Abstract
The virulence of Legionella pneumophila is dependent upon its capacity to acquire iron. To identify genes involved in expression of its siderophore, we screened a mutagenized population of L. pneumophila for strains that were no longer able to rescue the growth of a ferrous transport mutant. However, an unusual mutant was obtained that displayed a strong inhibitory effect on the feoB mutant. Due to an insertion in hmgA that encodes homogentisate 1,2-dioxygenase, the mutant secreted increased levels of pyomelanin, the L. pneumophila pigment that is derived from secreted homogentisic acid (HGA). Thus, we hypothesized that L. pneumophila-secreted HGA-melanin has intrinsic ferric reductase activity, converting Fe3+ to Fe2+, but that hyperpigmentation results in excessive reduction of iron that can, in the case of the feoB mutant, be inhibitory to growth. In support of this hypothesis, we demonstrated, for the first time, that wild-type L. pneumophila secretes ferric reductase activity. Moreover, whereas the hyperpigmented mutant had increased secreted activity, an lly mutant specifically impaired for pigment production lacked the activity. Compatible with the nature of HGA-melanins, the secreted ferric reductase activity was positively influenced by the amount of tyrosine in the growth medium, resistant to protease, acid precipitable, and heterogeneous in size. Together, these data represent the first demonstration of pyomelanin-mediated ferric reduction by a pathogenic bacterium.
Legionella pneumophila, an occupant of natural and human-made aquatic environments, is also the principal agent of Legionnaires' disease, a serious form of pneumonia that especially afflicts immunocompromised individuals (18, 37, 41). In water habitats, this gram-negative bacterium survives free, in biofilms, and as an intracellular parasite of protozoa (2, 62, 65); in the lung, it replicates in alveolar macrophages (67, 93, 95). Among the processes that promote L. pneumophila growth in both the environment and the mammalian host are Lsp type II protein secretion, Dot/Icm type IVB protein secretion, and the Lvh type IVA protein secretion system (8, 19, 27, 57, 84, 105). Other notable surface features of L. pneumophila include flagella, type IV pili, the major outer membrane protein porin, the Hsp60 chaperonin, and the Mip peptidylproline isomerase (9, 26, 39, 46, 49, 66, 88, 99, 108). In addition to exporting proteins and enzymes onto its surface or into the extracellular milieu and/or host cells, L. pneumophila also secretes a siderophore (legiobactin) that promotes iron uptake (3). Iron acquisition has long been regarded as a key aspect of L. pneumophila growth, intracellular infection, and virulence (12, 17, 82). Besides legiobactin, we have uncovered both FeoB ferrous iron transport, which is critical for bacterial growth in host cells and the lung, as well as the ccm, frgA, hbp, iraAB, and tat genes, which also promote L. pneumophila growth under low-iron conditions (20, 47, 68, 73, 81, 86, 87, 106, 107). The discovery of legiobactin has drawn further attention since, for many years, L. pneumophila was believed not to secrete siderophores (60, 83). Thus, we recently identified two genes (lbtAB) that promote the production and secretion of legiobactin and determined that siderophores are also expressed by other species of Legionella (3, 96).
Since bacteria generally have elaborate systems for siderophore production, secretion, and utilization (15, 43), we embarked upon a genetic screen to identify L. pneumophila genes besides lbtAB that are involved in legiobactin expression. In the course of this endeavor, we discovered that L. pneumophila secretes a ferric reductase activity. Interestingly, this activity was linked to the bacterium's pigment, a melanin that had previously been shown to be derived from homogentisic acid (HGA) and implicated in bacterial resistance to light (97). In L. pneumophila, the secretion of an iron-reducing pigment may contribute to an iron acquisition pathway that is complementary to the siderophore pathway.
MATERIALS AND METHODS
Bacterial strains.
L. pneumophila serogroup 1 strain 130b (American Type Culture Collection [ATCC] strain BAA-74, also known as strain AA100 or Wadsworth) served as the primary wild-type control, as it has in past iron acquisition studies (3, 32, 60, 86). Serogroup 1 strain JR32, a restriction-deficient, streptomycin-resistant derivative of wild-type Philadelphia 1, also served as a key positive control (113). Other wild-type L. pneumophila strains tested for pigment and ferric reductase included serogroup 4 strain Los Angeles 1 (ATCC 33156) and serogroup 5 strain Dallas 1E (ATCC 33216) (60). Mutants of strain 130b containing a kanamycin resistance cassette inserted into either feoB (NU269) or iraAB (NU216R) were previously described (86, 106), as was a mutant (JR32-1) containing a similar DNA insertion in the lly gene of strain JR32 (98, 113). Escherichia coli DH5α was used as the host for recombinant plasmids (Invitrogen, Carlsbad, CA).
Bacteriological media and growth experiments.
L. pneumophila strains were routinely cultured at 37°C on buffered charcoal yeast extract (BCYE) agar or in buffered yeast extract (BYE) broth (30). E. coli strains were routinely grown in LB medium (91). When appropriate, the following antibiotics were added to medium at the following final concentrations (in μg/ml): ampicillin, 100; gentamicin, 2.5 for L. pneumophila and 5 for E. coli; chloramphenicol, 3 for L. pneumophila and 30 for E. coli; kanamycin, 25 for L. pneumophila and 50 for E. coli. All chemicals, unless otherwise noted, were from Sigma (St. Louis, MO). In order to assess secreted activities, legionellae were also grown at 37°C in deferrated and nondeferrated chemically defined medium (CDM) as well as a variant of deferrated CDM that contained 250 mg/liter l-tyrosine (CDMP) (3). To monitor the extracellular growth of L. pneumophila strains, bacteria grown on BCYE agar were inoculated into BYE, CDM, or CDMP broth, and the optical density (OD) of the resulting cultures was determined at 660 nm using a DU720 spectrophotometer (Beckman, Fullerton, CA) (3). As an additional means to examine L. pneumophila growth under iron-limiting conditions, legionellae were tested, as before, for their ability to form colonies on BCYE gradient plates that contained either the ferric iron chelator deferroxamine mesylate at 20 or 60 μM or the ferrous iron chelator 2,2′-dipyridal at 300 or 600 μM (3, 81).
Construction and screening of an L. pneumophila mutant library.
Strain 130b bacteria were electroporated with mini-Tn10-containing pEH40, and transposon-containing transformants were selected on BCYE agar containing kanamycin (47, 68). The number and location of mini-Tn10 insertions within an L. pneumophila strain were determined by Southern hybridization and PCR analysis as previously described (68). Mutagenized legionellae were tested for their ability to stimulate the growth of a ferrous iron transport mutant, NU269, using our previously reported bioassay (3, 86). Briefly, isolated mutant colonies were streaked using a toothpick onto non-iron-supplemented BCYE agar onto which had been previously spread 105 CFU of NU269. Mutants that appeared defective were retested by spotting aliquots containing equivalent numbers of bacteria (in BYE) onto the NU269 indicator plate. As controls on each plate, we tested the stimulatory activity of the wild type and the negative activity associated with medium alone (3, 86).
DNA isolation, PCR, and DNA sequencing.
DNA was obtained from L. pneumophila as described previously (31), and plasmids were routinely isolated from E. coli using the Plasmid Mini Prep kit (Bio-Rad, Hercules, CA). All other DNA manipulations were performed using standard protocols (91). Oligonucleotide primers for sequencing or PCR were synthesized at Integrated DNA Technology (Coralville, IA). Standard PCR was performed using HIFI polymerase (Invitrogen). Inverse PCR was done as previously described (61). DNA samples for sequencing were labeled using the BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA), and sequencing was done at our Biotech Facility using an automated DNA sequencer (model 3100; Applied Biosystems). Sequences were analyzed with DNASTAR (DNASTAR, Inc., Madison, WI), and homology searches were done using BLAST programs at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/).
Complementation analysis.
In order to complement the lly and hmgA mutants, the Lly and HmgA coding sequences were amplified from 130b DNA by PCR using, respectively, the primer pairs Lly F (5′-TGATCCGAATGATCAGAGTGGA) and Lly R (5′ TGGCAGGTACATCACTGATT) and HmgA-F (5′-TTACGGAATTAGATCTTAGCCCAT) and HmgA-R (5′-ACTTTGCTCGAGCGGGTTG). The 1.4-kb and 1.5-kb fragments generated were ligated into pGemTEasy (Promega, Madison, WI). The resulting plasmid containing lly (i.e., pGEMlly) was then digested with Bcl1/Xho1, whereas the plasmid containing hmgA (i.e., pGEMhmgA) was digested with BamH1/Xho1 enzymes. The two different fragments containing Legionella DNA were then ligated into the complementation vector pMMB2002 (88) that had been digested with BamH1/Sal1. The final complementing plasmids were designated plly and phmgA and, in both cases, directional cloning placed each of the coding sequences under the control of the tac promoter from pMMB2002. Plasmids were ultimately introduced into L. pneumophila by electroporation, and their presence there was verified by PCR (21, 91).
Measurement of secreted pyomelanin and ferric reductase.
To monitor the presence of secreted pigment, bacteria were inoculated into 30 ml of BYE, CDM, or CDMP and then, after various periods of incubation, filter-sterilized culture supernatants were tested for their absorbance at 400 nm, as originally described for L. pneumophila (11, 85). Although some Legionella studies have monitored pigment production using 550 nm (5, 66, 112, 116), we found that the difference in absorbance between wild-type supernatants and pigment null mutant supernatants is more easily seen at the lower wavelength (see below). Additionally, studies examining similarly colored pigments in other bacteria routinely use wavelengths at or just below 400 nm (58, 90, 102, 115).
To assess ferric reductase activity, bacteria were inoculated into 30 ml of BYE, CDM, or CDMP and then, after various periods of incubation, filter-sterilized culture supernatants were tested in the ferrozine assay, as has been done for the study of proteins and pigments from a variety of bacteria and fungi (28, 50, 52, 53, 69, 72, 102). The assay solution typically consisted of 180 μl of supernatant, 25 mM Tris-HCl (pH 7.5), 160 μM ferrozine (Acros Organics, Geel, Belgium), and 120 μM ferric nitrate, all within a final volume of 250 μl, placed into the wells of a 96-well plate. The reaction proceeded for 1 h at room temperature and then the color change associated with ferrozine binding to ferrous iron was measured at 562 nm. In order to quantify the amount of ferric reductase activity in our samples, a standard curve was generated using known concentrations of ferrozine complexed with ferrous sulfate. In some experiments, ferric nitrate was replaced by an equimolar amount of ferric chloride, ferric ammonium citrate, or ferric pyrophosphate. In others, the influence of cofactors was determined by adding NADH or NADPH to the reaction mixture at a final concentration of 1 μM (52). To judge the effect of protein degradation on activity, the samples were preincubated with 1.5 mg/ml proteinase K or 0.01% sodium dodecyl sulfate for 30 min at room temperature. Finally, ferric reduction was also monitored in an assay using 1 mM bathophenanthroline disulfonate (BPDS) in place of ferrozine (50, 72, 103). BPDS is another colorimetric dye which is detected at 520 nm when complexed with ferrous iron.
All quantitative data were subjected to statistical analysis using Student's t test. P values of <0.05 were considered significant. The results presented are representative of at least three replicate experiments in which triplicate cultures for each strain were examined and found to have comparable levels of variation.
Acid precipitation and size filtration of pyomelanin.
In order to further investigate the relationship between the L. pneumophila pigment and ferric reduction, we partially purified the pyomelanin by acid precipitation using previously described methods (102). Following bacterial growth in 700 ml of CDMP to late stationary phase, culture supernatants were collected by centrifugation and filter sterilization. Full polymerization of secreted HGA was then achieved by shaking the supernatant, in baffled flasks, for 24 h at 37°C. The pH of the sample was lowered to 2 by addition of 6 N HCl. After precipitation at 4°C for 16 h, pigment was collected by centrifugation at 5,000 × g for 1 h. The resultant pellet was suspended in 7 ml of 25 mM Tris-HCl (pH 7.5) buffer and neutralized to pH 7 with 5 M KOH. The partially purified material was spectrophotometrically assessed for the presence of pigment, and 10-μl aliquots were tested in the ferrozine assay, as described above. In order to estimate the size of secreted activities, 100 μl of the partially purified material was added to 900 μl of buffer and then passed through Centricon centrifugal filters (regenerated cellulose) having molecular mass cutoffs of 3, 10, 30, or 50 kDa as per the manufacturer's specifications (Millipore, Billerica, MA). The various filtrates and retentates were brought to a volume of 1 ml with buffer, and then 180-μl aliquots were assayed for pigment and ferric reductase activity.
RESULTS
A hyperpigmented mutant of L. pneumophila inhibits the growth of a ferrous transport mutant.
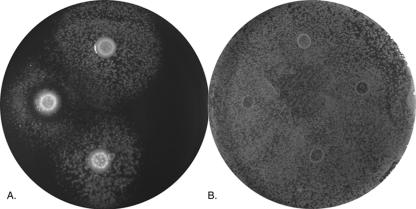
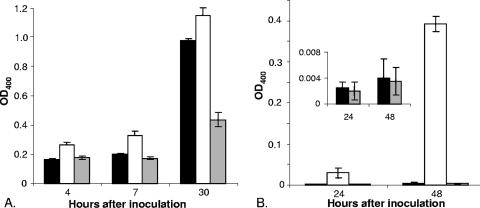
Previously, we observed that a ferrous transport (feoB) mutant of L. pneumophila strain 130b is defective for growth on BCYE agar that lacks its usual iron supplement (86). However, exposure to ferric iron, wild-type bacteria, or supernatants containing legiobactin restores the growth of the FeoB mutant (Fig. 1A, for growth stimulation by wild type) (3). Therefore, to identify L. pneumophila genes involved in the expression of legiobactin or other possible secreted Fe3+ chelators, we screened a mini-Tn10-mutagenized population of strain 130b for mutants that were unable to stimulate growth of the FeoB mutant on low-iron BCYE. In the course of examining the first ca. 800 mutants, a strain (NU326) was identified that produced an unusual zone of growth inhibition around itself (Fig. 1A). Since feoB mutant growth was seen beyond this ring of inhibition, we suspected that NU326 was still producing significant levels of legiobactin but overexpressing a higher molecular weight substance(s) that is inhibitory to the ferrous transport mutant. When it was grown in liquid medium, NU326 consistently displayed increased levels of brownish pigment in its culture supernatant (Fig. 2). This increase in pigment was most clearly manifested when strains were cultured in CDM, with a 100-fold increase in absorbance appearing after 48 h of incubation (Fig. 2B). The mutant did not, however, show a general alteration in growth in the liquid medium or on BCYE agar (data not shown).
FIG. 1.
Growth inhibition of a feoB mutant by a hyperpigmented mutant of L. pneumophila. Approximately 5 × 105 CFU of either an feoB mutant (A) or wild-type 130b (B) were spread onto BCYE agar lacking its standard ferric iron supplement and then aliquots of BYE medium containing either 5 × 103 CFU of wild-type 130b (top), hmgA mutant NU326 (left), or complemented NU326 (phmgA) (bottom) or no added bacteria (right) were spotted onto the plates. After 5 days of incubation at 37°C, the growth of the feoB mutant or strain 130b around the spots was recorded. The results are representative of at least three replicate experiments.
FIG. 2.
Levels of pigment in culture supernatants of wild-type and hmgA mutant L. pneumophila. Wild-type 130b (black bars), hmgA mutant NU326 (white bars), and complemented mutant NU326 (gray bars) were inoculated into standard BYE (A) or deferrated CDM (B) and then, after the indicated periods of incubation at 37°C, filter-sterilized, culture supernatants were assessed for pigment production as measured by their OD400 over that of medium controls. For ease of visualization, the inset in panel B repeats the data for the wild-type and complemented mutant strains with a smaller scale on the y axis. The data presented are the means and standard deviations from triplicate cultures for each strain. The results presented are representative of at least three replicate experiments in which triplicate cultures for each strain were examined and found to have a level of variation comparable to that presented here. When grown in CDM, the increases in pigment expression by the hmgA mutant NU326 were significant at all time points (P < 0.01). When grown in BYE, pigment expression by the mutant was consistently greater than that of wild type; however, the difference did not achieve the statistical significance seen with CDM cultures.
That L. pneumophila secretes a brown pigment was reported shortly after the discovery of the bacterium (6, 7, 35, 74, 78, 85, 104, 109). Early reports also showed that the production of the pigment is dependent upon l-tyrosine in the growth medium and is most apparent in bacteria experiencing slowed growth (6, 7, 11). It was later established that the pigment results from the spontaneous and oxidative polymerization of HGA, which is secreted into the supernatant (98). HGA, in turn, is made through the action of Lly, a p-hydroxyphenylpyruvate dioxygenase (40, 98, 113, 114). Thus, the pigment of L. pneumophila is a pyomelanin or HGA-melanin (71, 79, 90, 102, 110). To help determine if the altered pigmentation that we saw was due to changes in HGA-melanin, we identified the mutation in NU326. From inverse PCR and sequence analysis, the mini-Tn10 insertion in NU326 was mapped to a monocistronic open reading frame that is annotated in the L. pneumophila genome as homogentisate 1,2-dioxygenase (HmgA). In the sequenced strains Philadelphia, Paris, and Lens, hmgA is designated lpg1285, lpp1248, and lpl1248 (14, 16). HmgA, whether prokaryotic or eukaryotic, degrades HGA to 4-maleyl-acetoacetate and, therefore, in its absence HGA more readily oxidizes to benzoquinoneacetic acid, which then polymerizes to form pyomelanin (4, 33, 64, 92, 94). As an indication of its relatedness to like enzymes, L. pneumophila HmgA showed 65% similarity to Pseudomonas putida HmgA (4). When an intact plasmid copy of hmgA (i.e., phmgA) was introduced into NU326, there was a decrease in the OD400 of culture supernatants that reflected a level of pigment equal to wild type (Fig. 2). With this demonstration of complementation, we confirmed that the increased pigmentation of NU326 was due to increased levels of HGA-melanin in supernatants because of an inability to degrade HGA. Since NU326 (phmgA) stimulated the growth of the feoB mutant in a manner equivalent to wild type (Fig. 1A), we concluded that the growth inhibition by NU326 was also due to the loss of HmgA and its resultant increase in pigment production.
In contrast to its effect on the feoB mutant, the hmgA mutant did not inhibit the growth of wild-type 130b (Fig. 1B), a strain that can utilize either ferric or ferrous iron (86). Similarly, NU326 did not negatively influence the growth of lbtA and iraAB mutants of 130b (data not shown), strains that are only defective for ferric iron acquisition (3, 106). These data indicate that the inhibitory effect of NU326 is specific toward bacteria that lack Fe2+ transport and preferentially utilize Fe3+ as their iron source. Thus, it appeared that L. pneumophila hyperpigmentation inhibits ferrous transport mutants by removing available Fe3+ from the medium.
L. pneumophila supernatants contain ferric reductase activity.
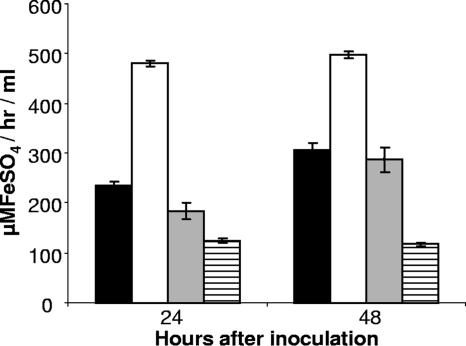
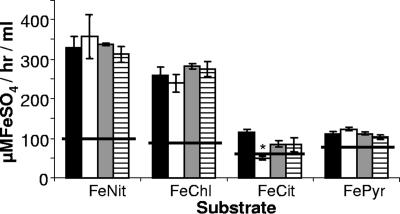
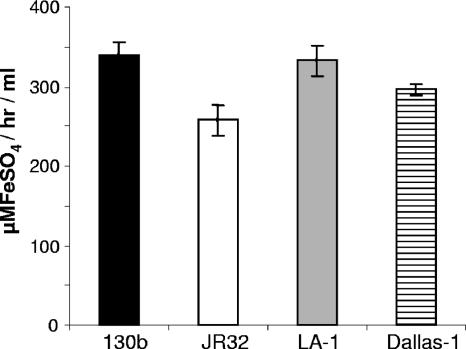
One hypothesis to explain our results would be that L. pneumophila HGA-melanin has intrinsic ferric reductase activity, converting Fe3+ to Fe2+, and that hyperpigmentation results in a rapid and/or uncontrolled reduction of iron that can, in some circumstances, be inhibitory to growth. As the first step toward addressing this hypothesis, wild-type 130b was grown in CDM, and then cell-free culture supernatants were tested for ferric reductase activity. Consistently, the supernatants displayed significant levels of ferric reductase activity as measured by the standard ferrozine assay (Fig. 3) or the standard BPDS assay (data not shown). Since the hmgA mutant, but not its complemented derivative, displayed a level of activity that was greater than that of parental 130b (Fig. 3), the newly found activity appeared to be due, at least in part, to HGA-melanin. In support of this hypothesis, the level of ferric reductase present in wild-type supernatants increased when the bacteria were grown in CDMP, a variant of CDM containing four times the usual amount of tyrosine; for example, in one of three representative experiments, CDMP cultures gave an activity of 296.4 ± 41.1 μMFeSO4/h/ml (mean ± standard deviation) versus the 174.6 ± 14.59 μMFeSO4/h/ml value obtained from parallel CDM cultures (P < 0.03). Previous reports indicated that L. pneumophila has both cytoplasmic and periplasmic ferric reductase activities (52, 53, 80). However, we concluded that these cell-associated enzymes contributed little to the activity that we were observing in culture supernatants, for three reasons. First, the supernatant readily reduced ferric nitrate and ferric chloride but had minimal activity against ferric citrate, a known substrate of the cellular enzymes (Fig. 4). Second, the supernatant activity was not increased by the inclusion of NADH or NADPH, molecules that are absolute cofactors for the cellular enzymes (Fig. 4). Third, the reduction of ferric nitrate and ferric chloride was not diminished when supernatants were treated with proteinase K, a protease that degrades the cellular enzymes (Fig. 4). Proteinase K treatment did lower activity against ferric citrate to the level of the medium control (Fig. 4), suggesting that there is a very small amount of cellular enzyme in the supernatants, which likely resulted from some spontaneous cell lysis. Since the reduction of ferric nitrate was not decreased by treatment of the supernatants with sodium dodecyl sulfate (data not shown), we further suspected that the secreted reductase activity was not due to any other protein enzyme. Taken together, these data indicate, for the first time, that L. pneumophila can secrete a ferric reductase activity. Since the activity was also observed in pigmented culture supernatants of serogroup 1 strain JR32, serogroup 4 strain ATCC 33156, and serogroup 5 strain ATCC 33216 (Fig. 5), we further concluded that secreted ferric reductase activity is common, if not conserved, within the L. pneumophila species.
FIG. 3.
Levels of ferric reductase activity in culture supernatants of wild-type and hmgA mutant L. pneumophila. Wild-type 130b (black bars), hmgA mutant NU326 (white bars), and complemented mutant NU326 (gray bars) were inoculated into deferrated CDM and then, after the indicated periods of incubation at 37°C, filter-sterilized, culture supernatants were assessed for the bulk reduction of ferric nitrate to Fe2+. The appearance of Fe2+:ferrozine was monitored at 562 nm and quantified by comparison to a standard curve of ferrous sulfate complexed with ferrozine. The background level of activity exhibited by the CDM is also depicted (hatched bars). The data presented are the means and standard deviations from triplicate cultures. The activities from all strains were significantly above the CDM control, and the activity exhibited by NU326 was significantly greater than that of the wild-type and complemented strain (P < 0.01).
FIG. 4.
Effects of substrates, cofactors, and protease treatment on ferric reductase activity. Following growth in CDMP, supernatants from wild-type 130b were tested for their ability to reduce ferric nitrate (FeNit), ferric chloride (FeChl), ferric ammonium citrate (FeCit), or ferric pyrophosphate (FePyr) (black bars). In parallel, the reduction reactions were carried out following either treatment of the supernatants with proteinase K (ProK) (white bars) or the addition of NADH (gray bars) or NADPH (hatched bars). The four horizontal lines depict the level of background activity exhibited by the medium-only negative control. The data presented are the means and standard deviations from triplicate cultures. The asterisk indicates a significant change in activity against ferric citrate following proteinase K treatment (P < 0.001).
FIG. 5.
Ferric reductase activity in culture supernatants of different strains of L. pneumophila. Strains 130b (black bar), JR32 (white bar), Los Angeles 1 (gray bar), and Dallas 1 (hatched bar) were grown in CDMP for 24 h and then filter-sterilized culture supernatants were tested for their ability to reduce ferric nitrate as described above. The horizontal line depicts the level of background activity exhibited by the medium control. The data are the means and standard deviations from triplicate cultures.
L. pneumophila HGA-melanin possesses ferric reductase activity.
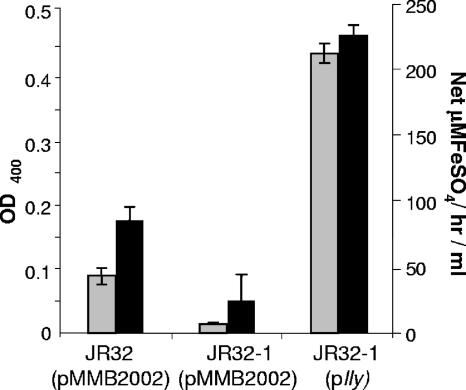
Since increases in the levels of ferric reductase activity in culture supernatants correlated with increased pigmentation, whether due to growth in tyrosine-supplemented medium or inactivation of hmgA, we suspected that the newly found activity is due to HGA-melanin. Thus, we obtained a mutant that is defective for pigment production and tested it in the ferrozine assay. JR32-1 is a previously described mutant of JR32 that contains an insertion in lly, the gene whose product degrades 4-hydroxyphenylpyruvate to yield HGA (97, 113). Thus, supernatants from JR32-1 lack both HGA and pyomelanin (98). As expected, JR32-1 did not exhibit pigmentation when it was grown in CDMP, and in support of our hypothesis, the lly mutant had significantly lowered levels of ferric reductase activity in its culture supernatants (Fig. 6). Both mutant phenotypes were complemented when the lly gene was expressed in trans (Fig. 6), confirming that lly and HGA promote the production of secreted ferric reductase activity.
FIG. 6.
Ferric reductase activity of a nonpigmented lly mutant L. pneumophila. Lly+ JR32 (pMMB2002), its Lly-negative derivative JR32-1 (pMMB2002), and a complemented JR32-1 (plly) were cultured in deferrated CDMP, and then filter-sterilized supernatants were assessed for pigmentation (gray bars) and ferric reductase activity (black bars) as described in the text. The background activities exhibited by the medium have been subtracted from the supernatant activities. The data are the means and standard deviations from triplicate cultures. The decreased activities exhibited by the lly mutant relative to both its parent and complemented derivative were significant (P < 0.01). Although the data presented for JR32 and JR32-1 derive from bacteria containing the cloning vector (pMMB2002) that was used to make plly, the same results were obtained when strains devoid of the vector were examined (data not shown).
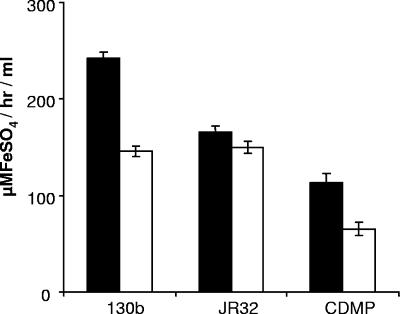
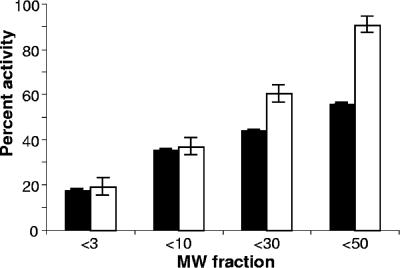
To further investigate the relationship between L. pneumophila HGA-melanin and ferric reductase activity, we partially purified pigment from strains 130b and JR32 using acid precipitation, as has been done for other microbial melanins (102), and then tested the resultant material in the ferrozine assay. For both strains, the acid-precipitated material retained most of the starting ferric reductase activity against ferric nitrate (Fig. 7). In an additional trial, the acid-precipitated material from strain 130b was also able to reduce ferric chloride to a comparable degree (data not shown). When the acid-precipitated material from strain 130b was subjected to size fractionation, we observed that both pigment absorbance and ferric reductase activity were present over a wide range (Fig. 8). Such a result is compatible with the heterogeneity that generally exists among microbial melanins (101, 111, 115). It also suggests that ferric reductase activity is not strictly associated with a particular size or form of HGA-melanin. It is also likely, though, that some of the ferric reductase activity observed in these various fractions is due to medium components that are also precipitated by the acid (Fig. 7). The strongest coincidence of pigment and ferric reductase activity occurred in the relatively low molecular mass range, e.g., the <3-kDa fraction contained 20% of both activities and the <10-kDa fraction had 40% of both (Fig. 8). In contrast, the <50-kDa fraction had 90% of the ferric reductase activity but only 55% of the pigment. Taken together, these data support the relationship between pigment and ferric reductase but suggest that not all forms of pigment, such as those fractionating as >50 kDa, retain ferric reductase activity.
FIG. 7.
Acid precipitation of the ferric reductase activity in culture supernatants of L. pneumophila. After culturing for 48 h in deferrated CDMP, a portion of the cell-free supernatants from wild-type strains 130b and JR32 was subjected to the acid precipitation protocol used to purify melanins. Untreated supernatants (black bars) and precipitated samples (white bars) were tested in the ferrozine assay using ferric nitrate as substrate. Also depicted is an uninoculated medium control that was similarly incubated and acid precipitated. The data presented are the means and standard deviations from triplicate cultures.
FIG. 8.
Size fractionation of pigment and ferric reductase activity of L. pneumophila. Aliquots of acid-precipitated material obtained from CDMP cultures of wild-type strain 130b were centrifuged through filter units having a molecular mass cutoff of 3, 10, 30, or 50 kDa, and then the pigmentation and ferric reductase activity in each filtrate were quantified as described in the text. The results are presented as the percentage of pigment (black bars) and ferric reductase activity (white bars) in each of the fractionated filtrates compared to the starting material. The data presented are the means and standard deviations from triplicate cultures.
Iron acquisition characteristics of L. pneumophila pigment mutants.
That wild-type L. pneumophila HGA-melanin provides ferric reductase activity suggests that the pigment might facilitate iron acquisition/assimilation by providing a ferrous iron source to growing bacteria and that a pigment mutant might therefore grow poorly under low-iron conditions. However, when the lly mutant was cultured in deferrated CDMP (above), it grew comparably to wild type. A similar result was obtained when the nonpigmented mutant was cultured on BCYE gradient plates containing various concentrations of iron chelators (data not shown). Conversely, the hyperpigmented hmgA mutant never exhibited increased growth in the low-iron medium compared to its wild type (data not shown). Although these various data indicate that HGA-melanin and secreted ferric reductase are not required for L. pneumophila growth under low-iron conditions, they do not prove that these factors are irrelevant to iron acquisition. For example, it is quite possible that an alternative iron acquisition system(s) compensates for the absence of HGA-mediated ferric reduction.
DISCUSSION
With the characterization of an hmgA mutant, we have confirmed that a complete HGA-melanin pathway is present and operational in L. pneumophila, i.e., tyrosine is converted to HGA via Lly, secreted HGA spontaneously polymerizes to HGA-melanin, and HGA is degraded by HmgA. The Legionella genus contains 51 species besides L. pneumophila, and 20 of these species have been shown to cause disease (34, 37, 56, 59, 76). Although not all Legionella species produce brown pigmentation when grown on tyrosine-containing media (34), limited testing indicates a correlation between melanin production and the presence of lly (10, 34). Hence, it is likely that legionellae, in addition to L. pneumophila, secrete HGA-melanin. Legionella is a member of a relatively large group of environmental bacteria that produce melanins, including species of Aeromonas, Azospirillum, Azotobacter, Bacillus, Burkholderia, Caulobacter, Hyphomonas, Klebsiella, Marinomonas, Microbulbifer, Micrococcus, Morganella, Mycobacterium, Proteus, Providencia, Pseudomonas, Ralstonia, Rhizobium, Serratia, Shewanella, Silicibacter, Sinorhizobium, Stenotrophomonas, Streptomyces, Vibrio, and Xanthomonas (1, 4, 24, 25, 33, 42, 44, 45, 48, 55, 58, 63, 64, 70, 75, 77, 79, 90, 100, 102, 110, 111, 115). Within this group, a variety of melanins are produced, including black and brown eumelanins, yellow-red pheomelanins, and brown allomelanins, which have as subsets the brown pyomelanins (HGA-melanin) and the black di/tetrahydroxynaphthelene melanins (71, 79, 110). Melanins are also broadly distributed among eukaryotic microorganisms, including fungi and protozoa (70, 79). Those bacteria that are known to specifically produce HGA-melanin are Caulobacter crescentus, Hyphomonas sp., Pseudomonas aeruginosa, P. putida, Ralstonia solanacearum, Shewanella algae, Shewanella colwelliana, Silicibacter pomeroyi, Sinorhizobium meliloti, Streptomyces avermitilis, Vibrio cholerae, and Xanthomonas campestris (4, 29, 33, 42, 45, 55, 64, 90, 102). Among the eukaryotic microbes, endogenous HGA-melanin production is thus far limited to the fungi Aspergillus nidulans and Yarrowia lipolytica (13, 36), although it has recently been shown that the fungus Cryptococcus neoformans can use HGA produced by others to achieve melanization (38). Thus, L. pneumophila, along with P. aeruginosa and V. cholerae, is one of the few known producers of HGA-melanin that is also a significant human pathogen.
In this study, we have also demonstrated that L. pneumophila HGA-melanin is responsible for a newly described secreted ferric reductase activity. Currently, there are two previous reports of an association between microbial melanins and ferric reduction. In the first case to be described, pathogenic C. neoformans elaborates a surface-associated eumelanin that was shown to reduce ferric hydroxyl-EDTA (51, 70, 72). A recent study reports that iron levels can modulate the transcriptional control of melanin biosynthesis in C. neoformans (54). As the second example of an iron-reducing melanin, the environmental, gram-negative bacterium S. algae secretes HGA-melanin that is capable of reducing ferric iron oxide (102). It is believed that this pyomelanin can also become cell associated and serve as an electron conduit in the terminal reduction of iron oxides (101). In addition to melanins, there are other methods of extracellular ferric reduction that exist in microorganisms. For example, there is a secreted blue pigment (pyocyanin) for P. aeruginosa, a cell-bound ferric reductase and secreted 3-hydroxyanthranilic acid for C. neoformans, extracellular and surface protein reductases for Mycobacterium paratuberculosis and Listeria monocytogenes, and secreted quinones for Geothrix fermentans and Shewanella oneidensis (22, 23, 28, 50, 69, 89). Since the lly mutant did not possess any residual secreted activity that was above the medium control, we suspect that all of the secreted ferric reductase activity observed for L. pneumophila is due to HGA-melanin. Supporting this viewpoint, in silico analysis of the L. pneumophila genome does not reveal any secreted proteins that are predicted to have ferric reductase activity (27). In sum, L. pneumophila is one of only a small number of microorganisms in which melanin secretion has been linked to ferric reduction and the only pathogen, thus far, in which HGA-melanin has been specifically implicated.
In the past, we and others defined a number of factors that promote L. pneumophila iron acquisition and/or growth under low-iron conditions (17). Arguably, the two factors that are most clearly involved in iron uptake are the legiobactin siderophore and the FeoB ferrous transport system (3, 86). Indeed, we recently hypothesized that these two represent the main pathways of iron uptake in L. pneumophila, i.e., one dedicated to ferric iron uptake and the other to ferrous iron assimilation (3). Given its capacity to promote extracellular ferric reduction, HGA-melanin is now proposed as yet another aspect of L. pneumophila iron acquisition/assimilation. We further propose that HGA-melanin operates within the FeoB pathway, for two reasons. First, the generation of ferrous iron by HGA-melanin would obviously have the potential to provide additional ferrous iron substrate for internalization across the cell envelope. Second, in preliminary experiments, the lly mutant exhibited increased CAS reactivity, suggesting that it might express elevated levels of legiobactin in order to compensate for the absent pigment (C. Chatfield and N. P. Cianciotto, unpublished results). Based upon the capacity of the lly mutant to still grow in iron-depleted media, iron assimilation via HGA-melanin would appear to not be required for L. pneumophila growth, at least under standard in vitro laboratory conditions. Given the similar behavior of feoB and lbtA mutants, the same has been concluded for iron assimilation via FeoB and legiobactin. Such results are entirely compatible with the widespread belief that bacteria have multiple pathways for iron acquisition. Since pigment mutants have not been described for S. algae, it is not possible to comment on the essentiality of the only other HGA-melanin implicated in iron reduction.
Our observations now bring to four the processes linked to the L. pneumophila HGA and HGA-melanin, i.e., the reduction of ferric iron and resistance to light by Lly+ L. pneumophila and the generation of turbomycin antibiotics and hemolytic activity by lly-containing E. coli clones (40, 97, 113). Given that as well as the many other processes linked to microbial melanins, including a variety of virulence traits (70, 71, 79), further work on the HGA-melanin of L. pneumophila is warranted. Finally, these findings in L. pneumophila have implications for the potential roles of melanins in other bacteria, especially other pathogens that are already known to produce HGA or other melanins, such as species of Burkholderia, Klebsiella, Proteus, Pseudomonas, Serratia, Stenotrophomonas, and Vibrio.
Acknowledgments
We thank members of the Cianciotto laboratory for many helpful discussions and Kimberly Allard for additional technical assistance. We also thank Michael Steinert and Jörg Hacker for sending us strain JR32 and its lly mutant derivative.
C.H.C. was partly supported by NIH Training Grant T32 AI-0007476-09. This work was funded by NIH grant AI34937 awarded to N.P.C.
Editor: D. L. Burns
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Agodi, A., S. Stefani, C. Corsaro, F. Campanile, S. Gribaldo, and G. Sichel. 1996. Study of a melanic pigment of Proteus mirabilis. Res. Microbiol. 147:167-174. [DOI] [PubMed] [Google Scholar]
- 2.Albert-Weissenberger, C., C. Cazalet, and C. Buchrieser. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell Mol. Life Sci. 64:432-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard, K. A., V. K. Viswanathan, and N. P. Cianciotto. 2006. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 188:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernandez, B. Galan, J. L. Garcia, E. Diaz, and B. Minambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186:5062-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachman, M. A., and M. S. Swanson. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect. Immun. 72:3284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baine, W. B., and J. K. Rasheed. 1979. Aromatic substrate specificity of browning by cultures of the Legionnaires' disease bacterium. Ann. Intern. Med. 90:619-620. [DOI] [PubMed] [Google Scholar]
- 7.Baine, W. B., J. K. Rasheed, J. C. Feeley, G. W. Gorman, and L. E. Casida. 1978. Effect of supplemental L-tyrosine on pigment production in cultures of the Legionnaires' disease bacterium. Curr. Microbiol. 1:93-94. [Google Scholar]
- 8.Bandyopadhyay, P., S. Liu, C. B. Gabbai, Z. Venitelli, and H. M. Steinman. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect. Immun. 75:723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellinger-Kawahara, C., and M. A. Horwitz. 1990. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J. Exp. Med. 172:1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender, L., M. Ott, A. Debes, U. Rdest, J. Heesemann, and J. Hacker. 1991. Distribution, expression, and long-range mapping of legiolysin gene (lly)-specific DNA sequences in legionellae. Infect. Immun. 59:3333-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg, J. D., J. C. Hoff, P. V. Roberts, and A. Matin. 1985. Growth of Legionella pneumophila in continuous culture. Appl. Environ. Microbiol. 49:1534-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrd, T. F., and M. A. Horwitz. 1989. Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J. Clin. Investig. 83:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carreira, A., L. M. Ferreira, and V. Loureiro. 2001. Brown pigments produced by Yarrowia lipolytica result from extracellular accumulation of homogentisic acid. Appl. Environ. Microbiol. 67:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 15.Challis, G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601-611. [DOI] [PubMed] [Google Scholar]
- 16.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 17.Cianciotto, N. P. 2007. Iron acquisition by Legionella pneumophila. Biometals 20:323-331. [DOI] [PubMed] [Google Scholar]
- 18.Cianciotto, N. P. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331-343. [DOI] [PubMed] [Google Scholar]
- 19.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 20.Cianciotto, N. P., P. Cornelis, and C. Baysse. 2005. Impact of the bacterial type I cytochrome c maturation system on different biological processes. Mol. Microbiol. 56:1408-1415. [DOI] [PubMed] [Google Scholar]
- 21.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowart, R. E. 2002. Reduction of iron by extracellular iron reductases: implications for microbial iron acquisition. Arch. Biochem. Biophys. 400:273-281. [DOI] [PubMed] [Google Scholar]
- 23.Cox, C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coyne, V. E., and L. al-Harthi. 1992. Induction of melanin biosynthesis in Vibrio cholerae. Appl. Environ. Microbiol. 58:2861-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cubo, M. T., A. M. Buendia-Claveria, J. E. Beringer, and J. E. Ruiz-Sainz. 1988. Melanin production by Rhizobium strains. Appl. Environ. Microbiol. 54:1812-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DebRoy, S., V. Aragon, S. Kurtz, and N. P. Cianciotto. 2006. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect. Immun. 74:5152-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DebRoy, S., J. Dao, M. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. USA 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deneer, H. G., V. Healey, and I. Boychuk. 1995. Reduction of exogenous ferric iron by a surface-associated ferric reductase of Listeria spp. Microbiology 141:1985-1992. [DOI] [PubMed] [Google Scholar]
- 29.Denoya, C. D., D. D. Skinner, and M. R. Morgenstern. 1994. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J. Bacteriol. 176:5312-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 34.Fang, G. D., V. L. Yu, and R. M. Vickers. 1989. Disease due to the Legionellaceae (other than Legionella pneumophila): historical, microbiological, clinical, and epidemiological review. Medicine 68:116-132. [DOI] [PubMed] [Google Scholar]
- 35.Feeley, J. C., G. W. Gorman, R. E. Weaver, D. C. Mackel, and H. W. Smith. 1978. Primary isolation media for Legionnaires' disease bacterium. J. Clin. Microbiol. 8:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Canon, J. M., and M. A. Penalva. 1995. Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J. Biol. Chem. 270:21199-21205. [DOI] [PubMed] [Google Scholar]
- 37.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frases, S., A. Salazar, E. Dadachova, and A. Casadevall. 2007. Cryptococcus neoformans can utilize the bacterial melanin precursor homogentisic acid for fungal melanogenesis. Appl. Environ. Microbiol. 73:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilmour, M. W., K. Bernard, D. M. Tracz, A. B. Olson, C. R. Corbett, T. Burdz, B. Ng, D. Wiebe, G. Broukhanski, P. Boleszczuk, P. Tang, F. Jamieson, G. Van Domselaar, F. A. Plummer, and J. D. Berry. 2007. Molecular typing of a Legionella pneumophila outbreak in Ontario, Canada. J. Med. Microbiol. 56:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodwin, P. H., and C. R. Sopher. 1993. Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentisic acid. Can. J. Microbiol. 40:28-34. [Google Scholar]
- 43.Grass, G. 2006. Iron transport in Escherichia coli: all has not been said and done. Biometals 19:159-172. [DOI] [PubMed] [Google Scholar]
- 44.Hawkey, P. M., A. McCormick, and R. A. Simpson. 1986. Selective and differential medium for the primary isolation of members of the Proteeae. J. Clin. Microbiol. 23:600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez-Romero, D., F. Solano, and A. Sanchez-Amat. 2005. Polyphenol oxidase activity expression in Ralstonia solanacearum. Appl. Environ. Microbiol. 71:6808-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heuner, K., and M. Steinert. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293:133-143. [DOI] [PubMed] [Google Scholar]
- 47.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodgson, D. A., P. Shaw, and L. Shapiro. 1984. Isolation and genetic analysis of Caulobacter mutants defective in cell shape and membrane lipid synthesis. Genetics 108:809-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman, P. S., M. Ripley, and R. Weeratna. 1992. Cloning and nucleotide sequence of a gene (ompS) encoding the major outer membrane protein of Legionella pneumophila. J. Bacteriol. 174:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homuth, M., P. Valentin-Weigand, M. Rohde, and G. F. Gerlach. 1998. Identification and characterization of a novel extracellular ferric reductase from Mycobacterium paratuberculosis. Infect. Immun. 66:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobson, E. S., and J. D. Hong. 1997. Redox buffering by melanin and Fe(II) in Cryptococcus neoformans. J. Bacteriol. 179:5340-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.James, B. W., W. S. Mauchline, P. J. Dennis, and C. W. Keevil. 1997. A study of iron acquisition mechanisms of Legionella pneumophila grown in chemostat culture. Curr. Microbiol. 34:238-243. [DOI] [PubMed] [Google Scholar]
- 53.Johnson, W., L. Varner, and M. Poch. 1991. Acquisition of iron by Legionella pneumophila: role of iron reductase. Infect. Immun. 59:2376-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung, W. H., A. Sham, R. White, and J. W. Kronstad. 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4:2282-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotob, S. I., S. L. Coon, E. J. Quintero, and R. M. Weiner. 1995. Homogentisic acid is the primary precursor of melanin synthesis in Vibrio cholerae, a Hyphomonas strain, and Shewanella colwelliana. Appl. Environ. Microbiol. 61:1620-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuroki, H., H. Miyamoto, K. Fukuda, H. Iihara, Y. Kawamura, M. Ogawa, Y. Wang, T. Ezaki, and H. Taniguchi. 2006. Legionella impletisoli sp. nov. and Legionella yabuuchiae sp. nov., isolated from soils contaminated with industrial wastes in Japan. Syst. Appl. Microbiol. 30:273-279. [DOI] [PubMed] [Google Scholar]
- 57.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 103:18745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagunas-Munoz, V. H., N. Cabrera-Valladares, F. Bolivar, G. Gosset, and A. Martinez. 2006. Optimum melanin production using recombinant Escherichia coli. J. Appl. Microbiol. 101:1002-1008. [DOI] [PubMed] [Google Scholar]
- 59.La Scola, B., R. J. Birtles, G. Greub, T. J. Harrison, R. M. Ratcliff, and D. Raoult. 2004. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 54:699-703. [DOI] [PubMed] [Google Scholar]
- 60.Liles, M. R., T. Aber Scheel, and N. P. Cianciotto. 2000. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 182:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mampel, J., T. Spirig, S. S. Weber, J. A. Haagensen, S. Molin, and H. Hilbi. 2006. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72:2885-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michiels, J., T. Van Soom, I. D'Hooghe, B. Dombrecht, T. Benhassine, P. de Wilde, and J. Vanderleyden. 1998. The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN, ptsN, and ptsA mutants. J. Bacteriol. 180:1729-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milcamps, A., and F. J. de Bruijn. 1999. Identification of a novel nutrient-deprivation-induced Sinorhizobium meliloti gene (hmgA) involved in the degradation of tyrosine. Microbiology 145:935-947. [DOI] [PubMed] [Google Scholar]
- 65.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73:5720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murata, T., A. Delprato, A. Ingmundson, D. K. Toomre, D. G. Lambright, and C. R. Roy. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8:971-977. [DOI] [PubMed] [Google Scholar]
- 68.Naylor, J., and N. P. Cianciotto. 2004. Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS Microbiol. Lett. 241:249-256. [DOI] [PubMed] [Google Scholar]
- 69.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nosanchuk, J. D., and A. Casadevall. 2003. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5:203-223. [DOI] [PubMed] [Google Scholar]
- 71.Nosanchuk, J. D., and A. Casadevall. 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 50:3519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nyhus, K. J., A. T. Wilborn, and E. S. Jacobson. 1997. Ferric iron reduction by Cryptococcus neoformans. Infect. Immun. 65:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Connell, W. A., E. K. Hickey, and N. P. Cianciotto. 1996. A Legionella pneumophila gene that promotes hemin binding. Infect. Immun. 64:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orrison, L. H., W. B. Cherry, C. B. Fliermans, S. B. Dees, L. K. McDougal, and D. J. Dodd. 1981. Characteristics of environmental isolates of Legionella pneumophila. Appl. Environ. Microbiol. 42:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Page, W. J., and S. Shivprasad. 1995. Iron binding to Azotobacter salinestris melanin, iron mobilization and uptake mediated by siderophores. Biometals 8:59-64. [DOI] [PubMed] [Google Scholar]
- 76.Park, M. Y., K. S. Ko, H. K. Lee, M. S. Park, and Y. H. Kook. 2003. Legionella busanensis sp. nov., isolated from cooling tower water in Korea. Int. J. Syst. Evol. Microbiol. 53:77-80. [DOI] [PubMed] [Google Scholar]
- 77.Pavan, M. E., S. L. Abbott, J. Zorzopulos, and J. M. Janda. 2000. Aeromonas salmonicida subsp. pectinolytica subsp. nov., a new pectinase-positive subspecies isolated from a heavily polluted river. Int. J. Syst. Evol. Microbiol. 50:1119-1124. [DOI] [PubMed] [Google Scholar]
- 78.Pine, L., J. R. George, M. W. Reeves, and W. K. Harrell. 1979. Development of a chemically defined liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 9:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plonka, P. M., and M. Grabacka. 2006. Melanin synthesis in microorganisms—biotechnological and medical aspects. Acta Biochim. Pol. 53:429-443. [PubMed] [Google Scholar]
- 80.Poch, M. T., and W. Johnson. 1993. Ferric reductases of Legionella pneumophila. Biometals 6:107-114. [DOI] [PubMed] [Google Scholar]
- 81.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reeves, M. W., L. Pine, S. H. Hutner, J. R. George, and W. K. Harrell. 1981. Metal requirements of Legionella pneumophila. J. Clin. Microbiol. 13:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reeves, M. W., L. Pine, J. B. Neilands, and A. Balows. 1983. Absence of siderophore activity in Legionella species grown in iron-deficient media. J. Bacteriol. 154:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ristroph, J. D., K. W. Hedlund, and S. Gowda. 1981. Chemically defined medium for Legionella pneumophila growth. J. Clin. Microbiol. 13:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossier, O., S. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ruebush, S. S., S. L. Brantley, and M. Tien. 2006. Reduction of soluble and insoluble iron forms by membrane fractions of Shewanella oneidensis grown under aerobic and anaerobic conditions. Appl. Environ. Microbiol. 72:2925-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruzafa, C., A. Sanchez-Amat, and F. Solano. 1995. Characterization of the melanogenic system in Vibrio cholerae, ATCC 14035. Pigment Cell Res. 8:147-152. [DOI] [PubMed] [Google Scholar]
- 91.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 92.Sanchez-Amat, A., C. Ruzafa, and F. Solano. 1998. Comparative tyrosine degradation in Vibrio cholerae strains. The strain ATCC 14035 as a prokaryotic melanogenic model of homogentisate-releasing cell. Comp. Biochem. Physiol. B 119:557-562. [DOI] [PubMed] [Google Scholar]
- 93.Sauer, J. D., J. G. Shannon, D. Howe, S. F. Hayes, M. S. Swanson, and R. A. Heinzen. 2005. Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by coinfection. Infect. Immun. 73:4494-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scazzocchio, C. 1997. Alkaptonuria: from humans to moulds and back. Trends Genet. 13:125-127. [DOI] [PubMed] [Google Scholar]
- 95.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA 102:4866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Starkenburg, S. R., J. M. Casey, and N. P. Cianciotto. 2004. Siderophore activity among members of the Legionella genus. Curr. Microbiol. 49:203-207. [DOI] [PubMed] [Google Scholar]
- 97.Steinert, M., H. Engelhard, M. Flugel, E. Wintermeyer, and J. Hacker. 1995. The Lly protein protects Legionella pneumophila from light but does not directly influence its intracellular survival in Hartmannella vermiformis. Appl. Environ. Microbiol. 61:2428-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steinert, M., M. Flugel, M. Schuppler, J. H. Helbig, A. Supriyono, P. Proksch, and P. C. Luck. 2001. The Lly protein is essential for p-hydroxyphenylpyruvate dioxygenase activity in Legionella pneumophila. FEMS Microbiol. Lett. 203:41-47. [DOI] [PubMed] [Google Scholar]
- 99.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanaka, T., L. Yan, and J. G. Burgess. 2003. Microbulbifer arenaceous sp. nov., a new endolithic bacterium isolated from the inside of red sandstone. Curr. Microbiol. 47:412-416. [DOI] [PubMed] [Google Scholar]
- 101.Turick, C. E., F. Caccavo, Jr., and L. S. Tisa. 2003. Electron transfer from Shewanella algae BrY to hydrous ferric oxide is mediated by cell-associated melanin. FEMS Microbiol. Lett. 220:99-104. [DOI] [PubMed] [Google Scholar]
- 102.Turick, C. E., L. S. Tisa, and F. Caccavo, Jr. 2002. Melanin production and use as a soluble electron shuttle for Fe(III) oxide reduction and as a terminal electron acceptor by Shewanella algae BrY. Appl. Environ. Microbiol. 68:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vartivarian, S. E., and R. E. Cowart. 1999. Extracellular iron reductases: identification of a new class of enzymes by siderophore-producing microorganisms. Arch. Biochem. Biophys. 364:75-82. [DOI] [PubMed] [Google Scholar]
- 104.Vickers, R. M., and V. L. Yu. 1984. Clinical laboratory differentiation of Legionellaceae family members with pigment production and fluorescence on media supplemented with aromatic substrates. J. Clin. Microbiol. 19:583-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vincent, C. D., and J. P. Vogel. 2006. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 61:596-613. [DOI] [PubMed] [Google Scholar]
- 106.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Viswanathan, V. K., S. Kurtz, L. L. Pedersen, Y. Abu-Kwaik, K. Krcmarik, S. Mody, and N. P. Cianciotto. 2002. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wagner, C., A. S. Khan, T. Kamphausen, B. Schmausser, C. Unal, U. Lorenz, G. Fischer, J. Hacker, and M. Steinert. 2007. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell. Microbiol. 9:450-462. [DOI] [PubMed] [Google Scholar]
- 109.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weiner, R. M. 1997. Biopolymers from marine prokaryotes. Trends Biotechnol. 15:390-394. [DOI] [PubMed] [Google Scholar]
- 111.Weiner, R. M., A. M. Segall, and R. R. Colwell. 1985. Characterization of a marine bacterium associated with Crassostrea virginica (the Eastern oyster). Appl. Environ. Microbiol. 49:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 113.Wintermeyer, E., M. Flugel, M. Ott, M. Steinert, U. Rdest, K. H. Mann, and J. Hacker. 1994. Sequence determination and mutational analysis of the lly locus of Legionella pneumophila. Infect. Immun. 62:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wintermeyer, E., U. Rdest, B. Ludwig, A. Debes, and J. Hacker. 1991. Characterization of legiolysin (lly), responsible for haemolytic activity, colour production and fluorescence of Legionella pneumophila. Mol. Microbiol. 5:1135-1143. [DOI] [PubMed] [Google Scholar]
- 115.Zughaier, S. M., H. C. Ryley, and S. K. Jackson. 1999. A melanin pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect. Immun. 67:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]