Abstract
Salmonella enterica is a facultative intracellular pathogen of worldwide importance and causes a spectrum of diseases depending on serovar- and host-specific factors. Oral infection of pigs with S. enterica serovar Typhimurium strain 4/74 produces acute enteritis but is rarely fatal, whereas serovar Choleraesuis strain A50 causes systemic disease with a high mortality rate. With a porcine ligated ileal loop model, we observed that systemic virulence of serovar Choleraesuis A50 is not associated with enhanced intestinal invasion, secretory responses, or neutrophil recruitment compared to serovar Typhimurium 4/74. The net growth in vivo of serovar Choleraesuis A50 and serovar Typhimurium 4/74 was monitored following oral inoculation of pigs with strains harboring pHSG422, which exhibits temperature-sensitive replication. Analysis of plasmid partitioning revealed that the enteric virulence of serovar Typhimurium 4/74 relative to that of serovar Choleraesuis A50 is associated with rapid replication in the intestinal wall, whereas systemic virulence of serovar Choleraesuis A50 is associated with enhanced persistence in intestinal mesenteric lymph nodes. Faster replication of serovar Typhimurium, compared to that of serovar Choleraesuis, in the intestinal mucosa was associated with greater induction of the proinflammatory cytokines tumor necrosis factor alpha, interleukin-8 (IL-8), and IL-18 as detected by reverse transcriptase PCR analysis of transcripts from infected mucosa. During replication in batch culture and porcine alveolar macrophages, transcription of genes encoding components of type III secretion systems 1 (sipC) and 2 (sseC) was observed to be significantly higher in serovar Typhimurium 4/74 than in serovar Choleraesuis A50, and this may contribute to the differences in epithelial invasion and intracellular proliferation. The rapid induction of proinflammatory responses by strain 4/74 may explain why pigs confine serovar Typhimurium infection to the intestines, whereas slow replication of serovar Choleraesuis may enable it to evade host innate immunity and thus disseminate by stealth.
Salmonella enterica subspecies enterica can be divided into more than 2,400 antigenically distinct serovars, some of which are important pathogens of humans and food-producing animals (50). The serovars can be divided into three broad classes (46). Ubiquitous serovars (e.g., Typhimurium) produce acute but self-limiting enteritis in a broad range of hosts, whereas host-specific serovars (e.g., Typhi) are associated with severe systemic disease in healthy outbred adults of a single species which may not involve diarrhea. Host-restricted serovars (e.g., Choleraesuis) are primarily associated with systemic disease in one host but may cause disease in a limited number of other species.
Clinical salmonellosis in pigs is mostly caused by serovars Choleraesuis and Typhimurium (36). Pigs infected with serovar Choleraesuis are lethargic and pyrexic and often have respiratory symptoms including pneumonia and coughing. Diarrhea may or may not be present, and cyanosis of the extremities is common. Gross lesions often include swollen mesenteric lymph nodes, enlargement of the spleen and liver, and congestion of the lungs. Mortality is high, particularly in intensively reared weaned pigs. In contrast, serovar Typhimurium typically causes watery diarrhea, anorexia, and pyrexia, but with a low mortality rate and little or no systemic involvement. Serovar Typhimurium infections may become persistent, involving asymptomatic excretion of the bacteria in the feces for several months postexposure. In a year-long randomized national abattoir survey in the United Kingdom, serovar Typhimurium was detected in the ceca of 11.1% of pigs presented for slaughter and on 2.1% of carcass swabs (15); thus, persistently infected pigs represent a significant reservoir of contamination of the food chain and environment. Serovar Choleraesuis was once the most frequently isolated serovar in pigs in the United Kingdom (41). While it is now rarely isolated in the United Kingdom, it remains a serious threat in the United States (36) and is also associated with sporadic cases of severe salmonellosis in humans (49; reviewed in reference 11).
The molecular mechanisms underlying the ability of S. enterica serovars to colonize the intestines of food-producing animals, and in some cases translocate to systemic sites, are poorly understood. Genome-wide mutagenesis has identified portfolios of serovar Typhimurium genes required for intestinal colonization of cattle (32, 44), chickens (32), pigs (8), and mice (10, 24, 28, 38, 44) and revealed that Salmonella uses both conserved and host-specific colonization factors. Variation in the repertoire, sequence, and expression of such factors may explain the differential virulence of S. enterica serovars. Type III secretion systems (T3SS) encoded by Salmonella pathogenicity island 1 (SPI-1) and SPI-2 play pivotal roles in the colonization of porcine intestines by serovar Typhimurium (5, 8), and mutation of a key SPI-1 regulator (hilA) is highly attenuating for serovar Choleraesuis following oral infection of pigs (29). Analysis of the complete genome sequence of serovar Choleraesuis strain SC-B67 recently revealed that it contains a higher proportion of deletions and pseudogenes than serovar Typhimurium LT2, particularly among genes involved in chemotaxis, signal transduction, and metabolism (12). The impact of such polymorphisms on the outcome of infections caused by serovars Typhimurium and Choleraesuis has received little attention.
The basis of systemic virulence of host-specific and -restricted serovars has so far not been definitively correlated with events during infection. No association could be detected between systemic virulence and the magnitude of intestinal invasion or induction of inflammatory or secretory responses by strains of host-restricted serovar Dublin in calves (34), Abortusovis in lambs (47), or Gallinarum in chickens (9). Indeed, strains of serovars that are systemically avirulent in these hosts following oral inoculation, including serovar Typhimurium, exhibited increased invasion (9, 34, 47) and elicited greater enteropathogenic responses (34, 47). Furthermore, although the natural resistance-associated macrophage protein is important in the control of systemic serovar Choleraesuis infection in pigs (48), strains of serovar Choleraesuis do not exhibit an ability to survive better in porcine primary alveolar macrophages compared to serovar Typhimurium and no significant differences in the induction of proinflammatory cytokines in cultured porcine alveolar macrophages by the serovars could be detected (53). It has been noted that host-restricted serovars persist better in mesenteric lymph nodes, indicating that bacterial net growth at intestinal sites may be an important determinant of systemic virulence (34, 47).
Here, we used the temperature-sensitive replicon pHSG422 (23) to study the net replication of S. enterica serovar Typhimurium and Choleraesuis strains of well-defined virulence following oral inoculation of pigs. Plasmid pHSG422 has a temperature-sensitive origin of replication that permits replication at 30°C or below but results in segregation of the plasmid between dividing cells at 37°C or higher. Thus, during infection at body temperature the plasmid is titrated out of the bacterial population with each round of replication. For a strain with a fast in vivo replication rate, more bacteria will be recovered but fewer will be plasmid bearing, whereas a slow-replicating strain will yield fewer bacteria, more of which would harbor pHSG422. While the proportion of bacteria carrying pHSG422 is an indication of the growth rate, the absolute number of cells carrying pHSG422 provides a measurement of killing (see reference 22). Plasmid pHSG422 has been used to study intracellular growth of Salmonella in murine dendritic cells (26) and the impact of mutation of SPI-2 (37), spv genes (21, 22), prior infection with mouse hepatitis virus (19), and the host Ity locus (2) on Salmonella replication in mice. The fate of the serovar Choleraesuis and Typhimurium strains in vivo was further dissected by analysis of intestinal invasion, induction of enteritis, and analysis of the transcription of host and bacterial genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Serovar Typhimurium 4/74 and serovar Choleraesuis A50 have been described previously, and their virulence in pigs is well defined (53). The behavior of these strains in cultured cells and animals is comparable to that of other strains of the same serovar (3, 34, 53), and the clinical symptoms they elicit are typical of field isolates. Plasmid pHSG422 (23) was kindly provided by T. Hashimoto-Gotoh, Kyoto, Japan, and was electroporated into the Salmonella strains with selection for resistance to ampicillin (100 μg/ml) and kanamycin (50 μg/ml) at 25°C. Plasmid carriage did not impair the growth of the strains at 25°C in vitro or their ability to invade cultured INT-407 cells or bovine ligated ileal mucosa (data not shown). Additional strains of serovars Typhimurium (12/75 and TML) and Choleraesuis (14/74) were used for analysis of sipC transcript and protein levels (53). Serovar Typhimurium strains 4/74 and 12/75 show nearly identical replication kinetics in porcine alveolar macrophages (53) and invasion and enteropathogenicity in calves (34). The serovar Choleraesuis A50 and 14/74 strains also behave similarly to each other in these models, albeit that they invade, persist, and induce enteritis at lower levels than the serovar Typhimurium strains. Serovar Typhimurium 4/74 sipB (1) and sipC (32) mutant strains have been described previously. Strains were cultured in Luria-Bertani (LB) medium with agitation and antibiotics as appropriate. Derivatives harboring pHSG422 were cultured to maintain a low copy number as previously described (21).
Construction of an aroA mutant of serovar Choleraesuis A50.
To establish that partitioning of pHSG422 can report differences in the net growth of Salmonella in pigs, a Tn10 insertion in aroA was transferred into serovar Choleraesuis A50 by phage P22 HT/int-mediated transduction as previously described (31). The mutant was verified by analysis of growth on M9 minimal medium with and without aromatic amino acids and examined for nonagglutination with 5% acriflavine-HCl to confirm normal expression of lipopolysaccharide.
Infection of pigs.
All animal experiments were performed in accordance with the Animals (Scientific Procedures) Act 1986 (license number 30/1998) with the approval of the local Ethical Review Committee. Large-White × Landrace male pigs aged ca. 8 weeks were obtained from a commercial supplier and housed in containment level 2 accommodations with access to antibiotic-free irradiated weaner pellet and water ad libitum. Pigs were confirmed to be culture negative for Salmonella prior to infection by overnight enrichment of rectal swabs in Rappaport broth (at 37°C) and selenite brilliant green broth (at 42°C), followed by plating to modified brilliant green agar (Oxoid, Basingstoke, United Kingdom). Stationary-phase 25°C LB-grown cultures of serovar Typhimurium 4/74/pHSG422, Choleraesuis A50/pHSG422, or Choleraesuis A50 aroA/pHSG422 were diluted to contain approximately 1 × 109 CFU and suspended in 10 ml antacid [5% (wt/vol) Mg(SiO3)3, 5% (wt/vol) NaHCO3, 5% (wt/vol) MgCO3 in H2O] and administered directly into the stomach of each pig immediately before the morning feed with a 10FG catheter. To validate the use of pHSG422 for analysis of net replication, seven pigs were infected with serovar Choleraesuis A50/pHSG422 and three pigs were infected with serovar Choleraesuis A50 aroA/pHSG422. To compare the net replication of the serovars, a total of 22 pigs were inoculated with serovar Typhimurium 4/74 and sacrificed at 24 h (9 pigs), 36 h (3 pigs), 48 h (7 pigs), and 72 h (3 pigs). Net replication was compared to that of serovar Choleraesuis A50 in a total of 20 pigs killed at 24 h (7 pigs), 36 h (3 pigs), 48 h (7 pigs), and 72 h (3 pigs). Rectal temperatures were recorded, and animals were monitored for clinical signs of disease twice daily. At postmortem examination, samples of mid-ileum, mid-colon, and associated mesenteric lymph nodes were aseptically excised and viable bacteria were enumerated by plating of serial 10-fold dilutions of homogenates of triplicate 1-g biopsy samples from each site onto MacConkey agar with and without ampicillin and kanamycin, as appropriate. Tissue samples were also recovered from the same sites for analysis of host and bacterial mRNAs (see below). The limit of accurate quantification was 2.0 log10 CFU/g. Samples containing fewer bacteria were enriched in Rappaport broth and selenite brilliant green broth as described above. The difference in the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g was calculated and provided a measurement of the titration of the plasmid from the bacterial population and hence the rate of replication (21, 22).
Quantification of intestinal invasion and induction of enteritis.
Ligated ileal loops were constructed in ca. 8-week-old Large-White × Landrace pigs following sedation by intramuscular administration of 2 mg/kg Stresnil (Janssen Animal Health, High Wycombe, United Kingdom), induction of anesthesia by intravenous administration of 18 mg/kg Saffan (Schering-Plough Animal Health, Welwyn Garden City, United Kingdom), and maintenance of anesthesia with isoflurane in oxygen via an endotracheal tube. A laparotomy was performed, and sequential 9-cm loops (invasion assays) or 6-cm loops (enteritis assays) with 1-cm spacers were constructed with surgical silk. Approximately 1 × 109 CFU of LB-grown mid-logarithmic-phase test strains were injected into appropriate loops (triplicate determinations in a semirandomized order), and the wound was repaired.
Invasion was quantified essentially as previously described (3), by injection of 4.5 ml of 300 μg/ml gentamicin in mucosal medium into distal ileal loops 2 h after inoculation with bacteria. Viable gentamicin-protected bacteria were then enumerated in tissues recovered after incubation in situ for a further 1 h (initial invasion) or 10 h (survival postinvasion) by plating of serial 10-fold dilutions of homogenates of triplicate 1-g biopsy samples from each loop blended separately for 30 s in 1% (vol/vol) Triton X-100 in phosphate-buffered saline onto MacConkey agar. The conditions used have been confirmed to result in efficient killing of an adherent noninvasive Escherichia coli strain (3, 52), and control loops were included to enumerate indigenous gentamicin-resistant bacteria.
Secretory responses were measured in triplicate mid-ileal loops for each strain and are defined by the ratio of the volume of fluid accumulated (V, ml) to loop length (L, cm). Neutrophil recruitment was quantified as a measurement of the induction of intestinal inflammatory responses by [111In]oxinate labeling of neutrophils purified from blood sampled from the vena cava at the outset of the experiment and reinjected within 1 to 2 h of loop inoculation, essentially as previously described (51). Neutrophil influx is defined as the ratio of gamma emission from 111In-labeled neutrophils within the combined mucosa and contents of test loops to that from loops filled with sterile medium as a negative control.
Analysis of porcine cytokine responses.
Samples of ileal and colonic mucosa were obtained at postmortem examination from the same sites at the same intervals postinoculation as plasmid partitioning was assessed. Tissues were collected and snap-frozen in liquid nitrogen with minimal delay after death and stored at −70°C until required. Total RNA was prepared from 1-g samples with TRI REAGENT (Sigma, St. Louis, MO) with an on-column DNase treatment step. RNA was confirmed to be intact and free of genomic DNA by agarose gel electrophoresis. Purified RNA was eluted in 100 μl RNase-free water and stored at −70°C. Primers and probes for the detection of porcine tumor necrosis factor alpha (TNF-α), interleukin-8 (IL-8), and IL-18 mRNAs and 28S rRNA by real-time reverse transcriptase PCR (RT-PCR) were designed by using the Sus scrofa genome sequence at intron-exon boundaries such that amplicons could only derive from mRNA (Table 1). RT-PCR was performed with the ABI PRISM 7700 sequence detection system (PE Applied Biosystems, Warrington, United Kingdom) with quantitative RT-PCR Mastermix (Eurogentec, Seraing, Belgium) and an initial cycle of 50°C for 2 min, 60°C for 30 min, and 95°C for 5 min, followed by 40 cycles of 94°C for 20 s and 59°C for 1 min. To generate standard curves for the cytokine- and 28S rRNA-specific reactions, RNA was serially diluted from 10−1 to 10−5 in sterile, RNase-free water. Each RT experiment contained three no-template controls, test samples, and a log10 dilution series and was performed in triplicate with replicates prepared on different days. Results are expressed as the threshold cycle value (Ct), the cycle at which the change in reporter dye (Rn) passes a significant threshold. To account for variation in sampling and RNA preparation, the Ct values for the cytokine-specific product for each sample were standardized by using the Ct value of the 28S rRNA product for the same sample. Regression analysis of the mean values of six replicate RT-PCRs for the log10-diluted RNA was used to generate standard curves. Corrected Ct values were calculated, and results were then expressed as 40-Ct values as previously described (25), 40 being the maximum number of amplification cycles in the assay. Hence, the higher the 40-Ct value, the greater the amount of specific mRNA in a particular sample.
TABLE 1.
Primers and probes used for detection of porcine 28S rRNA and TNF-α, IL-8, and IL-18 mRNAs by real-time RT-PCR
| Primer or probe | Sequence 5′-3′a |
|---|---|
| 28S rRNA.fwd | GCTCCACGGGAGGTTTCTG |
| 28S rRNA.rev | GGTACACCTGTCAAACGGTAACG |
| 28S rRNA.probe | (FAM)-CTCCCTGAGCTCGCCTTAGGACACCT-(TAMRA) |
| TNF-α.fwd | AAGGACTCAGATCATCGTCTCAAAC |
| TNF-α.rev | CGGCTTTGACATTGGCTACA |
| TNF-α.probe | (FAM)-CGTGGGCGACGGGCTTATCTGA-(TAMRA) |
| IL-8.fwd | AGTTTTCCTGCTTTCTGCAGCT |
| IL-8.rev | TGGCATCGAAGTTCTGCACT |
| IL-8.probe | (FAM)-ACTCTTGCCAGAACTGCAGCCTCACA-(TAMRA) |
| IL-18.fwd | TCCTTTTCATTAACCAGGGACATC |
| IL-18.rev | GGTCTGAGGTGCATTATCTGAACA |
| IL-18.probe | (FAM)-CAGAATCAGGCATATCCTCAAACACGGCT-(TAMRA) |
FAM, 6-carboxyfluoroscein; TAMRA, 6-carboxytetramethylrhodamine.
Preparation and infection of porcine alveolar macrophages.
Alveolar macrophages were isolated from healthy ca. 8-week-old Large-White × Landrace pigs by bronchoalveolar lavage as previously described (53). Macrophages were cultured in RPMI 1640 medium buffered with 2 g/liter sodium bicarbonate and supplemented with 18 mM HEPES buffer, 2 mM l-glutamine, and 5% (vol/vol) fetal calf serum. They were inoculated with a multiplicity of infection of 100:1 of nonopsonized, LB-grown stationary-phase bacteria. This relatively high multiplicity of infection was used in studies to define the serovar Typhimurium transcriptome in murine macrophage-like cells and was required to yield adequate bacterial mRNA for subsequent analysis (18). The cells were incubated at 37°C in a humidified 5% CO2 atmosphere and harvested for RNA extraction at 4, 8, and 24 h after infection.
Analysis of the transcription of T3SS-1 and -2 genes.
Total RNA from LB-grown bacteria and infected macrophages was extracted with TRI REAGENT and treated in solution with Turbo DNase (Ambion, Inc., Austin, TX), followed by on-column DNase treatment with RNase-free DNase. A conserved housekeeping gene (yejA) was used as an internal standard and is expressed at constant basal levels in mid-logarithmic- and stationary-phase LB cultures following exposure to pH 3.0 and 10.0 low magnesium and phosphate levels and upon infection of epithelial and murine macrophage-like cells (18; J. Hinton, personal communication). Primers and probes used for amplification of sipC, sseC, and yejA are listed in Table 2. RT-PCR was performed as described for porcine cytokine mRNAs. Control reaction mixtures omitting RT or RNA were included, and three independent biological replicates were performed. The data from repeated experiments were pooled and analyzed as previously described (30). The Ct values for test genes were normalized to the Ct value of the internal standard (yejA) amplified from the corresponding sample. The sequences of the target genes are 100% identical in the sequenced genomes of serovars Typhimurium and Choleraesuis; therefore, differences in detection of the transcripts are likely to be real, rather than due to sequence polymorphisms in the primer or probe targets or template accessibility owing to secondary-structure variations. To quantify transcription, the 2ΔΔ−Ct method (30) was used for data analysis and transcription was reported as n-fold induction normalized to the internal standard and relative to the uninfected control at time zero. The limit of detection of transcripts from LB-grown bacteria was determined by comparing the mean Ct value against the number of viable bacteria. Bacterial mRNAs could be detected over a linear range of 108 to 101 CFU/ml for both serovar Choleraesuis A50 and serovar Typhimurium 4/74, with Ct values ranging from 13 to 28.
TABLE 2.
Primers and probes used for detection of mgtC, sipC, sseC, and yejA mRNAs by real-time RT-PCR
| Primer or probe | Sequence 5′-3′a |
|---|---|
| sipC.fwd | GGACGAAGCCCGTGAAAGT |
| sipC.rev | TGCTCTCCATTGTTTTCAGCATT |
| sipC.probe | (FAM)-CCTGAATCAGGCTGGTCGATTTACG TG-(TAMRA) |
| sseC.fwd | GCAGGTTGTGCAGGAATGGT |
| sseC.rev | TGGTCAGCACCGCACATC |
| sseC.probe | (FAM)-TGCCGTTTCGGCTCCGGCT-(TAMRA) |
| yejA.fwd | TCAGCGATCCGCTTTCAAC |
| yejA.rev | GCCCATTTTCCACTGAGTAATCC |
| yejA.probe | (FAM)-CCGCCCTTAGCCAGCGGGCCA AT-(TAMRA) |
FAM, 6-carboxyfluoroscein; TAMRA, 6-carboxytetramethylrhodamine.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
Total and secreted Salmonella proteins were prepared as previously described (55) and were resolved by 4 to 15% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were either stained with Gel Code Blue (Pierce Biotechnology, Rockport, IL) or transferred to nitrocellulose membrane and probed with SipC-specific murine monoclonal antibody (clone F569AC6; E. Galyov, Institute for Animal Health, Compton, Newbury, Berkshire, United Kingdom) detected with horseradish peroxidase-conjugated anti-mouse immunoglobulin G as previously described (55).
Statistical analysis.
A general linear model (Minitab) was used to determine whether the mean log10 total number of CFU/g or the difference between the log10 total number of CFU/g and the log10 number of plasmid-bearing CFU/g was statistically significant between time intervals and tissues sampled. RT-PCR data were analyzed by Student's t test with P < 0.05 taken to be significant.
RESULTS
Systemic virulence of serovar Choleraesuis A50 in pigs is not associated with enhanced intestinal invasion, secretory responses, or neutrophil recruitment.
We previously described the outcome of oral infection of age-matched pigs with strains of serovars Choleraesuis and Typhimurium (53); however, it is not known if the systemic virulence of serovar Choleraesuis in pigs is associated with enhanced invasion of mucosal surfaces or intestinal pathology. With a porcine ligated ileal loop model, the number of viable gentamicin-protected bacteria was determined at 3 h postinoculation of distal ileal loops with serovar Choleraesuis A50, serovar Typhimurium 4/74, and an isogenic serovar Typhimurium 4/74 sipB mutant. The magnitude of invasion by the sipB mutant was significantly lower than that of invasion by the parent strain (Fig. 1A; P < 0.05), confirming the key role of T3SS-1 in invasion of porcine intestinal mucosa and indicating that recovered bacteria are not derived from the intestinal lumen (P < 0.05). The number of gentamicin-protected serovar Typhimurium 4/74 CFU was significantly higher than that of serovar Choleraesuis A50 CFU (Fig. 1A; P < 0.05). Serovar Typhimurium 4/74 also elicited significantly higher levels of fluid accumulation and neutrophil recruitment 12 h postinoculation of mid-ileal loops compared to serovar Choleraesuis A50 (Fig. 1B and C; P < 0.05), indicating that the systemic virulence of the latter strain is not associated with enhanced invasion or induction of enteritis.
FIG. 1.
Systemic virulence of serovar Choleraesuis (SCS) A50, relative to that of serovar Typhimurium (STm) 4/74, in pigs is not associated with enhanced intestinal invasion or enteritis. Panel A shows the magnitude of intestinal invasion by strains of S. enterica serovars Choleraesuis and Typhimurium 3 h after inoculation of porcine distal ileal loops, with the data representing the mean of triplicate determinations in two pigs ± the standard error of the mean (SEM). The dotted line represents the limit of accurate bacterial quantification. The magnitudes of secretory and inflammatory responses induced by the strains 12 h after inoculation of porcine mid-ileal loops are shown in panels B and C, respectively. Data in panels B and C represent the means of triplicate determinations in two pigs ± the standard errors of the means. The secretory response is reported as the ratio of the volume of fluid accumulated (V, ml) to the loop length (L, cm). Neutrophil influx is the ratio of the total gamma radiation emitted from 111In-labeled neutrophils from test loops to that emitted from control loops. *, P < 0.05.
Plasmid partitioning detects reduced net replication of a serovar Choleraesuis A50 aroA mutant following oral inoculation of pigs relative to the parent strain.
To confirm that partitioning of pHSG422 could accurately report the replication rate of Salmonella in the porcine intestines, pigs were inoculated orally with serovar Choleraesuis A50/pHSG422 (seven pigs) or serovar Choleraesuis A50 aroA/pHSG422 (three pigs) grown under conditions permissive for plasmid replication. Forty-eight hours postinoculation, plasmid-bearing and -nonbearing salmonellae were enumerated in homogenates of ileal and colonic mucosa and associated mesenteric lymph nodes. The difference in the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g was calculated and provides a measurement of the titration of the plasmid from the bacterial population and hence the rate of replication (21, 22). The serovar Choleraesuis A50 parent strain was found in overall higher numbers than the isogenic aroA mutant at the tissue sites examined (Fig. 2; total bar height; P values denoted by asterisks). In addition, the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g was greater with the wild type at the sites examined than with the aroA mutant (Fig. 2; difference in height of solid bar compared to total bar height; P values denoted by triangles), indicating that the plasmid had been titrated out of the population at a faster rate owing to the increased net replication of the wild-type strain.
FIG. 2.
Analysis of plasmid partitioning detects differences in the net replication of serovar Choleraesuis (SCS) A50 aroA mutant and parent strains 48 h after oral inoculation of pigs. Triplicate samples of each tissue type were taken, and means were calculated to give a value per animal. Each bar represents the mean bacterial count derived from seven pigs infected with serovar Choleraesuis A50 harboring pHSG422 and three pigs infected with serovar Choleraesuis A50 aroA/pHSG422 ± the standard error of the mean. The solid bars represent the number of bacteria carrying pHSG422, and the hashed bars represent total bacterial numbers. Triangles denote the statistical significance of the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g between the two strains (P < 0.1, 0.05, and 0.01 for one, two, and three triangles, respectively), and asterisks denote statistically significant differences in the log10 total number of CFU/g between strains (P < 0.1, 0.05, 0.01, and 0.001 for one, two, three, and four asterisks, respectively).
Analysis of the net replication of serovar Choleraesuis A50 and serovar Typhimurium 4/74 following oral inoculation of pigs.
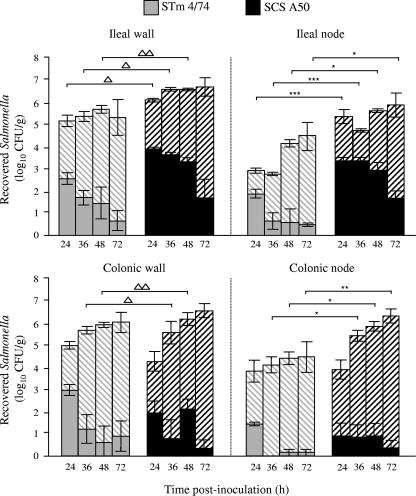
To measure the net replication of strains representing the two serovars following oral inoculation of pigs, a total of 42 pigs were challenged with pHSG422-bearing derivatives of serovar Typhimurium 4/74 (22 pigs) or Choleraesuis A50 (20 pigs) cultured at 25°C. Pigs were then sacrificed in groups of three to nine at 24, 36, 48, or 72 h postinoculation, and the plasmid-bearing and -nonbearing salmonellae were enumerated as described above. In the ileal and colonic mucosa, the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g was significantly greater at 36 and 48 h postinoculation for serovar Typhimurium 4/74 than for serovar Choleraesuis A50, indicating that the replication rate was higher (Fig. 3; P values denoted by triangles). Significantly higher numbers of serovar Choleraesuis A50 CFU were recovered from ileal lymph nodes at all time points and from colonic lymph nodes at 36 to 72 h postinoculation compared to serovar Typhimurium 4/74 (Fig. 3; P values denoted by asterisks). Fewer plasmid-bearing cells of serovar Choleraesuis A50 were recovered from colonic lymph nodes compared to ileal lymph nodes, and it is possible that this reflects differences in the rate of replication at the two sites; however, at both sites for all but one time point, greater numbers of serovar Choleraesuis pHSG422-bearing cells were recovered compared to serovar Typhimurium 4/74-bearing cells (Fig. 3). Thus, the enteric virulence of serovar Typhimurium 4/74 following oral inoculation of pigs is associated with rapid replication in the intestinal mucosa whereas the systemic virulence of serovar Choleraesuis A50 is associated with enhanced persistence in mesenteric lymph nodes. Despite inoculation with comparable doses, by 24 h postinoculation of pigs a significant difference in the level of fecal excretion of the two strains was observed, with serovar Typhimurium 4/74 being excreted at 7.3 ± 0.4 log10 CFU/g compared to serovar Choleraesuis A50/pHSG422 at just 4.1 ± 0.4 log10 CFU/g (P < 0.05).
FIG. 3.
Net replication of serovar Typhimurium (STm) 4/74 and serovar Choleraesuis (SCS) A50 in intestinal mucosa and lymph nodes associates with their differential virulence. Recoveries of total and plasmid-bearing serovar Choleraesuis A50/pHSG422 and serovar Typhimurium 4/74/pHSG422 at intervals of 24, 36, 48, and 72 h after oral inoculation of pigs are shown. Triplicate samples were taken from each tissue, and means were calculated to give a value per animal. Each bar represents the mean bacterial count derived from three to nine pigs per strain (see Materials and Methods) at each time point ± the standard error of the mean. The solid bars represent the number of bacteria carrying pHSG422, and the hashed bars represent the total bacterial numbers. Gray bars represent serovar Typhimurium 4/74, and black bars serovar Choleraesuis A50. Triangles denote the statistical significance of the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g between the two strains (P < 0.1, 0.05, and 0.01 for one, two, and three triangles, respectively), and asterisks denote statistically significant differences in total bacterial numbers between strains (P < 0.1, 0.05, and 0.01 for one, two, and three asterisks, respectively).
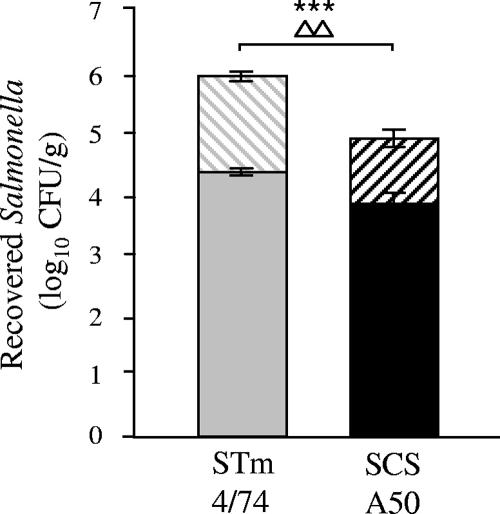
Serovar Typhimurium 4/74 replicates more rapidly than serovar Choleraesuis A50 in ileal mucosa early after bacterial invasion.
To probe replication rates earlier in the course of infection, porcine mid-ileal loops were inoculated with pHSG422-bearing derivatives of serovar Choleraesuis A50 and serovar Typhimurium 4/74 cultured at 25°C. Gentamicin was added 1 h after loop inoculation to kill extracellular bacteria, and the plasmid-bearing and -nonbearing salmonellae were enumerated at 12 h postinoculation as described above. The data indicate that serovar Typhimurium 4/74 reached statistically significantly higher numbers than serovar Choleraesuis in the ileal mucosa in the 12 h after loop inoculation (P < 0.01; Fig. 4), thereby confirming observations in orally challenged animals (Fig. 3). Though comparable numbers of plasmid-bearing cells of the two strains were recovered, the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g was significantly greater for serovar Typhimurium 4/74 than for serovar Choleraesuis (P < 0.05), implying a faster rate of replication. The data also define the growth kinetics of the strains in the same model and for the same duration as inflammatory and secretory responses were measured (above).
FIG. 4.
Serovar Typhimurium (STm) 4/74 replicates more rapidly than serovar Choleraesuis (SCS) A50 in porcine ileal mucosa early after infection. Recoveries of total and plasmid-bearing serovar Choleraesuis A50/pHSG422 and serovar Typhimurium 4/74/pHSG422 12 h after intraluminal inoculation of porcine mid-ileal loops are shown. Gentamicin was added to each loop after 1 h to kill extracellular bacteria. Each bar represents the mean bacterial count derived from triplicate biopsy samples in triplicate loops from two pigs ± the standard error of the mean. The solid bars represent the number of bacteria carrying the pHSG422, and the hashed bars represent total bacterial numbers. Gray bars represent serovar Typhimurium 4/74, and black bars represent serovar Choleraesuis A50. Triangles denote the statistical significance of the difference between the log10 total number of CFU/g and the log10 number of pHSG422-bearing CFU/g between the two strains (P < 0.05), and asterisks denote significant difference between the total bacterial numbers (P < 0.01).
Faster replication of serovar Typhimurium in the intestinal wall is associated with enhanced induction of porcine proinflammatory cytokines early after infection.
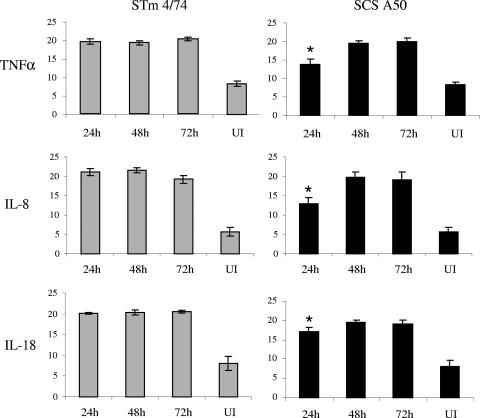
The transcription of genes encoding the porcine proinflammatory cytokines TNF-α, IL-8, and IL-18 was assessed 24, 48, and 72 h after oral inoculation of pigs by using biopsy samples of the same ileal mucosa as used to assess net replication by plasmid partitioning (as described above). Real-time RT-PCR assays were developed, and they revealed strong induction of transcription of each cytokine relative to 28S rRNA at each time interval compared to ileal mucosa from age-matched control uninoculated pigs (Fig. 5). At 24 h postinoculation, serovar Typhimurium 4/74 elicited significantly greater levels of transcription of TNF-α, IL-8, and IL-18 compared to serovar Choleraesuis A50 (Fig. 5; P < 0.01 for TNF-α and IL-8 and <0.05 for IL-18). By 48 and 72 h postinoculation, there were no significant differences in the levels of TNF-α, IL-8, or IL-18 mRNA in ileal mucosa infected with either strain.
FIG. 5.
Serovar Typhimurium (STm) 4/74 induces elevated levels of proinflammatory cytokine mRNAs compared to serovar Choleraesuis (SCS) early after oral inoculation of pigs. Data represent the mean 40-Ct values (a measurement of the abundance of cytokine mRNAs) for porcine TNF-α, IL-8, and IL-18 mRNAs in ileal mucosa 24, 48, and 72 h after oral inoculation of pigs with serovar Typhimurium 4/74 or serovar Choleraesuis A50 or mock infection relative to that for 28S rRNA. The mean was derived from independent determinations from at least three pigs at each time interval ± the standard error of the mean. *, P < 0.05 for elevated transcript levels induced by serovar Typhimurium 4/74 compared to those induced by serovar Choleraesuis A50 at 24 h postinfection. UI, uninfected.
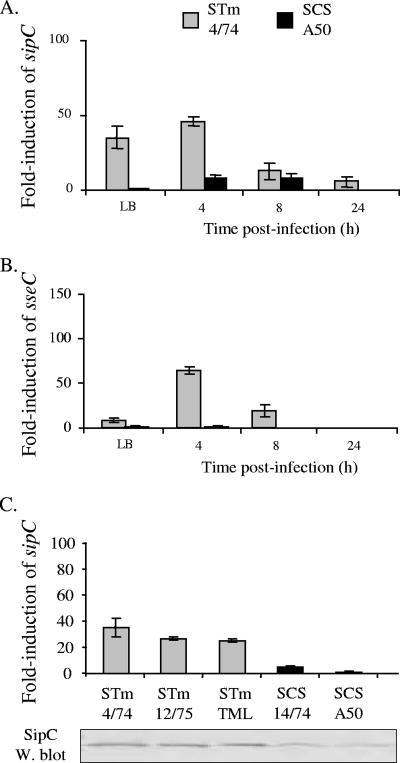
Enhanced epithelial invasion and intracellular proliferation of serovar Typhimurium 4/74 may be associated with elevated expression of T3SS-1 and -2 genes relative to serovar Choleraesuis A50.
To probe the possible basis of enhanced invasion and net growth of serovar Typhimurium 4/74 in porcine ileal tissue relative to serovar Choleraesuis A50, we examined the transcription of genes encoding translocon components of T3SS-1 (sipC) and -2 (sseC), since both systems are known to play key roles in the colonization of porcine intestines by serovar Typhimurium (8). The role of the T3SS-1 translocon was confirmed in this study by the reduced invasion of a serovar Typhimurium 4/74 sipB mutant in porcine ligated ileal loops (Fig. 1). Real-time RT-PCR assays for sipC and sseC mRNA were developed, and n-fold induction was measured relative to the housekeeping gene yejA. Despite being able to detect mRNA of porcine cytokines in RNA extracted from ileal mucosa from pigs inoculated orally with serovar Typhimurium 4/74 or Choleraesuis A50, we were unable to amplify bacterial RNA from the same samples (data not shown). However, during mid-logarithmic-phase growth in LB broth and following infection of porcine primary alveolar macrophages, the n-fold induction of sipC and sseC (relative to yejA) was significantly greater in serovar Typhimurium 4/74 compared to that in serovar Choleraesuis A50 (Fig. 6A and B; P < 0.05). Furthermore, the level of transcription of sipC was found to be consistently higher in three serovar Typhimurium strains compared to two serovar Choleraesuis strains and the elevated level of sipC mRNA was mirrored by the presence of higher levels of SipC protein in the secreted fraction of serovar Typhimurium strains, as detected by Western blotting with a SipC-specific monoclonal antibody (Fig. 6C).
FIG. 6.
Type III secretion-associated genes are expressed at a greater level in serovar Typhimurium (STm) 4/74 compared to serovar Choleraesuis (SCS) A50 during batch culture and infection of porcine alveolar macrophages. n-Fold induction of sipC (panel A) and sseC (panel B) relative to yejA in serovar Typhimurium 4/74 (gray bars) and serovar Choleraesuis (black bars) during mid-logarithmic-phase growth in LB broth (A600, ca. 0.7) and 4, 8, and 24 h postinfection of primary porcine alveolar macrophages is shown. The n-fold induction of sipC mRNA relative to yejA during mid-logarithmic-phase growth in LB was also measured for different serovar Typhimurium and Choleraesuis strains (panel C). Secreted proteins prepared from the same cultures as used for RNA extraction were probed for SipC protein by Western (W.) blotting with a SipC-specific monoclonal antibody (panel C, bottom). All P values for n-fold differences, <0.05.
DISCUSSION
Differences in the outcome of infection of pigs by S. enterica serovars Typhimurium and Choleraesuis are well defined in the field (reviewed in reference 50) and in experimental studies (3, 53). Toward an understanding of the differential virulence of these serovars, we dissected aspects of serovar Typhimurium 4/74 and serovar Choleraesuis A50 infections in pigs. These strains have been extensively characterized following oral inoculation of pigs (53) and infection of porcine alveolar macrophages (53) and in bovine oral challenge and ligated ileal loop models (34). Their behavior in such models is comparable to that of other strains of the same serovar; thus, as far is reasonably practicable given the costly nature of large-animal experimentation, they may be considered representative of the serovar.
Consistent with studies on host-restricted S. enterica serovar Dublin in cattle (34), serovar Abortusovis in lambs (47), and serovar Gallinarum in chickens (9), systemic virulence of serovar Choleraesuis in pigs was not associated with enhanced invasion of ileal mucosa or greater induction of intestinal inflammatory responses compared to serovar Typhimurium 4/74. These data support previous observations that serovar Choleraesuis A50 causes less destruction of porcine intestinal mucosa than does serovar Dublin 3246, as assessed by electron microscopy of ileal mucosa 3 h after loop inoculation (3), even though serovar Choleraesuis A50 is more virulent than serovar Dublin 3246 following oral inoculation of pigs (53). We therefore hypothesized that the differential virulence of serovar Choleraesuis A50 and serovar Typhimurium 4/74 may be a function of distinct replication or killing kinetics in the intestinal mucosa and associated mesenteric lymph nodes.
Net bacterial replication in vivo was measured by analysis of the partitioning of a plasmid that exhibits temperature-sensitive replication. The system was confirmed to be suitable for this purpose by comparison of a serovar Choleraesuis aroA mutant and its parent strain bearing pHSG422 following oral inoculation of pigs. Significantly fewer aroA mutant bacteria were recovered from most intestinal sites, and the difference between the log10 total number of CFU/g and the log10 number of plasmid-bearing CFU/g was significantly lower for the aroA mutant than for the parent strain by 48 h postinoculation, implying a slower rate of replication. These data are consistent with the known role of aromatic amino acid biosynthesis in Salmonella replication in vivo (33). Since the system had been validated, plasmid partitioning was used to measure the net growth of serovar Typhimurium 4/74 and serovar Choleraesuis A50 following oral inoculation of pigs by sacrificing groups of at least 3 pigs at intervals of 24, 36, 48, and 72 h postinfection. The data indicated that serovar Typhimurium 4/74 replicates faster in the ileal and colonic mucosa than does serovar Choleraesuis A50, whereas strain A50 reached higher numbers in draining mesenteric lymph nodes than did strain 4/74. The faster replication rate of serovar Typhimurium 4/74 in intestinal mucosa was associated with elevated levels of porcine proinflammatory cytokines at 24 h after oral inoculation, elevated secretory and inflammatory responses in mid-ileal loops, and elevated fecal excretion relative to that of serovar Choleraesuis A50. In contrast, replication at a slower rate and elevated persistence in mesenteric lymph nodes are associated with the systemic virulence of serovar Choleraesuis A50.
The differences in the magnitudes of induction of proinflammatory cytokine mRNAs detected herein following oral inoculation of pigs with strains of serovars Typhimurium and Choleraesuis are broadly in agreement with those detected in porcine mesenteric lymph nodes in a recent study by Uthe et al. (45). Compared to uninfected control animals, serovar Typhimurium significantly induced IL-8 transcription in ileocecal nodes early after oral infection (24 and 48 h) whereas IL-8 was initially repressed at 8 to 24 h after serovar Choleraesuis infection and then observed to be induced at later times (2 and 7 days) (45). In that study and in the present one, it is possible that differences in earlier transcriptional responses to the strains that may influence the outcome of infection were missed. Studies using the porcine jejunal epithelial cell line IPEC-J2 have indicated that a strain of serovar Typhimurium elicits significantly higher levels of IL-8 protein and mRNA than serovar Choleraesuis at 3 and 6 h after invasion (39, 40). Conflicting data exist on the induction of IL-8 mRNA at just 1.5 h after invasion of IPEC-J2 cells, with one study detecting no difference between the serovars (39) and another reporting significantly elevated levels of IL-8 in response to serovar Typhimurium (40). Cytokine responses were not correlated with the magnitude of invasion or net bacterial replication in these studies.
One may speculate that the rapid replication of serovar Typhimurium would trigger inflammatory responses earlier following infection and of a greater magnitude, which may confine the infection locally to the intestinal mucosa, whereas slow replication of serovar Choleraesuis may enable it to evade the activation of host innate immunity, thus facilitating dissemination by stealth. Differences in the level of induction of proinflammatory responses likely also explain why serovar Typhimurium 4/74 elicits stronger enteropathogenic responses in ligated loops and following oral inoculation than does serovar Choleraesuis A50. It remains unclear if the elevated fecal excretion of serovar Typhimurium 4/74 compared to serovar Choleraesuis A50 shortly after oral inoculation of pigs with comparable doses results from faster replication in the intestinal wall and/or lumen or reflects differences in the frequency and/or consistency of passed fecal matter.
The connection between the bacterial replication rate and disease outcome is reinforced by the identification of serovar Typhimurium mutants that overgrow in macrophages because of their inability to stimulate host nitric oxide synthase but are attenuated in BALB/c mice (17). These and other findings (reviewed in reference 43) imply that control of net replication by Salmonella may confer a survival advantage. In an attempt to dissect the molecular mechanisms that dictate the differential growth rates of serovar Typhimurium 4/74 and serovar Choleraesuis A50, we examined the transcription of genes encoding translocon components of T3SS-1 and T3SS-2. Both systems are known to play key roles in colonization of the porcine intestines by serovar Typhimurium (5, 8; Fig. 1A), and their activity has been correlated with the progression of typhoid fever in murine models. T3SS-1-injected effector proteins stimulate rearrangements of the subcortical actin cytoskeleton that facilitate bacterial invasion; however, T3SS-1 also contributes to bacterial growth in epithelial cells (42) and the T3SS-1 effector SopB stimulates nitric oxide production and remains active long after bacterial entry (16). T3SS-1 expression in vivo is also correlated with induction of a lysosomal repair response that can control Salmonella infection (35). T3SS-2 facilitates intracellular proliferation by modulating the trafficking of the Salmonella-containing vacuole. It is induced in the intestinal lumen (6), and contextual regulation of T3SS-2 is important in the progression of typhoid fever following oral dosing of mice with serovar Typhimurium (14).
In the strains of well-defined virulence used, transcription of sipC and sseC was consistently higher in serovar Typhimurium 4/74 than in serovar Choleraesuis A50 during mid-logarithmic-phase growth in vitro and following infection of porcine primary alveolar macrophages. We were unable to measure transcript levels in the same tissues as plasmid partitioning was assessed, presumably as they were present below the limit of detection since RNA extracted from the tissues yielded amplicons for porcine cytokine mRNA. Immunohistochemistry with antibodies directed against T3SS-1 and -2 effector proteins or epitope-tagged variants thereof also failed to detect expression of the proteins in vivo, presumably owing to the small quantities present (data not shown). Transcription of sipC was consistently higher in serovar Typhimurium strains that show behavior comparable to that of strain 4/74 in porcine macrophages (53), and the difference from serovar Choleraesuis strains was also reflected in the quantities of SipC protein produced.
In addition to differences in the levels of transcription of the genes for T3SS-1 and -2, our analysis of the genomes of sequenced S. enterica serovars reveals that the genes encoding the effector proteins SopA, SlrP, SseI, and SspH2 are pseudogenes in serovar Choleraesuis SC-B67 and SseK2 is absent, yet the genes are intact in serovar Typhimurium LT2. Of these, SopA is known to be important in the induction of enteritis by serovar Dublin in bovine ligated ileal loops (54). We confirmed that SopA is a pseudogene in serovar Choleraesuis A50 by nucleotide sequencing and Western blot analysis with a SopA-specific monoclonal antibody (data not shown). Such polymorphisms in the repertoire, sequence, and expression of T3SS-related genes may explain the reduced invasion and inflammatory potential of serovar Choleraesuis; however, it is likely that other factors are involved. Of 151 pseudogenes in the serovar Choleraesuis SC-B67 genome, 64 were predicted to be metabolism related (12). Furthermore, the shdA gene, which encodes a fibronectin-binding protein required for Peyer's patch colonization and fecal shedding in mice (27) and enteric virulence early after oral inoculation of pigs (4), is a pseudogene in SC-B67 relative to LT2. It is equally possible that such defects may explain the reduced replication rate of serovar Choleraesuis compared to serovar Typhimurium in intestinal epithelia; however, a requirement exists to confirm that the polymorphisms also exist between the two strains examined herein. Mutational attrition of several predicted regulators has also been described previously in SC-B67 (12); however, the PhoPQ two-component system known to play a key role in intracellular proliferation (reviewed in references 20 and 43) appears to be intact in the sequenced genomes. Polynucleotide phosphorylase, which has been proposed to alter the balance between acute infection and persistence by modulating SPI-1 and -2 genes (13), is also identical in strains SC-B67 and LT2.
The importance of T3SS in the translocation of serovar Choleraesuis from the porcine intestines remains ill defined. A serovar Choleraesuis hilA mutant defective in the regulation of T3SS-1 was less able to colonize the intestines and disseminate to the liver and spleen following oral inoculation of pigs, but not following intraperitoneal infection (29). The role played by T3SS-2 in dissemination from the porcine intestines is unknown. In recent studies with calves, early translocation of serovar Dublin from the intestines to mesenteric lymph nodes, the liver, and the spleen was found to occur independently of T3SS-2 (G. D. Pullinger et al., submitted for publication); thus, caution is required in interpreting the importance of variations in the expression or sequence of T3SS-related genes until their precise contribution to pathogenesis is known.
As elegantly reviewed in reference 43, the rate of intracellular proliferation of Salmonella is strictly dependent on the cell type, occurring rapidly in immortalized epithelial and macrophage lines, whereas in dendritic cells and fibroblasts a homogeneous population in a nonreplicating state exists. It is unclear if the differences in net replication of the S. enterica strains examined here reflect the targeting of different cell populations. Understanding of cell tropism is of key importance when probing the roles of bacterial factors in net growth, since mutation of some systems can have different effects in different cell types. For example, SPI-2 is dispensable for survival and intracellular proliferation in dendritic cells even though it is induced (26) and mutation of PhoPQ enhances growth in fibroblasts but inhibits it in other cell types (7). The data reported here indicate that differences in replication rate and host innate immune activation by S. enterica serovars may be under bacterial control and have profound implications for disease progression.
Acknowledgments
We gratefully acknowledge the support of the Biotechnology & Biological Sciences Research Council (grant 201/S14753) and the Department for the Environment, Food and Rural Affairs (grant OZ0319).
We thank T. Hashimoto-Gotoh for the kind gift of pHSG422.
Editor: F. C. Fang
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Bakshi, C. S., V. P. Singh, M. W. Wood, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin, W. H., Jr., P. Hall, S. J. Roberts, and D. E. Briles. 1990. The primary effect of the Ity locus is on the rate of growth of Salmonella typhimurium that are relatively protected from killing. J. Immunol. 144:3143-3151. [PubMed] [Google Scholar]
- 3.Bolton, A. J., M. P. Osborne, T. S. Wallis, and J. Stephen. 1999. Interaction of Salmonella choleraesuis, Salmonella dublin and Salmonella typhimurium with porcine and bovine terminal ileum in vivo. Microbiology 145:2431-2441. [DOI] [PubMed] [Google Scholar]
- 4.Boyen, F., F. Pasmans, E. Donne, F. van Immerseel, E. Morgan, C. Adriaensen, J. P. Hernalsteens, T. S. Wallis, R. Ducatelle, and F. Haesebrouck. 2006. The fibronectin binding protein ShdA is not a prerequisite for long term faecal shedding of Salmonella typhimurium in pigs. Vet. Microbiol. 115:284-290. [DOI] [PubMed] [Google Scholar]
- 5.Boyen, F., F. Pasmans, F. van Immerseel, E. Morgan, C. Adriaensen, J. P. Hernalsteens, A. Decostere, R. Ducatelle, and F. Haesebrouck. 2006. Salmonella Typhimurium SPI-1 genes promote intestinal but not tonsillar colonization in pigs. Microbes Infect. 8:2899-2907. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. F., B. A. Vallance, B. K. Coombes, Y. Valdez, R. A. Coburn, and B. B. Finlay. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog. 1:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano, D. A., M. Martinez-Moya, M. G. Pucciarelli, E. A. Groisman, J. Casadesus, and F. Garcia-Del Portillo. 2001. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69:6463-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnell, S., A. Bowen, E. Morgan, D. J. Maskell, T. S. Wallis, and M. P. Stevens. 2007. Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis. Microbiology 153:1940-1952. [DOI] [PubMed] [Google Scholar]
- 9.Chadfield, M. S., D. J. Brown, S. Aabo, J. P. Christensen, and J. E. Olsen. 2003. Comparison of intestinal invasion and macrophage response of Salmonella Gallinarum and other host-adapted Salmonella enterica serovars in the avian host. Vet. Microbiol. 92:49-64. [DOI] [PubMed] [Google Scholar]
- 10.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu, C. H., L. H. Su, and C. Chu. 2004. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin. Microbiol. Rev. 17:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, C. H., P. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y. Y. Chou, H. S. Wang, and Y. S. Lee. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements, M. O., S. Eriksson, A. Thompson, S. Lucchini, J. C. Hinton, S. Normark, and M. Rhen. 2002. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 99:8784-8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 102:17460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, R. H., R. Dalziel, J. C. Gibbens, J. W. Wilesmith, J. M. Ryan, S. J. Evans, C. Byrne, G. A. Paiba, S. J. Pascoe, and C. J. Teale. 2004. National survey for Salmonella in pigs, cattle and sheep at slaughter in Great Britain (1999-2000). J. Appl. Microbiol. 96:750-760. [DOI] [PubMed] [Google Scholar]
- 16.Drecktrah, D., L. A. Knodler, K. Galbraith, and O. Steele-Mortimer. 2005. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell. Microbiol. 7:105-113. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, S., J. Bjorkman, S. Borg, A. Syk, S. Pettersson, D. I. Andersson, and M. Rhen. 2000. Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell. Microbiol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 19.Fallon, M. T., W. H. Benjamin, Jr., T. R. Schoeb, and D. E. Briles. 1991. Mouse hepatitis virus strain UAB infection enhances resistance to Salmonella typhimurium in mice by inducing suppression of bacterial growth. Infect. Immun. 59:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulig, P. A., and T. J. Doyle. 1993. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect. Immun. 61:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulig, P. A., T. J. Doyle, M. J. Clare-Salzler, R. L. Maiese, and H. Matsui. 1997. Systemic infection of mice by wild-type but not Spv− Salmonella typhimurium is enhanced by neutralization of gamma interferon and tumor necrosis factor alpha. Infect. Immun. 65:5191-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 24.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, S., T.-Y. Poh, N. Bumstead, and P. Kaiser. 2007. Re-evaluation of the chicken MIP family of chemokines and their receptors suggests that CCL5 is the prototypic MIP family chemokine, and that different species have developed different repertoires of both the CC chemokines and their receptors. Dev. Comp. Immun. 31:72-86. [DOI] [PubMed] [Google Scholar]
- 26.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 27.Kingsley, R. A., K. van Amsterdam, N. Kramer, and A. J. Baumler. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtensteiger, C. A., and E. R. Vimr. 2003. Systemic and enteric colonization of pigs by a hilA signature-tagged mutant of Salmonella choleraesuis. Microb. Pathog. 34:149-154. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative transcription data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.McFarland, W. C., and B. A. D. Stocker. 1987. Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3:129-141. [DOI] [PubMed] [Google Scholar]
- 32.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 33.O'Callaghan, D., D. Maskell, F. Y. Liew, C. S. Easmon, and G. Dougan. 1988. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attenuation, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 56:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulin, S. M., P. R. Watson, A. R. Benmore, M. P. Stevens, P. W. Jones, B. Villarreal-Ramos, and T. S. Wallis. 2002. Analysis of Salmonella enterica serotype-host specificity in calves: avirulence of S. enterica serotype Gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infect. Immun. 70:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy, D., D. R. Liston, V. J. Idone, A. Di, D. J. Nelson, C. Pujol, J. B. Bliska, S. Chakrabarti, and N. W. Andrews. 2004. A process for controlling intracellular bacterial infections induced by membrane injury. Science 304:1515-1518. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz, K. J. 1999. Salmonellosis, p. 535-551. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Disease of swine, 8th ed. Iowa State University Press, Ames.
- 37.Shea, J. E., C. R. Beuzon, C. Gleeson, R. Mundy, and D. W. Holden. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun. 67:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skjolaas, K. A., T. E. Burkey, S. S. Dritz, and J. E. Minton. 2006. Effects of Salmonella enterica serovars Typhimurium (ST) and Choleraesuis (SC) on chemokine and cytokine expression in swine ileum and jejunal epithelial cells. Vet. Immunol. Immunopathol. 111:199-209. [DOI] [PubMed] [Google Scholar]
- 40.Skjolaas, K. A., T. E. Burkey, S. S. Dritz, and J. E. Minton. 2007. Effects of Salmonella enterica serovar Typhimurium, or serovar Choleraesuis, Lactobacillus reuteri and Bacillus licheniformis on chemokine and cytokine expression in the swine jejunal epithelial cell line, IPEC-J2. Vet. Immunol. Immunopathol. 115:299-308. [DOI] [PubMed] [Google Scholar]
- 41.Sojka, W. J., C. Wray, J. Shreeve, and A. J. Benson. 1977. Incidence of Salmonella infection in animals in England and Wales 1968-1974. J. Hyg. Camb. 78:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43-54. [DOI] [PubMed] [Google Scholar]
- 43.Tierrez, A., and F. García-del Portillo. 2005. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell. Microbiol. 7:901-909. [DOI] [PubMed] [Google Scholar]
- 44.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uthe, J. J., A. Royaee, J. K. Lunney, T. J. Stabel, S. H. Zhao, C. K. Tuggle, and S. M. Bearson. 2007. Porcine differential gene expression in response to Salmonella enterica serovars Choleraesuis and Typhimurium. Mol. Immunol. 44:2900-2914. [DOI] [PubMed] [Google Scholar]
- 46.Uzzau, S., D. J. Brown, T. Wallis, S. Rubino, G. Leori, S. Bernard, J. Casadesus, D. J. Platt, and J. E. Olsen. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzzau, S., G. S. Leori, V. Petruzzi, P. R. Watson, G. Schianchi, D. Bacciu, V. Mazzarello, T. S. Wallis, and S. Rubino. 2001. Salmonella enterica serovar-host specificity does not correlate with the magnitude of intestinal invasion in sheep. Infect. Immun. 69:3092-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Diemen, P. M., M. B. Kreukniet, L. Galina, N. Bumstead, and T. S. Wallis. 2002. Characterisation of a resource population of pigs screened for resistance to salmonellosis. Vet. Immunol. Immunopathol. 88:183-196. [DOI] [PubMed] [Google Scholar]
- 49.Vugia, D. J., M. Samuel, M. M. Farley, R. Marcus, B. Shiferaw, S. Shallow, K. Smith, and F. J. Angulo. 2004. Invasive Salmonella infections in the United States, FoodNet, 1996-1999: incidence, serotype distribution, and outcome. Clin. Infect. Dis. 38(Suppl. 3):S149-S156. [DOI] [PubMed] [Google Scholar]
- 50.Wallis, T. S., and P. A. Barrow. July 2005, posting date. Chapter 8.6.2.1. Salmonella epidemiology and pathogenesis in food-producing animals. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://ecosal.org. [DOI] [PubMed]
- 51.Wallis, T. S., S. M. Paulin, J. S. Plested, P. R. Watson, and P. W. Jones. 1995. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect. Immun. 63:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson, P. R., S. M. Paulin, A. P. Bland, P. W. Jones, and T. S. Wallis. 1995. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect. Immun. 63:2743-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, P. R., S. M. Paulin, P. W. Jones, and T. S. Wallis. 2000. Interaction of Salmonella serotypes with porcine macrophages in vitro does not correlate with virulence. Microbiology 146:1639-1649. [DOI] [PubMed] [Google Scholar]
- 54.Wood, M. W., M. A. Jones, P. R. Watson, A. M. Siber, B. A. McCormick, S. Hedges, R. Rosqvist, T. S. Wallis, and E. E. Galyov. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell. Microbiol. 2:293-303. [DOI] [PubMed] [Google Scholar]
- 55.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]