Abstract
The 6-kDa early secreted antigenic target ESAT-6 and the 10-kDa culture filtrate protein CFP-10 of Mycobacterium tuberculosis are secreted by the ESX-1 system into the host cell and thereby contribute to pathogenicity. Although different studies performed at the organismal and cellular levels have helped to explain ESX-1-associated phenomena, not much is known about how ESAT-6 and CFP-10 contribute to pathogenesis at the molecular level. In this study we describe the interaction of both proteins with lipid bilayers, using biologically relevant liposomal preparations containing dimyristoylphosphatidylcholine (DMPC), dimyristoylphosphatidylglycerol, and cholesterol. Using floatation gradient centrifugation, we demonstrate that ESAT-6 showed strong association with liposomes, and in particular with preparations containing DMPC and cholesterol, whereas the interaction of CFP-10 with membranes appeared to be weaker and less specific. Most importantly, binding to the biomembranes no longer occurred when the proteins were present as a 1:1 ESAT-6·CFP-10 complex. However, lowering of the pH resulted in dissociation of the protein complex and subsequent protein-liposome interaction. Finally, cryoelectron microscopy revealed that ESAT-6 destabilized and lysed liposomes, whereas CFP-10 did not. In conclusion, we propose that one of the main features of ESAT-6 in the infection process of M. tuberculosis is the interaction with biomembranes that occurs after dissociation from its putative chaperone CFP-10 under acidic conditions typically encountered in the phagosome.
Mycobacterium tuberculosis is one of the most successful human pathogens, infecting nearly one-third of the world's population. Among the various factors that contribute, it is certainly the bacterium's ability to multiply and persist within professional phagocytic cells that is of primary importance (30).
The extended RD1 region of M. tuberculosis encodes ESX-1, a novel protein secretion system involved in the immunogenicity and pathogenicity. The system, which is absent from the attenuated vaccines Mycobacterium bovis BCG and Mycobacterium microti (4, 6, 24), is responsible for the export of the 6-kDa early secreted antigenic target ESAT-6 and the 10-kDa culture filtrate protein CFP-10. ESX-1 is present in the saprophyte Mycobacterium smegmatis (9), where it acts in DNA uptake (11), which suggests that pathogenic mycobacteria might have adapted an ancestral conjugation system for protein secretion, which is required for survival and multiplication in the host cell.
The importance of ESX-1 proteins for pathogenicity was shown by reintroduction of the extended RD1 region into BCG (34), deletion of RD1 from M. tuberculosis (23), signature-tagged and insertional mutagenesis (8, 40, 46), and targeted gene deletions (5, 15, 18). Several effects related to pathogenicity have been found to be associated with the expression of ESX-1 in M. tuberculosis and/or Mycobacterium marinum, a fish pathogen that harbors an ESX-1 system similar to that of M. tuberculosis (14, 27). These include suppression of proinflammatory responses (46), interaction with Toll-like receptor 2 (33) and/or syntenin (42), cytotoxicity and/or cytolysis (10, 12, 18), necrosis (19), phagosome maturation arrest (49), and granuloma formation (52).
To further explore the involvement of ESAT-6 and CFP-10 in these processes, we have elaborated different approaches to investigate the interaction of ESAT-6 and CFP-10 with biomembranes, which constitute important potential interaction partners, once the bacterium is engulfed by professional phagocytic cells.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and recombinant ESAT-6 (rESAT-6) purification.
The BCG::RD1-2F9 strain, used to isolate culture filtrates containing wild-type ESAT-6 and CFP-10, has been previously described (34). Cell-free protein extracts were prepared from early-log-phase cultures of M. tuberculosis strain H37Rv, used for purification of native ESAT-6 (nESAT-6), or BCG::RD1-2F9 Pasteur strains grown in Sauton's medium. Culture filtrates were concentrated using Millipore filters with a 3-kDa cutoff.
Escherichia coli strain BL21(DE3) was transformed with expression vector pMRLB7 or pMRLB46 for the expression of His-tagged ESAT-6 or CFP-10, respectively. Cells were grown in 1 liter of LB medium supplemented with ampicillin (100 μg/ml) at 37°C. When the cultures reached an optical density at 600 nm of 0.6, isopropyl-β-d-galactopyranoside (IPTG) was added to a final concentration of 1 mM. After 3 h of incubation at 37°C, cells were harvested by centrifugation, and cell pellets were washed once with protein extraction buffer containing 100 mM sodium phosphate buffer and 150 mM NaCl (pH 7) and resuspended in 50 ml of protein extraction buffer supplemented with a cocktail of protease inhibitors (Complete; Roche Diagnostics GmbH, Mannheim, Germany). The cells were lysed by sonication (Branson Sonifier 250). Lysates were centrifuged at 11,500 × g for 10 min at 4°C, and the pellets were resuspended in 1 ml of 8 M urea, 100 mM sodium phosphate, and 300 mM NaCl, pH 8. The denatured lysates were cleared by centrifugation at 89,000 × g; stepwise dialyzed in phosphate-buffered saline (PBS) containing 3 M, 1 M, and 0 M urea for refolding; and loaded onto a 2-ml Ni2+-nitrilotriacetic acid-agarose column equilibrated with PBS. The proteins were eluted in PBS with 250 mM imidazole. The purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining (data not shown).
Purification of nESAT-6.
Sauton culture filtrates were concentrated under 2 bars of nitrogen pressure using an ultrafiltration membrane with a 3-kDa cutoff and a diameter of 150 mm (Millipore, Bedford, MA) and then washed three times with H2O containing 4% butanol (to reach a resistivity of lower than 30 μS) and lyophilized. The lyophilized samples were rehydrated with a buffer containing 100 mM NaCl and 50 mM Na2HPO4; the pH was adjusted to 7.1 with 1 M KH2PO4. The samples were centrifuged for 1 h at 20,000 × g and then passed through a 0.22-μm filter and applied to a Superdex G75 (Amersham Pharmacia Biotech) column after the addition of 4% butanol. The ESAT-6 containing fractions were collected, pooled, washed, concentrated by ultrafiltration as described above, and applied to the strong basic ion exchange column R15 Q (Amersham Pharmacia Biotech). Protein was eluted in a 20 mM Tris-HCl (pH 8.1) buffer with a linear NaCl gradient (0 to 150 mM). The ESAT-6-containing fractions were collected, pooled, desalted, and further purified by reversed-phase high-pressure liquid chromatography (Amersham Pharmacia Biotech) in an ammonium acetate buffer (20 mM). Protein was eluted from the column with a linear acetonitrile (CH3CN) gradient (0 to 90%). The ESAT-6-containing fractions were collected and pooled, and acetonitrile was evaporated using a Rotavapor. The high-pressure liquid chromatography purification was repeated twice with C4 silica-based reversed-phase columns with two different acetonitrile gradients (45 to 55% and 48.5 to 51%). The evaporated samples were analyzed by N-terminal amino acid sequencing and mass spectroscopy (matrix-assisted laser desorption ionization and electrospray ionization) to confirm the purity. All purified nESAT-6 samples were lyophilized and stored at −20°C, as was also done between the different purification steps to maintain the stability of the protein.
Preparation of liposomes.
The standard preparations of liposomes used in this study were made of dimyristoylphosphatidylcholine (DMPC) (Rhöne-Poulenc Rorer, Köln, Germany), dimyristoylphosphatidylglycerol (DMPG) (a gift from Lipoïd GmbH, Ludwigshafen, Germany), and cholesterol (Sigma, St. Louis, MO) in a 4:1:1 molar ratio. All lipids were dissolved in CHCl3-methanol (2:1, vol/vol) and mixed in a round-bottom flask. The solvent was evaporated using a Rotavapor and subsequently flushed with nitrogen for 30 min. The lipid film was hydrated in 50 mM Tris and 150 mM NaCl, pH 7.4 (TBS), by shaking with glass beads and subsequently extruded four times through a 400-nm filter. The phospholipid concentration of the liposomes was determined by the colorimetric method of Rouser et al. (38). The mean particle size and size distribution were determined by dynamic light scattering with a Malvern 4700 system (Malvern Ltd., Malvern, United Kingdom).
Floatation gradient centrifugation.
Purified proteins (5 μg) or culture filtrate (100 μg) was incubated for 10 min at 37°C in the absence or presence of 10 mM liposomes in a final volume of 100 μl and subjected to floatation analysis using a discontinuous sucrose density gradient as previously described by others (16). After incubation, the samples were mixed with 80% sucrose in TBS. Onto this suspension 2 ml of 60% sucrose, 1.5 ml of 50%, and 500 μl of TBS without sucrose were layered. The gradient was centrifuged at 115,000 × g for 4 h at 4°C. Fractions (1 ml) were taken from the top, trichloroacetic acid precipitated, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15%) and immunoblotting using monoclonal anti-ESAT-6 (AntibodyShop, Statens Serum Institute, Denmark) or polyclonal anti-CFP-10 antibodies (34).
To investigate the influence of ESAT-6·CFP-10 complex formation on the interaction with lipids, we incubated equimolar amounts (500 pmol) of rESAT-6 and CFP-10 for 10 min at room temperature as previously described by Renshaw et al. (37). To test the influence of the pH on the dissociation of the ESAT-6·CFP-10 complex and subsequent lipid association, the above-described floatation gradient centrifugation was repeated using three different buffers: 50 mM Na acetate and 150 mM NaCl (pH 4), 50 mM Na acetate and 150 mM NaCl (pH 5), and 50 mM NaH2PO4 and 150 mM NaCl (pH 6). The buffer of the concentrated culture filtrates was changed using Microcon centrifugal filter devices with a cutoff of 3 kDa (Millipore, Bedford, MA).
Surface plasmon resonance assays.
The PentaHis monoclonal antibody (QIAGEN) was covalently coupled to a CM5 sensor chip on a Biacore 2000 instrument at 25°C to a level of 10,000 resonance units. rCFP-10 (200 nM) was then captured noncovalently to a level of 850 resonance units on the penta-His surface in 20 mM phosphate, 50 mM NaCl, and 1 mM EDTA (pH 6.5), followed by nESAT-6 (1 μM), and then 10 mM sodium acetate at different pHs (5.5, 5.0, 4.5, or 4.0) was injected for 1 minute at 20 μl·min−1 during the dissociation phase. Control experiments were performed on a CFP-10 surface devoid of ESAT-6.
Cryoelectron microscopy.
Fifty microliters of DMPC-, DMPG-, and cholesterol-containing liposomes (molar ratio, 4:1:1) was incubated with 500 pmol rESAT-6, nESAT-6, or rCFP-10 protein (protein/phospholipid ratio of around 1:25,000) for 10 min at 37°C in TBS, pH 7.4. For cryoelectron microscopy, 4 μl of sample was applied to homemade holey carbon grids and vitrified in liquid ethane using a cryofixation device obtained from F. Livolant and A. Leforestier. The specimens were transferred to a Gatan 626 DH cryoholder and examined with an FEG 2010F electron microscope (Jeol, Tokyo, Japan) operating at 200 kV. Images were recorded on Kodak SO163 film or using a 1,024-by-1,024 charge-coupled device Gatan camera (Gatan 694 slow-scan charge-coupled device camera) at a nominal magnification of 25,000 with 2.0 μm of underfocus under low-dose conditions. The negative control (liposomes without protein) was included in every experiment to exclude artifacts. Images were made from three or four independent experiments per sample.
RESULTS
Interaction of ESAT-6 and CFP-10 with lipid bilayers.
To study the potential interaction of ESAT-6 and CFP-10 with biomembranes, we incubated extruded liposomes with purified ESAT-6 and CFP-10 proteins or with concentrated culture filtrate of BCG::RD1 and subjected these mixtures to floatation gradient centrifugation. To reflect a biologically relevant situation, the liposomes used in this assay contained phosphatidylcholine (PC), phoshphatidylglycerol (PG), and cholesterol in a ratio of 4:1:1. Zwitterionic phosholipid PC is the main component of biological membranes and, together with the negatively charged phospholipid PG, belongs to the surfactant lipids produced by alveolar type II cells (51), while cholesterol is an important component of phagosomal membranes (13, 22).
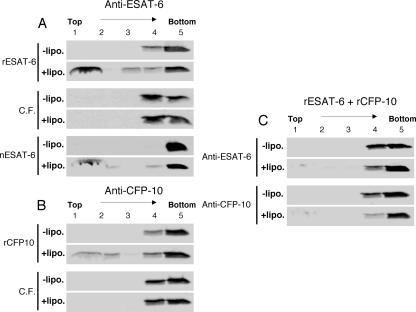
Floatation gradient centrifugation is a method in which the protein-lipid interaction is measured by the presence of the protein in the liposome-containing top fraction. As shown in Fig. 1A and B, both purified recombinant proteins were found in the upper fractions after incubation with liposomes, indicating interaction of ESAT-6 or CFP-10 with lipid bilayers. In parallel, nESAT-6 from M. tuberculosis without a hexahistidine tag showed a similar floatation pattern, indicating that the interaction was independent of the tag (Fig. 1A).
FIG. 1.
Binding of purified recombinant, native, or culture filtrate-derived (C.F.) ESAT-6 and CFP-10 to liposomes studied by liposome floatation analysis. (A and B) Protein samples were incubated for 10 min at 37°C in the presence (+lipo.) or absence (−lipo.) of liposomes and subsequently applied to a sucrose density gradient. (C) Equal quantities of rESAT-6 and rCFP-10 were incubated for 5 min at room temperature and subsequently for 10 min in the presence or absence of liposomes. Gradients were collected in five fractions from the top (1 to 5), trichloroacetic acid precipitated, and analyzed by immunoblotting using a monoclonal antibody for ESAT-6 (anti-ESAT-6) or affinity-purified polyclonal CFP-10 antibodies (anti-CFP-10).
In contrast, no cofloatation with liposomes of either ESAT-6 or CFP-10 was found when concentrated BCG::RD1 culture filtrate was tested (Fig. 1A and B). To further evaluate this observation, equal amounts of purified ESAT-6 and CFP-10 proteins were incubated to allow the formation of a 1:1 protein complex prior to the addition of liposomes, and this mixture was subjected to floatation analysis. As depicted in Fig. 1C, hardly any of the ESAT-6 or CFP-10 was detected in the top fractions of this assay, confirming that ESAT-6 and CFP-10 do not associate with the lipid bilayer when they occur as a 1:1 complex.
Complex dissociation and subsequent lipid interaction induced by pH shift.
Previous structural analyses revealed that the tight interaction between ESAT-6 and CFP-10 is stabilized by two salt bridges (36). As pH changes are known to be key factors inside the phagosome, we tested the protein-lipid interaction at different pH levels. First, we repeated the floatation experiments with concentrated culture filtrates at pH 6, pH 5, and pH 4 instead of pH 7.4. While ESAT-6 and CFP-10 were not found in the top fractions of the assay mixture when the experiments were performed at pH 5 or pH 6, both proteins were in the top fractions at pH 4 (Fig. 2). These results suggest that between pH 5 and pH 4, which is close to the pI of ESAT-6 and CFP-10 (≈ 4.5), the complex dissociates, allowing the released monomeric ESAT-6 or CFP-10 molecules to interact with the biomembranes.
FIG. 2.
Liposome floatation analysis using BCG::RD1-derived culture filtrates in buffers with different pHs. Culture filtrates of BCG::RD1 were incubated with liposomes in the appropriate buffers for 10 min at 37°C, applied to sucrose density gradients, and processed for immunoblotting.
Most interestingly, surface plasmon resonance assays using a Biacore 2000 instrument further confirmed that while the ESAT-6·CFP-10 complex dissociated slowly at pH 6.5 (koff = 9 × 10−4 s−1; half-life ≈ 13 min), 1-minute pulses of lower-pH solutions induced a sudden disruption of an increasing proportion of the complexes (30% at pH 5.5, 57% at pH 5.0, 84% at pH 4.5, and 99% at pH 4.0) (Fig. 3), clearly indicating that the ESAT-6·CFP-10 complex dissociates at acidic pH values that are encountered in the phagosome.
FIG. 3.
Determination of the effect of acidic pH on the stability of the CFP-10·ESAT-6 complex by surface plasmon resonance (SPR). nESAT-6 was injected at pH 6.5 on a CFP-10 surface from time t0 to t1. The complex was allowed to dissociate spontaneously at pH 6.5 until time t2, when 1-minute pulses at different pHs were applied (until t3), leading to a partial or total disruption of the complexes. RU, resonance units (1 RU ≈ 1 pg·mm−2).
Specific lipids or lipid complexes involved in ESAT-6 and CFP-10 interaction.
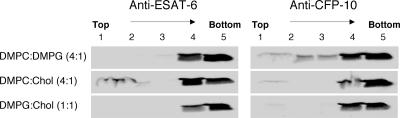
To determine the specificity of the protein-liposome interactions, we tested three different liposome preparations derived from the standard preparation, DMPC-DMPG-cholesterol (4:1:1). Each of the three preparations combined two of the three lipids previously used in the standard preparation. The different combinations retained the molecular ratio used in the standard preparation: DMPC-DMPG (4:1), DMPC-cholesterol (4:1), and DMPG-cholesterol (1:1). Figure 4 shows that for binding of ESAT-6, the presence of PC and cholesterol was important, while the interaction of CFP-10 with these liposomes appeared to be weaker and less specific.
FIG. 4.
Liposome floatation analysis with liposomes containing different lipids. To analyze the specificity of lipid interaction, rESAT-6 or rCFP-10 was incubated with DMPC-DMPG (4:1), DMPC-cholesterol (4:1), or DMPG-cholesterol (1:1) liposomes; applied to sucrose gradients; and processed for immunoblotting.
Effect of the interactions of ESAT-6 and CFP-10 with liposomal membranes.
Cryoelectron microscopy is a very powerful method to visualize potential changes in the shape of cellular structures. Therefore, the liposomes were incubated with various ESAT-6 or CFP-10 protein preparations and subjected to cryoelectron microscopy analysis. As shown in Fig. 5, the addition of small quantities of ESAT-6 to the liposomes (protein/phosholipid ratio, 1:2.5 × 104) resulted in rigidified liposomal membranes and fragments derived from disrupted liposomes. At higher magnification, one can clearly see that the first changes in the lipid membrane, due to the addition of ESAT-6, consist of nicks, which progress over time to cause lysis of the liposomes. While these effects were observed for both the rESAT-6 and the nESAT-6, such a phenomenon was never observed when the liposomes were incubated with CFP-10. Indeed, as shown in Fig. 5B, after incubation with CFP-10 the liposomes remained completely round and intact, similar to the controls without addition of protein. These observations were highly reproducible and were independent of the individual ESAT-6 and CFP-10 preparations that were used.
FIG. 5.
Cryoelectron microscopy analysis of liposomes with or without ESAT-6 or CFP-10. Typical electron cryomicrographs of liposomes are shown. (A and B) Liposomes alone (A) and liposomes incubated with rCFP-10 (B), where no lysis was observed. (C to F) Liposomes incubated with nESAT-6 (C, E, and F) or rESAT-6 (D), all showing rigidification of the membranes and lysis of the liposomes. Bars, 100 nm (A, B, and D) and 50 nm (C, E, and F).
DISCUSSION
Recent studies on ESAT-6 and ESX-1 have highlighted the involvement of this novel secretion system in the pathogenesis and immunogenicity of M. tuberculosis. One of the ultimate aims now is to elucidate the actual role of ESAT-6 and CFP-10 in these processes. The two proteins have been shown to form a 1:1 complex in vitro (37), and nuclear magnetic resonance structure analysis of the complex revealed two similar helix-turn-helix hairpin structures formed by the individual proteins, which lie antiparallel to each other and form a four-helix bundle (36). Overall, no clear structure-function relationship could be deduced from this analysis; however, a few possibilities could be excluded or classified as unlikely. No significant cleft or enzyme active site was identified, suggesting a noncatalytic role for these proteins. Furthermore, interaction with nucleic acids is unlikely due to the absence of basic patches, and the electrostatic surface analysis argued against a pore-forming role of the complex (36).
The high content of α-helical structure and the hydrophobic nature of both ESAT-6 and CFP-10 led us to look more closely into a potential interaction of ESAT-6 and CFP-10 with lipid bilayers, bearing in mind the principles of binding and spontaneous membrane insertion of α-helical proteins or peptides (20, 43).
Interestingly, ESAT-6 bound specifically to PC- and cholesterol-containing liposomes, while less specificity was found for CFP-10, indicating that the interaction with the lipid bilayers might be different for the two proteins. This is in accordance with the difference we observed in the effects on liposomes as measured by cryoelectron microscopy and agrees with results of Hsu et al., who found disruption of artificial lipid bilayers with ESAT-6 but not with CFP-10 (18). Furthermore, Meher et al. found an increase of the α-helical content of ESAT-6 but not of CFP-10 (29), suggesting that ESAT-6 is inserted into the lipid bilayer, while CFP-10 might only remain loosely attached. The two long helices in the ESAT-6 hairpin structure formed by residues Phe8 to Trp43 and Glu49 to Ala79 are indeed twice the length required to insert into a lipid bilayer. A number of studies showed the importance of cholesterol in protein-lipid interactions in bacterial infection processes (2, 13, 17). Although our results suggest that both cholesterol and PC are needed for membrane interaction of ESAT-6, under different experimental conditions, the presence of PC alone has shown some effect (18, 29).
We recently demonstrated by copurification studies that ESAT-6 and CFP-10 are present in the form of a complex in the cytosol and the culture filtrate of tubercle bacilli, before and after secretion, respectively (3). However, it remains unclear what happens to secreted ESAT-6 and CFP-10 proteins under natural circumstances when tubercle bacilli reside within the phagosome of a macrophage, the key interface between M. tuberculosis and its host. At first glance, the observation that ESAT-6 and CFP-10 individually interact with lipid bilayers (Fig. 1) suggests that these proteins might act at the phagosomal membrane. However, as clearly shown in Results, interaction with liposomes was not observed when ESAT-6 and CFP-10 formed a complex. These observations can be explained by the fact that hydrophobic residues in ESAT-6 and CFP-10 that are involved in lipid binding become hidden inside the four-helical-bundle structure upon complex formation (3, 36). The results also agree with recent findings from Meher et al., who measured conformational changes of ESAT-6 in the presence of lipid vesicles and found that complex formation interfered with potential lipid binding (29). The presence of the complex might be important to keep ESAT-6 and CFP-10 water soluble under physiological conditions and to avoid aspecific hydrophobic interactions with the cytoplasmic membrane inside the bacterium. Another important aspect might be the biochemical stability (29) and the resistance of the complex against protease activity (26).
However, as demonstrated by two different methods (Fig. 2 and 3), we show here for the first time that the ESAT-6·CFP-10 complex dissociates at acidic pH, allowing ESAT-6 and CFP-10 to interact with phospholipid membranes, and it is possible that the pH dependent effect is further enhanced by posttranslational modifications (31). This behavior might have important implications for the fate of M. tuberculosis in professional phagocytes. While it is well established that M. tuberculosis can abolish or retard acidification of the phagosome (39), M. tuberculosis-infected cells also contain acidified phagosomes or phagolysosomes under certain circumstances (21, 41, 47). This is in agreement with the observation that ESAT-6 shows superior T-cell activation compared to the ESAT-6·CFP-10 complex (26). An attractive hypothesis in this respect is that CFP-10 might function as a chaperone that is uncoupled from ESAT-6 when the bacteria undergo acidic stress. Further support for the role of CFP-10 as a potential chaperone-like protein comes from Champion et al., who reported that only CFP-10 and not ESAT-6 contains the C-terminal signal sequence-like fragment that is needed for interaction with membrane protein Rv3871 of the ESX-1 secretion machinery (7). This could mean that CFP-10 is involved in the transport and protection of ESAT-6 until it reaches the phagosomal compartment.
It should also be mentioned that M. marinum can escape from phagosomes (44). This ability has been linked to the activity of ESAT-6 (12). Although for tubercle bacilli the infection mechanisms might be quite different, our data combined with data for M. marinum suggest that ESAT-6 from M. tuberculosis might exhibit similar properties. Several lines of evidence support this hypothesis. We have previously shown that certain single point mutations in ESAT-6 cause attenuation of M. tuberculosis, emphasizing that ESAT-6 is one of the key players in this phenomenon (3). Guinn and colleagues have reported that ESAT-6 deletion mutants of M. tuberculosis could multiply within macrophage-like cells lines but were unable to spread to uninfected macrophages (15). Hsu and colleagues described that ESAT-6 deleted mutants show reduced tissue invasiveness due to lacking cytolytic activity (18), and very recently it was suggested that ESAT-6 induces apoptosis linked to a potential pore-forming activity of ESAT-6. This subject also relates to a controversial study that reported that M. tuberculosis H37Rv and H37Ra can escape from fused vesicles into the cytosol of the macrophage as the infection progresses, whereas ESAT-6-lacking BCG did not (28). More recently, a similar effect was observed when responses to major histocompatibility complex molecules were investigated. M. tuberculosis, in contrast to BCG or an M. tuberculosis ESX-1 transposon mutant, was translocated from the phagosomes to the cytosol in human dendritic cells and macrophages at later time points of the infection (50a).
Altogether, these findings suggest that the ESX-1 system, which is essential for the in vivo growth of M. tuberculosis, might provide the bacterium with the tools necessary for eventually escaping from the phagosomal compartment of professional phagocytic cells. The ensuing access to the cytosol would then also explain the strikingly higher recruitment and activation of CD8+ T cells found in the lungs of mice that were aerosol infected with M. tuberculosis H37Rv or BCG::RD1 compared to uninfected or BCG-challenged mice (25). Indeed, there is more and more evidence from immunologic studies that ESX-1 proteins gain access to the cytosol of the host cell (45). Such an effect could have important consequences for the design of novel antituberculosis vaccines that show increased CD8+-mediated responses (35).
Finally, it should be mentioned that ESAT-6 and CFP-10 belong to the family of WXG100 proteins (32), which are widely distributed among actinobacteria and gram-positive bacteria (32). In M. tuberculosis some of them are encoded in ESX loci that show similarity in gene content and operon structure to ESX-1 (14, 50). In contrast to ESAT-6 and CFP-10, these other Esx protein couples (e.g., Rv0287/88) do not show the coiled-coil motif characteristic of ESAT-6 and CFP-10 (3), nor do they contain the salt bridges that stabilize the ESAT-6·CFP-10 complex (36). Thus, it remains to be determined whether they form four-helical-bundle structures. However, in a recent study, it was shown that ESX-5 was involved in the secretion of protein PPE41 (1), which is organized with its binding partner in a four-helical bundle (48). Hence, altogether, the pH-dependent interaction of ESAT-6 with biomembranes presented here not only helps to elucidate the role of this protein in the infection process of M. tuberculosis but also opens new perspectives for the study of ESX systems in a much wider range.
Acknowledgments
We thank F. Livolant and A. Leforestier for the use of the cryofixation device developed in the Laboratoire de Physique des Solides, CNRS, URA 8502, Université Paris-Sud, Orsay, France, and C. Snel and L. Marsollier for their assistance and helpful discussions. We acknowledge the kind gift of DMPG from Lipoïd GmbH, Ludwigshafen, Germany.
This work received support from the Institut Pasteur (GPH-5), the European Commision, contract LHSP-CT-2005-018923 (NM4TB), the ACI Microbiologie (MIC0311), and the Association Française Raoul Follereau. Vectors pMRLB7 and pMRLB46 were received as part of NIH NIAID contract no. HHSN266200400091C, entitled “Tuberculosis Vaccine Testing and Research Materials,” which was awarded to Colorado State University.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Abdallah, A. M., T. Verboom, F. Hannes, M. Safi, M. Strong, D. Eisenberg, R. J. Musters, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, J. Luirink, and W. Bitter. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62:667-679. [DOI] [PubMed] [Google Scholar]
- 2.Bonev, B. B., R. J. Gilbert, P. W. Andrew, O. Byron, and A. Watts. 2001. Structural analysis of the protein/lipid complexes associated with pore formation by the bacterial toxin pneumolysin. J. Biol. Chem. 276:5714-5719. [DOI] [PubMed] [Google Scholar]
- 3.Brodin, P., M. I. de Jonge, L. Majlessi, C. Leclerc, M. Nilges, S. T. Cole, and R. Brosch. 2005. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J. Biol. Chem. 280:33953-33959. [DOI] [PubMed] [Google Scholar]
- 4.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin, P., L. Majlessi, L. Marsollier, M. I. de Jonge, D. Bottai, C. Demangel, J. Hinds, O. Neyrolles, P. D. Butcher, C. Leclerc, S. T. Cole, and R. Brosch. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. Dos Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 104:5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632-1636. [DOI] [PubMed] [Google Scholar]
- 8.Collins, D. M., B. Skou, S. White, S. Bassett, L. Collins, R. For, K. Hurr, G. Hotter, and G. W. de Lisle. 2005. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 73:2379-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Converse, S. E., and J. S. Cox. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 187:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrick, S. C., and S. L. Morris. The ESAT6 protein of Mycobacterium tuberculosis induces apoptosis of macrophages by activating caspase expression. Cell. Microbiol. 9:1547-1555. [DOI] [PubMed]
- 11.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 101:12598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677-1693. [DOI] [PubMed] [Google Scholar]
- 13.Gatfield, J., and J. Pieters. 2000. Essential role for cholesterol in entry of mycobacteria into macrophages. Science 288:1647-1650. [DOI] [PubMed] [Google Scholar]
- 14.Gey Van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guinn, K. I., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodel, A., S. J. An, N. J. Hansen, J. Lawrence, B. Wasle, M. Schrader, and J. M. Edwardson. 2001. Cholesterol-dependent interaction of syncollin with the membrane of the pancreatic zymogen granule. Biochem. J. 356:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe, D., and R. A. Heinzen. 2006. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell. Microbiol. 8:496-507. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junqueira-Kipnis, A. P., R. J. Basaraba, V. Gruppo, G. Palanisamy, O. C. Turner, T. Hsu, W. R. Jacobs, Jr., S. A. Fulton, S. M. Reba, W. H. Boom, and I. M. Orme. 2006. Mycobacteria lacking the RD1 region do not induce necrosis in the lungs of mice lacking interferon-gamma. Immunology 119:224-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Killian, J. A. 2003. Synthetic peptides as models for intrinsic membrane proteins. FEBS Lett. 555:134-138. [DOI] [PubMed] [Google Scholar]
- 21.Kusner, D. J., and J. A. Barton. 2001. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome-lysosome fusion. J. Immunol. 167:3308-3315. [DOI] [PubMed] [Google Scholar]
- 22.Lafont, F., and F. G. van der Goot. 2005. Oiling the key hole. Mol. Microbiol. 56:575-577. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majlessi, L., P. Brodin, R. Brosch, M. J. Rojas, H. Khun, M. Huerre, S. T. Cole, and C. Leclerc. 2005. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J. Immunol. 174:3570-3579. [DOI] [PubMed] [Google Scholar]
- 26.Marei, A., A. Ghaemmaghami, P. Renshaw, M. Wiselka, M. Barer, M. Carr, and L. Ziegler-Heitbrock. 2005. Superior T cell activation by ESAT-6 as compared with the ESAT-6-CFP-10 complex. Int. Immunol. 17:1439-1446. [DOI] [PubMed] [Google Scholar]
- 27.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 28.McDonough, K. A., Y. Kress, and B. R. Bloom. 1993. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect. Immun. 61:2763-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meher, A. K., N. C. Bal, K. V. Chary, and A. Arora. 2006. Mycobacterium tuberculosis H37Rv ESAT-6-CFP-10 complex formation confers thermodynamic and biochemical stability. FEBS J. 273:1445-1462. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, L., and J. Pieters. 2005. The Trojan horse: survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol. 15:269-276. [DOI] [PubMed] [Google Scholar]
- 31.Okkels, L. M., E. C. Muller, M. Schmid, I. Rosenkrands, S. H. Kaufmann, P. Andersen, and P. R. Jungblut. 2004. CFP10 discriminates between nonacetylated and acetylated ESAT-6 of Mycobacterium tuberculosis by differential interaction. Proteomics 4:2954-2960. [DOI] [PubMed] [Google Scholar]
- 32.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 33.Pathak, S. K., S. Basu, K. K. Basu, A. Banerjee, S. Pathak, A. Bhattacharyya, T. Kaisho, M. Kundu, and J. R. Basu. 2007. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat. Immunol. 8:610-618. [DOI] [PubMed] [Google Scholar]
- 34.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 35.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533-539. [DOI] [PubMed] [Google Scholar]
- 36.Renshaw, P. S., K. L. Lightbody, V. Veverka, F. W. Muskett, G. Kelly, T. A. Frenkiel, S. V. Gordon, R. G. Hewinson, B. Burke, J. Norman, R. A. Williamson, and M. D. Carr. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24:2491-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, R. G. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 277:21598-21603. [DOI] [PubMed] [Google Scholar]
- 38.Rouser, G., S. Fkeischer, and A. Yamamoto. 1970. Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5:494-496. [DOI] [PubMed] [Google Scholar]
- 39.Russell, D. G. 2003. Phagosomes, fatty acids and tuberculosis. Nat. Cell Biol. 5:776-778. [DOI] [PubMed] [Google Scholar]
- 40.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaible, U. E., S. Sturgill-Koszycki, P. H. Schlesinger, and D. G. Russell. 1998. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J. Immunol. 160:1290-1296. [PubMed] [Google Scholar]
- 42.Schumann, G., S. Schleier, I. Rosenkrands, N. Nehmann, S. Halbich, P. F. Zipfel, M. I. de Jonge, S. T. Cole, T. Munder, and U. Mollmann. 2006. Mycobacterium tuberculosis secreted protein ESAT-6 interacts with the human protein syntenin-1. Centr. Eur. J. Biol. 1:183-202. [Google Scholar]
- 43.Seelig, J. 2004. Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta 1666:40-50. [DOI] [PubMed] [Google Scholar]
- 44.Stamm, L. M., J. H. Morisaki, L. Y. Gao, R. L. Jeng, K. L. McDonald, R. Roth, S. Takeshita, J. Heuser, M. D. Welch, and E. J. Brown. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 198:1361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanley, S. A., J. E. Johndrow, P. Manzanillo, and J. S. Cox. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178:3143-3152. [DOI] [PubMed] [Google Scholar]
- 46.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 100:13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stober, C. B., D. A. Lammas, C. M. Li, D. S. Kumararatne, S. L. Lightman, and C. A. McArdle. 2001. ATP-mediated killing of Mycobacterium bovis bacille Calmette-Guerin within human macrophages is calcium dependent and associated with the acidification of mycobacteria-containing phagosomes. J. Immunol. 166:6276-6286. [DOI] [PubMed] [Google Scholar]
- 48.Strong, M., M. R. Sawaya, S. Wang, M. Phillips, D. Cascio, and D. Eisenberg. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:8060-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan, T., W. Lee, D. C. Alexander, S. Grinstein, and J. Liu. 2006. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell. Microbiol. 8:1417-1429. [DOI] [PubMed] [Google Scholar]
- 50.Tekaia, F., S. V. Gordon, T. Garnier, R. Brosch, B. G. Barrell, and S. T. Cole. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 79:329-342. [DOI] [PubMed] [Google Scholar]
- 50a.van der Wel, N., D. Hava, D. Houben, D. Fluitsma, M. van Zon, J. Pierson, M. Brenner, and P. J. Peters. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287-1298. [DOI] [PubMed] [Google Scholar]
- 51.Veldhuizen, R., K. Nag, S. Orgeig, and F. Possmayer. 1998. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta 1408:90-108. [DOI] [PubMed] [Google Scholar]
- 52.Volkman, H. E., H. Clay, D. Beery, J. C. Chang, D. R. Sherman, and L. Ramakrishnan. 2004. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2:e367. [DOI] [PMC free article] [PubMed] [Google Scholar]