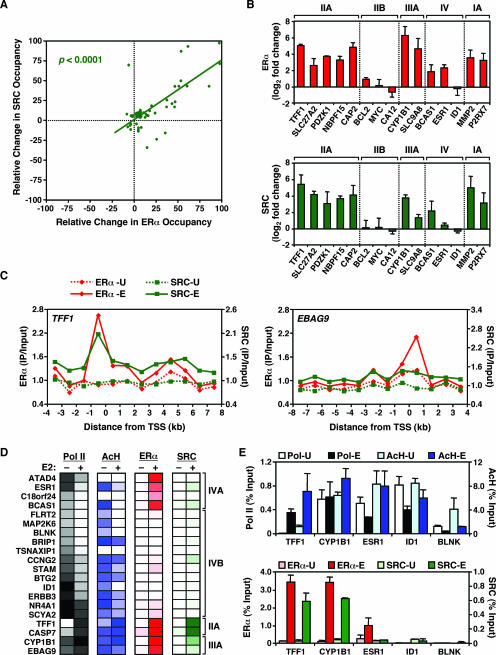
In our ChIP-chip analyses, we also identified a significant correlation between ERα binding and SRC recruitment at the promoters of E2-stimulated genes upon E2 treatment (Table 2). Although SRC was not located at all ERα-bound promoters, our results suggest that SRC binding is dependent on ERα binding, since the vast majority of the SRC-bound promoters were also bound by ERα (Fig. 5A and D; also see Fig. S5 at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Again, these results were confirmed for more than 25 promoters by ChIP-qPCR (Fig. 5B; also data not shown). Interestingly, although some of the SRC-recruiting promoters that we identified have been reported previously in gene-specific assays (e.g., the TFF1, CAP2, and CYP1B1 promoters [30]), we also identified and confirmed a number of novel SRC target promoters (e.g., SLC27A2, MMP2, PRUNE, and SERPINA1) where the recruitment of SRC correlated with the recruitment of ERα. A similar correspondence between ERα binding and SRC recruitment was also evident in ChIP-chip tiling assays with the TFF1, EBAG9, c-Myc, and CASP7 genes (Fig. 5C; also see Fig. S1C and F at the URL noted above) and in ChIP-chip assays with distal enhancers (Table 1), in that SRC binding was observed only in regions where ERα was bound.
FIG. 5.
ERα recruitment correlates with SRC recruitment to the promoters of E2-stimulated, but not E2-repressed, genes. (A) Correlation between E2-dependent ERα recruitment and SRC recruitment to gene promoters. The relative change in factor occupancy is defined as the log2 change (n-fold) in occupancy upon E2 treatment as a percentage of the maximum log2 change (n-fold) observed for each factor. The correlation coefficient is 0.78 (P < 0.0001). All array elements with significant ChIP enrichment ratios (P < 0.05) were included in the analysis. (B) Validation by ChIP-qPCR of the ChIP-chip results for selected promoters from the estrogen-regulated promoter array. Results for ERα (upper graph; red) and SRC (lower graph; green) are shown. The class IIA, IIB, IIIA, IV, and IA promoters are as shown in Fig. 2C. Each bar represents the mean + SEM for at least three separate determinations. (C) ChIP-chip tiling for the TFF1 and EBAG9 genes. Occupancy, expressed as ChIP enrichment ratios (IP/input), for ERα and SRC throughout the indicated genomic region is shown in the absence (-U) or presence (-E) of E2. “0” represents the TSS; regions upstream of the TSS are indicated by negative numbers. (D) ChIP-chip data showing Pol II, ERα, and SRC occupancy, as well as AcH levels, for a set of E2-repressed promoters before and after E2 treatment. Data for all class IV promoters, as well as for some class IIA and class IIIA promoters, are shown. The intensity of the color in each group indicates the strength of ChIP-chip signal, with white indicating no signal. (E) Validation by ChIP-qPCR of the ChIP-chip results for selected promoters from panel D. The light- and dark-colored bars for each factor indicate ChIP enrichment for untreated (-U) and E2-treated (-E) cells, respectively. Each bar represents the mean + SEM for at least three separate determinations.
Interestingly, SRC binding was not detected at the promoters of E2-repressed genes (i.e., class IV, reduced Pol II upon E2 treatment) irrespective of ERα binding status (Fig. 5D). These results were confirmed for a subset of the class IV promoters (Fig. 5E; see ESR1, ID1, and BLNK). In addition to the correlation between SRC and ERα at the promoters of E2-stimulated genes, a significant, but weaker, correlation between SRC occupancy and AcH levels was also observed (Table 2; correlation coefficient = 0.42857 [P = 0.0257]). For more than half of the promoters that we screened by ChIP-qPCR, E2-regulated SRC occupancy corresponded with AcH levels (Fig. 4B, bottom, and 5B, bottom; also data not shown). These results are consistent with the known role of SRC in recruiting HATs to target genes (25, 32). Collectively, our results suggest that although the E2-dependent recruitment of SRCs to activated promoters requires ERα, promoter-bound liganded ERαs are not capable of recruiting SRCs at E2-repressed genes in the same cells. Future studies will investigate the mechanisms determining whether or not a promoter-bound liganded ERα can recruit SRCs.
Relationship of RNA expression to factor occupancy and histone modification at E2-regulated promoters.
To evaluate the roles of factor recruitment and histone acetylation in the transcription of E2 target genes, we performed a gene expression microarray analysis in MCF-7 cells in the presence or absence of E2 treatment using Affymetrix U133A 2.0 microarrays (representing ∼14,500 well-characterized human genes). A short treatment of 3 h was used to enrich for genes directly regulated by E2 and not through secondary effects. The cell growth and treatment conditions were similar to those used for the ChIP-chip analyses. By use of stringent selection criteria (P < 0.05; regulation [n-fold], ≥2), 122 genes were identified as stimulated by E2 and 95 genes were identified as inhibited by E2 (Fig. 6A).
To determine if the regulation of gene expression by E2 was associated with factor occupancy or histone acetylation at promoters, we performed correlation analysis with the data from our expression microarray and ChIP-chip experiments (Table 2). The strongest and most significant correlations with RNA expression were Pol II recruitment (correlation coefficient = 0.45 [P < 0.0001]) and AcH levels (correlation coefficient = 0.44 [P < 0.0001]) (Fig. 6B; also see Fig. S6A at http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). These correlations were even more pronounced for genes in classes I through IV when gene expression was measured by qPCR (see Fig. S6B at the URL noted above). The relationships between Pol II occupancy, AcH levels, and gene expression held for genes apparently controlled by distal EREs (Table 1). Interestingly, although a weaker, but significant, correlation between ERα recruitment and gene expression was observed (correlation coefficient = 0.21 [P = 0.0487]), no significant correlation between SRC recruitment and mRNA expression was observed (Table 2). This latter result indicates that the recruitment of SRC alone is not sufficient to predict the mode of gene regulation across the set of E2-regulated genes.
To validate the expression microarray results and further analyze the expression of genes where Pol II and/or ERα binding was observed (i.e., classes I through IV), we measured RNA expression in MCF-7 cells by reverse transcription-qPCR after 1 or 3 h of treatment with E2 (Fig. 6C; also see Fig. S6C at the URL noted above). As expected, the genes in classes IIA and IIB (i.e., genes showing Pol II recruitment to their promoters upon E2 treatment) were stimulated by E2, whereas genes in class IV (i.e., genes showing dismissal of Pol II from their promoters upon E2) were inhibited by E2. Gene regulation after a 3-h E2 treatment correlated more strongly with Pol II recruitment and AcH levels than did gene regulation after a 1-h E2 treatment (see also Fig. S6C at the URL noted above). Interestingly, all genes in class IA (i.e., genes showing ERα recruitment to promoters upon E2, but no Pol II binding) were regulated by E2 (Fig. 6C). As noted above, for these genes, the detection of Pol II may fall below the limit of sensitivity of our ChIP-chip assay or, alternatively, Pol II may not be detected at the promoters due to rapid movement into the body of the gene. Finally, genes in class IIIA (i.e., genes showing ERα recruitment to promoters upon E2, and constitutively high occupancy by Pol II) were also regulated by E2 (Fig. 6C), suggesting that ERα regulates the transcriptional activity of constitutively bound Pol II at these promoters.
E2-dependent regulation of constitutively bound Pol II at target promoters.
To examine how E2 signaling may be regulating the activity of constitutively bound Pol IIs that are present at target promoters prior to E2 induction, we assayed by ChIP-qPCR for the presence of ERα, Pol II, and phospho-Pol II (i.e., phospho-Ser2 in the heptapeptide repeat of the Pol II C-terminal domain). We focused on two genes: the class IIA TFF1 gene, which does not have a constitutively bound Pol II, and the class IIIA CYP1B1 gene, which has a constitutively bound Pol II. Specifically, we assayed ∼200 bp upstream and downstream of the TSS as well as more than ∼1,400 bp downstream of the TSS. As expected, E2-dependent recruitment of ERα was observed for the upstream regions of both the TFF1 gene and CYP1B1 (Fig. 7A and B, top panels; −235 and −216 regions, respectively). The TFF1 gene showed little or no Pol II at the promoter prior to E2 treatment but did show recruitment of Pol II to the promoter, phosphorylation of Ser2, and movement of Pol II into the body of the gene after E2 treatment (Fig. 7A, middle two panels). In contrast, for CYP1B1, Pol II was present at the promoter prior to E2 treatment but was in a predominantly hypophosphorylated form (Fig. 7B, middle two panels; −216). After E2 treatment, Pol II was phosphorylated at Ser2 and moved into the body of the gene (Fig. 7B, middle two panels; +214 and +1412). The Pol II and Pol II phospho-Ser2 ChIP signatures that we observed with CYP1B1 are consistent with the presence of a paused Pol II at the promoter (6, 44). In fact, nuclear run-on experiments, which measure the density of Pol II at specific regions of a gene (34), confirmed the presence of a previously unidentified paused Pol II in the CYP1B1 promoter region (L. J. Core, M. Kininis, J. T. Lis, and W. L. Kraus, unpublished data).
To explore the E2-dependent regulation of the CYP1B1 promoter-proximally paused Pol II in more detail, we compared the localizations of NELF at the CYP1B1 and TFF1 promoters before and after E2 treatment, as monitored by ChIP for the NELFA subunit. NELF is a coregulatory complex involved in the establishment of paused Pol IIs at promoters (44). Interestingly, in the absence of E2 treatment, we observed strong binding of NELFA to the CYP1B1 promoter (i.e., the −216 probe) but not the TFF1 promoter, suggesting a role for the NELF complex in establishing the paused Pol II at the CYP1B1 promoter (Fig. 7, bottom panels). After treatment with E2, we observed a redistribution of NELFA, resulting in a reduction of NELFA levels at the CYP1B1 promoter and an increase immediately downstream of the TSS (Fig. 7B, bottom panel). This redistributed pattern of NELFA is consistent with the known roles of NELF in regulating the Pol II release from the promoter-proximal pause and in controlling the magnitude of a rapid transcriptional response (2, 44). Together, these results suggest that liganded ERα directs the release from pausing at the CYP1B1 promoter by triggering a redistribution of NELF.
Collectively, our results indicate that the CYP1B1 promoter contains a constitutively bound promoter-proximally paused Pol II in conjunction with NELF. Upon E2 treatment, the Pol II becomes phosphorylated and is released from the promoter to transcribe through the CYP1B1 gene. Using a similar approach based on our ChIP-chip data, we identified a number of other E2-regulated genes, including the c-Myc and c-Fos genes, which are also likely to have promoter-proximally paused Pol IIs (data not shown). To our knowledge, this is the first report of E2 signaling regulating the elongation activity, and not the recruitment, of Pol II at target genes. The presence of paused Pol IIs at E2-regulated promoters may allow for rapid transcriptional responses upon E2 treatment.
DISCUSSION
In the studies described herein, we used ChIP-chip to examine the E2-regulated localization of Pol II, ERα, SRC, and AcH at a set of target promoters that control the expression of more than half of the early E2-regulated transcriptome in MCF-7 cells. We defined a set of 58 direct E2 target genes based on E2-dependent changes in Pol II promoter occupancy and gene expression (i.e., class II, IV, and IIIA promoters) (see http://mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm), as opposed to inferences from bioinformatics and statistical analyses or correlations with RNA expression alone. As noted above, many of these direct E2 target genes exhibit interesting modes of regulation and biological activities, some of which may be relevant to onset and proliferation of breast cancers (e.g., UGT2B15, CYP1B1, and PRUNE; references 17, 22, and 47).
Our ChIP-chip experiments extend beyond previous ERα genomic localization experiments by (i) examining the ligand-dependent localization of a non-DNA-binding nuclear receptor cofactor (i.e., SRC), (ii) examining the genomic location of four different types of factors simultaneously (i.e., ERα [a DNA-binding factor], SRC [a non-DNA-binding coregulator], histone acetylation [a histone mark], and Pol II), (iii) determining the patterns of localization in both the presence and the absence of E2, and (iv) correlating genomic localization with functional output, namely, Pol II recruitment and transcription from target promoters. Our results have revealed global features of E2-regulated gene expression and allowed us to classify the estrogen-regulated promoters in MCF-7 cells based on factor binding, histone modification, and transcriptional output. Furthermore, the examination of both E2-treated and untreated conditions has revealed novel aspects of ligand-regulated gene expression by ERα and provided new mechanistic insights. Finally, our bioinformatic analyses coupled with gene-specific ChIP-qPCR have allowed us to identify novel genes likely regulated by ERα through AP-1 and link our results to the biology of estrogen signaling through the ERα/AP-1 tethering pathway (Fig. 3D and E). A growing body of published genomic analyses indicates that groups of genes share a limited number of general regulatory mechanisms that control their expression (4, 8, 26). Our studies simultaneously examining the genomic localization of four different factors have allowed us to reveal these common patterns and explore the regulatory mechanisms that they represent.
Combinatorial ChIP-chip analyses reveal global features of E2-regulated gene expression.
Our ChIP-chip analyses have revealed some previously unrecognized global features of E2-regulated gene expression, which are likely to extend to steroid hormone-regulated gene expression in general. First, we found a strong positive correlation between occupancy by Pol II and the levels of AcH (Table 2 and Fig. 4). This finding fits well with the observation that acetylation of H3 and H4 is generally associated with a more open, transcriptionally permissive chromatin structure (12) and has been implicated in gene regulation by many different nuclear receptors (19, 27). Second, we found a strong positive correlation between the E2-dependent recruitment of ERα and the recruitment of SRC at the promoters of E2-stimulated, but not E2-repressed, genes (Table 2 and Fig. 5A, B, and C). SRC proteins bind directly to liganded ERα and play an important role in recruiting histone-modifying coactivators, such as p300, CBP, PCAF, CARM1 (coactivator-associated arginine methyltransferase 1), and PRMT (protein arginine N-methyltransferase) (19, 32). In this regard, we also observed a significant positive correlation between SRC occupancy and AcH levels, consistent with the known role of SRC in recruiting HATs to target promoters (19, 32). Note, however, that the recruitment of SRC alone is not sufficient to predict the mode of gene regulation across the set of E2-regulated genes. Third, we found that for the set of ERα-binding promoters, the presence of an AP-1 site was more likely in the absence of an ERE sequence (Fig. 3B), a result consistent with recent bioinformatic and ChIP-chip analyses (10). ERα action through DNA-bound AP-1 (i.e., Fos/Jun heterodimers in the “tethering” pathway [28]) represents an alternate mechanism for the E2-dependent regulation of ERE-less promoters. Finally, as discussed in more detail in the next section, we found that many direct E2 target genes contain ERα-binding sites within their proximal promoter regions.
Many direct E2 target genes contain ERα-binding sites within their proximal promoter regions.
A recent study using genome-wide ERα and Pol II ChIP-chip in E2-treated MCF-7 cells, as well as microarray analyses of E2-dependent gene expression, concluded that only a small percentage of E2-regulated genes are regulated by ERα binding to the promoter-proximal region, with the rest being regulated by ERα binding at distal enhancers (10). This study, however, did not include an untreated (i.e., −E2) condition, so the role of E2 in regulating Pol II binding could not be assessed directly. A more direct approach to study E2-regulated transcription is to limit the analysis to confirmed direct E2 target genes (i.e., genes showing E2-dependent changes in Pol II promoter occupancy and gene expression), such as those in classes II, IV, and IIIA in our analyses. In this regard, we found that about 36 percent of these direct E2 target genes (21 out of 58) show ERα binding within a 1-kb fragment encompassing the proximal promoter (Fig. 1C and 2C and associated text; also see the URL noted above). Thus, a considerably higher percentage of direct E2 target genes may contain promoter-proximal ERα-binding sites than previously suggested. In this regard, a recent study using sensitive ChIP DNA selection and ligation technology identified a fraction of ERα-binding promoters considerably higher than that identified in previous studies (29). Interestingly, about half of the promoter-proximal ERα-binding sites that we identified contain an ERE-like sequence, whereas the other half contain a variety of other potential factor binding elements, including AP1-binding sites and half EREs (Fig. 3B, C, and D and associated text). The role of ERα binding to distal enhancers has yet to be determined, although results with estrogen and androgen receptor suggest that proximal and distal enhancers may collaborate though a looping mechanism (9, 48).
ChIP-chip analyses with both untreated and E2-treated cells reveal new mechanistic insights into E2-regulated gene expression.
Previous ERα ChIP-chip analyses with MCF-7 cells examined the genomic localization of ERα and Pol II in the presence, but not the absence, of E2 (9, 10, 29, 31). In our ChIP-chip experiments, we examined both the untreated and E2-treated conditions during the genomic discovery phase, which allowed us to examine on a global scale the various mechanisms controlling E2-regulated transcription. Specifically, comparisons between the untreated and E2-treated conditions in MCF-7 cells allowed us to identify the following: (i) a set of promoters constitutively bound by ERα (Fig. 2A), (ii) a set of E2-repressed genes with low occupancy by SRC (Fig. 5D and E), and (iii) a set of E2-regulated genes with constitutively bound and possibly promoter-proximally paused Pol IIs (Fig. 7A and B). Thus, our genomic analyses have yielded new information about E2-dependent gene-specific regulatory mechanisms and allowed us to classify E2-regulated genes based on these modes of regulation.
The class IV genes from our analysis represent a set of direct E2-repressed target genes that were revealed only by comparing the −E2 and +E2 conditions. For these genes, Pol II leaves the promoter and expression is reduced upon E2 treatment (Fig. 5D and E and 6C). Although E2 acting through ERα has historically been associated with the stimulation of gene transcription, recent microarray expression studies have indicated that more than half of the E2-regulated transcriptome may be repressed in response to E2, depending on the cell type and length of treatment (18, 33, 38). Our results have provided new insights into the underlying mechanisms of repression, namely, the absence of SRC binding at E2-repressed genes. As noted above, SRC proteins play an important role in recruiting the histone-modifying enzymes that help to promote the formation of more open, transcriptionally permissive chromatin (19, 32). In the absence of SRC recruitment, one would expect the chromatin to remain closed and transcriptionally repressed. In this regard, a recent detailed analysis of CCNG2, a class IV gene in our studies, showed that E2-dependent repression of the gene is associated with the recruitment of the nuclear receptor corepressor NCoR and the histone deacetylase 1 (46). The lack of SRC recruitment to the class IV genes is striking and suggests that E2-dependent SRC recruitment is not compatible with transcriptional repression. The means by which E2-bound ERα fails to bind and recruit SRC to specific repressed genes, while at the same time actively recruiting SRC to stimulated genes in other regions of the genome, is an open question that will be addressed in future studies.
Comparison of the −E2 and +E2 conditions also revealed a set of genes that are likely to have promoter-proximally paused Pol IIs at their promoters in the absence of E2 treatment. These include promoters with elevated levels of Pol II prior to E2 treatment that either stay the same (e.g., class III) or increase (e.g., class II) upon E2 treatment (Fig. 1C). CYP1B1 is one promoter that shows some of the hallmarks of a paused Pol II, namely, elevated Pol II and NELF levels at the promoter before signal induction and an increase in Pol II Ser2 phosphorylation upon release from pausing after signal induction. The establishment of a paused Pol II at a target promoter involves a variety of factors, including NELF, a multisubunit factor (44). Interestingly, a recent study has shown that COBRA1 (cofactor of BRCA1 [breast cancer-associated protein 1]), an integral subunit of NELF, interacts with ERα and modulates the transcription of E2 target genes (2). The activity of COBRA1 with ERα at a promoter containing a paused Pol II, however, has not yet been examined. Although a small number of signal-regulated promoters with paused Pol IIs in mammalian cells have been identified and characterized, this mechanism of transcriptional regulation has not previously been associated with E2-regulated genes. The presence of a paused Pol II may provide a means by which cells can initiate a rapid transcriptional response to E2. In the case of CYP1B1, which encodes a cytochrome P450 monooxygenase that metabolizes E2 and has been implicated in breast and endometrial carcinogenesis (47), a paused Pol II may be part of a rapid metabolic feedback response in E2 target tissues. Future studies will determine which factors are involved in the activation of promoter paused Pol IIs by E2 signaling and the biological significance of this mode of regulation.
Acknowledgments
We acknowledge and thank the following: John Lis and members of the Kraus and Lis labs for critical reading of the manuscript; Rick Young, Thomas Volkert, and the Whitehead Institute Center for Microarray Technology for providing the primer sequences used to produce the promoter array; Brian Dynlacht, Duncan Odom, Jonna Frasor, Joshua Stender, Leighton Core, and members of the Kraus lab for technical advice and helpful discussions; Wei Wang, David Lin, and Jie Zhao from the Cornell University Microarray Core Facility for advice and technical support for the ChIP-chip and expression microarray experiments; and Benita Katzenellenbogen for providing MCF-7 cells.
This work was supported by grants from the NIH/NIDDK (DK058110) and the Cornell Center of Vertebrate Genomics to W.L.K., predoctoral fellowships from the Department of Defense Breast Cancer Research Program to M.K. (BC050755) and G.D.I. (BC050806), and a postdoctoral fellowship from the New York State Health Research Science Board to T.Z. (C020912).
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Acevedo, M. L., K. C. Lee, J. D. Stender, B. S. Katzenellenbogen, and W. L. Kraus. 2004. Selective recognition of distinct classes of coactivators by a ligand-inducible activation domain. Mol. Cell 13:725-738. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar, S. E., J. L. Sun, A. L. Blair, C. A. Moskaluk, Y. Z. Lu, Q. N. Ye, Y. Yamaguchi, A. Mukherjee, D. M. Ren, H. Handa, and R. Li. 2004. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 18:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrau, J. C., L. van de Pasch, P. Lijnzaad, T. Bijma, M. G. Koerkamp, J. van de Peppel, M. Werner, and F. C. Holstege. 2006. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22:179-192. [DOI] [PubMed] [Google Scholar]
- 4.Blais, A., and B. D. Dynlacht. 2005. Constructing transcriptional regulatory networks. Genes Dev. 19:1499-1511. [DOI] [PubMed] [Google Scholar]
- 5.Bodescot, M., and O. Brison. 1996. Characterization of new human c-myc mRNA species produced by alternative splicing. Gene 174:115-120. [DOI] [PubMed] [Google Scholar]
- 6.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23:7628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83:349-360. [DOI] [PubMed] [Google Scholar]
- 8.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, A. S., V. X. Jin, M. Fan, L. T. Smith, S. Liyanarachchi, P. S. Yan, Y. W. Leu, M. W. Chan, C. Plass, K. P. Nephew, R. V. Davuluri, and T. H. Huang. 2006. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell 21:393-404. [DOI] [PubMed] [Google Scholar]
- 12.Clayton, A. L., C. A. Hazzalin, and L. C. Mahadevan. 2006. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23:289-296. [DOI] [PubMed] [Google Scholar]
- 13.Couse, J. F., and K. S. Korach. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev. 20:358-417. [DOI] [PubMed] [Google Scholar]
- 14.Deroo, B. J., and K. S. Korach. 2006. Estrogen receptors and human disease. J. Clin. Investig. 116:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20:2513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forus, A., A. D'Angelo, J. Henriksen, G. Merla, G. M. Maelandsmo, V. A. Florenes, S. Olivieri, B. Bjerkehagen, L. A. Meza-Zepeda, F. del Vecchio Blanco, C. Muller, F. Sanvito, J. Kononen, J. M. Nesland, O. Fodstad, A. Reymond, O. P. Kallioniemi, G. Arrigoni, A. Ballabio, O. Myklebost, and M. Zollo. 2001. Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas—a possible mechanism for altering the nm23-H1 activity. Oncogene 20:6881-6890. [DOI] [PubMed] [Google Scholar]
- 18.Frasor, J., J. M. Danes, B. Komm, K. C. Chang, C. R. Lyttle, and B. S. Katzenellenbogen. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562-4574. [DOI] [PubMed] [Google Scholar]
- 19.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 20.Gromak, N., S. West, and N. J. Proudfoot. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 26:3986-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guenther, M. G., R. G. Jenner, B. Chevalier, T. Nakamura, C. M. Croce, E. Canaani, and R. A. Young. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. USA 102:8603-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington, W. R., S. Sengupta, and B. S. Katzenellenbogen. 2006. Estrogen regulation of the glucuronidation enzyme UGT2B15 in estrogen receptor-positive breast cancer cells. Endocrinology 147:3843-3850. [DOI] [PubMed] [Google Scholar]
- 23.Jin, V. X., Y. W. Leu, S. Liyanarachchi, H. Sun, M. Fan, K. P. Nephew, T. H. Huang, and R. V. Davuluri. 2004. Identifying estrogen receptor alpha target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 32:6627-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kel, A., O. Kel-Margoulis, V. Babenko, and E. Wingender. 1999. Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J. Mol. Biol. 288:353-376. [DOI] [PubMed] [Google Scholar]
- 25.Kim, M. Y., S. J. Hsiao, and W. L. Kraus. 2001. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, W. L., and J. Wong. 2002. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur. J. Biochem. 269:2275-2283. [DOI] [PubMed] [Google Scholar]
- 28.Kushner, P. J., D. A. Agard, G. L. Greene, T. S. Scanlan, A. K. Shiau, R. M. Uht, and P. Webb. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74:311-317. [DOI] [PubMed] [Google Scholar]
- 29.Kwon, Y. S., I. Garcia-Bassets, K. R. Hutt, C. S. Cheng, M. Jin, D. Liu, C. Benner, D. Wang, Z. Ye, M. Bibikova, J. B. Fan, L. Duan, C. K. Glass, M. G. Rosenfeld, and X. D. Fu. 2007. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc. Natl. Acad. Sci. USA 104:4852-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labhart, P., S. Karmakar, E. M. Salicru, B. S. Egan, V. Alexiadis, B. W. O'Malley, and C. L. Smith. 2005. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc. Natl. Acad. Sci. USA 102:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguere. 2005. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 102:11651-11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leo, C., and J. D. Chen. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Levenson, A. S., K. M. Svoboda, K. M. Pease, S. A. Kaiser, B. Chen, L. A. Simons, B. D. Jovanovic, P. A. Dyck, and V. C. Jordan. 2002. Gene expression profiles with activation of the estrogen receptor alpha-selective estrogen receptor modulator complex in breast cancer cells expressing wild-type estrogen receptor. Cancer Res. 62:4419-4426. [PubMed] [Google Scholar]
- 34.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:347-356. [DOI] [PubMed] [Google Scholar]
- 35.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martini, P. G., and B. S. Katzenellenbogen. 2003. Modulation of estrogen receptor activity by selective coregulators. J. Steroid Biochem. Mol. Biol. 85:117-122. [DOI] [PubMed] [Google Scholar]
- 37.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 38.Monroe, D. G., B. J. Getz, S. A. Johnsen, B. L. Riggs, S. Khosla, and T. C. Spelsberg. 2003. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J. Cell. Biochem. 90:315-326. [DOI] [PubMed] [Google Scholar]
- 39.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson, S., S. Makela, E. Treuter, M. Tujague, J. Thomsen, G. Andersson, E. Enmark, K. Pettersson, M. Warner, and J. A. Gustafsson. 2001. Mechanisms of estrogen action. Physiol. Rev. 81:1535-1565. [DOI] [PubMed] [Google Scholar]
- 41.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Lone, R., M. C. Frith, E. K. Karlsson, and U. Hansen. 2004. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 18:1859-1875. [DOI] [PubMed] [Google Scholar]
- 43.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557-567. [DOI] [PubMed] [Google Scholar]
- 45.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 46.Stossi, F., V. S. Likhite, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2006. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J. Biol. Chem. 281:16272-16278. [DOI] [PubMed] [Google Scholar]
- 47.Tsuchiya, Y., M. Nakajima, and T. Yokoi. 2005. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 227:115-124. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19:631-642. [DOI] [PubMed] [Google Scholar]
- 49.White, J. H., I. Fernandes, S. Mader, and X. J. Yang. 2004. Corepressor recruitment by agonist-bound nuclear receptors. Vitam. Horm. 68:123-143. [DOI] [PubMed] [Google Scholar]
- 50.Wu, J., L. T. Smith, C. Plass, and T. H. Huang. 2006. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 66:6899-6902. [DOI] [PubMed] [Google Scholar]