Abstract
The response regulator PhoP is part of the PhoQ/PhoP two-component system involved in responses to depletion of extracellular Mg2+. Here, we report the crystal structures of the receiver domain of Escherichia coli PhoP determined in the absence and presence of the phosphoryl analog beryllofluoride. In the presence of beryllofluoride, the active receiver domain forms a twofold symmetric dimer similar to that seen in structures of other regulatory domains from the OmpR/PhoB family, providing further evidence that members of this family utilize a common mode of dimerization in the active state. In the absence of activating agents, the PhoP receiver domain crystallizes with a similar structure, consistent with the previous observation that high concentrations can promote an active state of PhoP independent of phosphorylation.
During infection, pathogenic bacteria differentially regulate the expression of specific sets of genes to adapt to conditions within the host cell, an environment that differs substantially from the extracellular milieu. The PhoQ/PhoP two-component system that responds to pH and the concentration of extracytoplasmic Mg2+ is used by several organisms to distinguish between the environment inside (typically low Mg2+) and that outside (typically high Mg2+) the host cell (12). PhoP, a member of the large family of OmpR/PhoB response regulators, regulates the transcription of key virulence genes essential for invasion of host cells and for growth and survival in macrophages in several gram-negative pathogenic species, including Salmonella enterica (13, 29), Shigella flexneri (31), Yersinia pestis (35), and Neisseria meningitidis (18). The PhoQ/PhoP system controls the expression of genes involved in the modification of many components in the bacterial cell envelope and is implicated in conferring resistance to the host immune system and to several cationic antimicrobial peptides (41) by regulating either modification of lipids in the lipopolysaccharide (14) or expression of extracytoplasmic proteases that are capable of cleaving antimicrobial peptides. These properties make the PhoQ/PhoP two-component system an attractive target for vaccine and antimicrobial-drug development. In addition to direct regulation at a number of promoters, PhoP also regulates other two-component systems and regulatory proteins indirectly at the transcriptional (RstA/RstB) (30), posttranscriptional (SsrB/SpiR) (3), and posttranslational (PmrA/PmrB) (20) levels. Thus, the PhoQ/PhoP system is a central component of a highly networked signal transduction pathway.
The PhoQ-PhoP system was first identified in S. enterica serovar Typhimurium by isolation of a mutant that resulted in elevated expression of a nonspecific phosphatase (23), which gave it the misleading identifier “Pho,” a designation often used for the Escherichia coli PhoR-PhoB or the Bacillus subtilis PhoR-PhoP phosphate regulons that control phosphate assimilation in response to phosphate starvation. The PhoQ-PhoP system has been extensively studied in Salmonella, in which it regulates more than 40 genes, including the PhoP-activated genes (pag) and the PhoP-repressed genes (prg). The PhoQ/PhoP system is also present in several nonpathogenic gram-negative bacteria, suggesting that it plays a role in fundamental responses to Mg2+ starvation. A number of PhoP-regulated genes have been found to be species specific and to confer unique properties upon the bacterium.
E. coli PhoP is a 223-residue protein containing a 120-residue regulatory domain joined by a 5-residue linker to a 98-residue C-terminal DNA-binding effector domain. The regulatory domain of PhoP, also referred to as a receiver domain, can be modified at a conserved aspartate by a phosphoryl group from the histidine protein kinase PhoQ. Phosphorylation of the regulatory domain modulates the activity of the effector domain to bind to DNA and regulate transcription. Phosphorylation promotes binding of PhoP to tandemly arranged binding sites known as PhoP boxes (21), consisting of two direct hexanucleotide repeats, (T/G)GTTTA, separated by 5 nucleotides. The archetypal PhoP promoters, like the well-characterized high-affinity Mg2+ transporter promoter mgtA, harbor a PhoP box in place of the −35 region, but other promoters have been identified in which the PhoP box is found in an opposite orientation and at various distances from the RNA polymerase binding site (54).
In this study, we report the crystal structures of the regulatory domain of E. coli PhoP (PhoPN) determined in the absence and presence of a noncovalent activating agent, the phosphoryl analog beryllofluoride (52). In the crystal structures, both the unactivated and the activated regulatory domains dimerize with twofold symmetry using the face formed by helix α4, strand β5, and helix α5, similar to dimers of other activated regulatory domains of OmpR/PhoB family transcription factors (1, 47, 48). Although the overall conformations of the unactivated and activated PhoP regulatory domains are similar, the structures are not identical, and the differences support a role for phosphorylation in stabilizing the active conformation. The twofold symmetry of the regulatory-domain dimer, together with the assumption of a head-to-tail orientation of DNA-binding domains dictated by the tandem arrangement of PhoP box binding sites, is consistent with a model for the active transcription factor dimer in which the regulatory and effector domains are tethered by flexible linkers. The absence of an intramolecular domain interface precludes direct transmission of an activation signal between the regulatory and DNA-binding domains. This suggests that the role of the phosphorylated regulatory domain in the active state may be limited to its function as a dimerization motif.
MATERIALS AND METHODS
Bacterial strains and plasmids.
DNA encoding full-length PhoP was amplified by PCR from E. coli genomic DNA. The sequence encoding the regulatory-domain PhoPN was amplified by PCR from the full-length PhoP plasmid pEF25. The DNA fragments were inserted into NdeI and BamHI restriction sites of the T7-based vector pJES307 (46) to yield plasmids pEF25 and pEF31 for expression of full-length PhoP and PhoPN, respectively. The plasmids were transformed into E. coli strain DH5α (Invitrogen), amplified, purified, and subsequently transformed into BL21(DE3) (Invitrogen) for expression of unlabeled proteins and into B834(DE3)-pLysS cells (8) for the expression of selenomethionine-derivatized protein in which methionyl residues were replaced with selenomethionyl residues. The PhoPN expression vector encodes a 121-residue protein consisting of residues 1 to 120 from the original PhoP sequence and an added Gln residue at the C terminus.
Overexpression and purification.
B834(DE3)-pLysS cells, transformed with the plasmid pEF31, were grown in M9 minimal medium, and selenomethionine-derivatized PhoPN was expressed using a protocol adapted from Hendrickson et al. (16) and described elsewhere (37), with the exception that the cells were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside and incubated overnight at 20°C. For unlabeled PhoP and PhoPN proteins, the transformed BL21(DE3) cells were grown to mid-logarithmic phase at 37°C in Luria-Bertani medium supplemented with ampicillin at a final concentration of 100 mg/liter. Overexpression was then induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for 3 h at 37°C. All subsequent steps were carried out at 4°C.
For purification of PhoPN, the cells were harvested by centrifugation for 30 min at 5,000 × g and then washed and resuspended in buffer A containing 50 mM Tris-HCl, pH 8.0. The cell suspension was lysed by sonication, and the lysate was centrifuged at 35,000 × g for 1 h to remove cell debris. The supernatant was then subjected to fractionation at 30% saturated (NH4)2SO4, which precipitated >95% of PhoPN, along with other proteins. The precipitate was collected and solubilized in buffer A and then dialyzed overnight against buffer A. The protein was filtered and applied to 5-ml or 70-ml HiTrap Q HP columns (GE Healthcare) preequilibrated with buffer A. PhoPN was eluted using a three-column volume gradient from 0 to 1.0 M NaCl. The fractions containing PhoPN were pooled, concentrated, and loaded on a Superdex 75 26/60 (GE Healthcare) column preequilibrated with 50 mM Tris-HCl, pH 8.0, 0.1 M NaCl. Fractions containing PhoPN were pooled, and the purity was assessed by Coomassie blue staining subsequent to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The protein was >95% pure. The same procedure was used for selenomethionine-derivatized PhoPN purification, with the exception that 10 mM β-mercaptoethanol was added to solutions at all steps to prevent the oxidation of selenium.
For full-length PhoP, cells expressing PhoP were lysed by sonication as described above, and the supernatant was subjected to fractionation at 60% saturated (NH4)2SO4, which precipitated >95% of PhoP, along with other proteins. The precipitate was collected and solubilized in buffer A and then dialyzed overnight against buffer A. The protein was filtered and applied to 5-ml or 70-ml HiTrap Q HP anion-exchange columns (GE Healthcare) preequilibrated with buffer B (20 mM Tris-HCl, pH 8, 50 mM NaCl). Unbound proteins were removed with 2 column volumes of buffer B, and bound proteins were eluted with a 0 to 25% gradient of buffer C (buffer B with 2 M NaCl) in 6 column volumes. PhoP eluted in the first major peak between 5% and 20% buffer C. Fractions containing PhoP were pooled and applied to a 16/10 HiLoad Phenyl Sepharose column (GE Healthcare) preequilibrated with buffer D [20 mM Na/K phosphate, pH 6.8, and 1 M (NH4)2SO4]. The column was washed with 3 column volumes of buffer D, followed by elution with a 0 to 80% gradient of 20 mM Na/K phosphate, pH 6.8, over 20 column volumes. Fractions containing PhoP were pooled and loaded on a Superdex 75 26/60 (GE Healthcare) size exclusion column equilibrated with 50 mM Tris-HCl, pH 8.0, 100 mM NaCl. PhoP elutes at a volume consistent with a monomer. Fractions containing PhoP were pooled, and the purities of the fractions were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 15% polyacrylamide gel. The protein was >95% pure.
Activation of PhoPN and intact PhoP.
For size exclusion chromatography, PhoPN and PhoP were phosphorylated in the presence of 50 mM ammonium hydrogen phosphoramidate (synthesized by the method of Sheridan et al. [40]) and 20 mM MgCl2 for 30 min at room temperature. The proteins were >80% phosphorylated, as assessed by a shift in mobility on a Vydac reverse-phase protein C4 high-performance liquid chromatography (HPLC) column (catalog no. 214TP5415). The column was equilibrated in 98% solution A (0.1% trifluoroacetic acid)/2% solution B (90% acetonitrile, 0.1% trifluoroacetic acid) and eluted with a gradient of 2 to 82% solution B at a flow rate of 1 ml/min for 40 min. After activation, the proteins were transferred to 4°C to reduce the rate of dephosphorylation due to hydrolysis of the phosphoaspartate bond. For crystallization, PhoPN was activated by adding 5.3 mM BeCl2 (Sigma-Aldrich; product no. 201197), 35 mM NaF, and 7 mM MgCl2 to 1 mg/ml protein and concentrated using an Amicon ultracentrifugal filtration device with a molecular mass cutoff of 10 kDa (Millipore, Billerica, MA).
Crystallization of unactivated and activated PhoPN.
Initial crystallization trials for unactivated (protein in the absence of activating agents) and activated PhoPN were performed using a commercially available PEG/Ion Grid Screen (Hampton Research, CA), and more than half of the conditions containing polyethylene glycol (PEG) 3350 produced crystals. After optimization, crystals were obtained by the hanging-drop method for selenomethionine-derivatized PhoPN and BeF3−-activated selenomethionine-derivatized PhoPN by mixing equal volumes of protein solution (30 mg/ml PhoPN in 50 mM Tris-HCl, pH 8.0, 0.1 M NaCl with or without activating agents as described above) and reservoir solution (22 to 25% PEG 3350, 0.2 M NaSCN) and incubating the mixture at room temperature. These conditions generated crystals with good morphology under both the unactivated and activated conditions. Crystals appeared in 1 to 2 days. The unactivated selenomethionine-derivatized PhoPN crystals were cryoprotected by quickly passing the crystals through a solution containing 30% PEG 3350, 0.2 M NaSCN, 7.5% ethylene glycol, and 4% glycerol and flash-freezing them directly in a 100 K nitrogen cryostream. The activated selenomethionine-derivatized crystals were derivatized with Pt and cryoprotected as described below.
Data collection and structure determination of activated PhoPN.
Single-wavelength anomalous-dispersion (SAD) data were collected at beamline X4A at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) at the selenium peak wavelength based on X-ray absorption fine-structure (XAFS) scans, but no selenium sites could be found due to a weak anomalous signal. The selenomethionine-derivatized PhoPN crystals were then soaked in 2 to 10 mM K2PtCl4 by adding a cryoprotectant solution containing 5.3 mM BeF3−, 6.7 mM MgCl2, 35 mM NaF, 30% PEG 3350, 0.2 M NaSCN, 7.5% ethylene glycol, 15% glycerol, 5% sucrose, and 4 to 20 mM K2PtCl4 directly to the drops containing the crystals (the cryoprotectant drop was placed next to the drop containing the crystal, and a small liquid channel was made with a pipette tip to mix the two drops, which were then incubated in the dark for ∼20 to 50 min). The crystal used for data collection was soaked in 2.5 mM K2PtCl4 for 30 to 40 min and flash-frozen on a nylon loop directly in the cryostream. SAD data were collected at the platinum peak wavelength (1.0714 Å), based on an XAFS scan, using an oscillation range of 1°. The data, processed and scaled with Denzo and Scalepack (34), showed an excellent anomalous signal. The crystals belong to the space group P64 with unit cell dimensions as follows: a = b = 103.12 Å and c = 31.01 Å (Table 1) , corresponding to one molecule of activated PhoPN per asymmetric unit. The intensities were transformed into structure factors using the Crystallography and NMR System (CNS) (5). Two Pt sites were clearly identified using the SAD phasing protocol in the CNS software suite using data to 2.4 Å, and a third was found by computing two maps and choosing the highest peak common to both maps; the first map was an anomalous-difference Fourier map generated by combining the SAD phases from the two sites and the anomalous-difference structure factors, and the second map was a gradient map. Enantiomorph ambiguity was resolved by testing both the original heavy-atom configuration and its inverse. Phase ambiguity was resolved with maximum likelihood density modification by solvent flipping in CNS, which yielded excellent quality electron density maps. These maps were used to make an initial model using XtalView (28). This was followed by a round of torsion angle simulated annealing in CNS using the maximum likelihood target function and the experimental phase information and anisotropic bulk solvent correction to remove errors in the geometry due to manual building. The structure was then refined by iterative cycles of maximum likelihood, simulated annealing, and temperature factor refinement and manual rebuilding by inspection of the SigmaA-weighted phase-combined 2Fo − Fc and Fo − Fc (the observed and calculated structure factor amplitudes) electron density maps after every cycle. Water molecules were added at positive-difference Fourier peaks greater than 3σ using the automatic water pick script in CNS, and after they were manually verified, more water molecules were added. The model was refined to a final resolution of 1.9 Å with R/Rfree values of 0.19/0.22. It contains 121 amino acids; three Pt, one Mg2+, one Be, and three F atoms; and 109 water molecules (Table 1). The data quality and structure stereochemistry were validated using the programs SFCHECK (49), PROCHECK (24), and MolProbity (26). From PROCHECK analysis, 96% of the residues appear in the most favorable regions, 3% occur in allowed regions, and 1% in generously allowed regions.
TABLE 1.
Data collection and refinement statistics
| Statistic | Value
|
|
|---|---|---|
| Unactivated PhoPN | Active PhoPN | |
| Data collection | ||
| Wavelength (Å) | 1.07217 | 1.07140 |
| Space group | P65 | P64 |
| Cell dimensions | ||
| a, b, c (Å) | 102.64, 102.64, 62.64 | 103.12, 103.12, 31.01 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 30.00-2.45 (2.64-2.54)a | 30.00-1.76 (1.98-1.90) |
| Rsymb | 0.11 (0.44) | 0.10 (0.48) |
| I /σI | 13.46 (2.45) | 11.50 (2.02) |
| Completeness (%) | 99.5 (100) | 97.2 (92.4) |
| Redundancy | 5.0 (3.3) | 3.9 (2.5) |
| Refinement | ||
| Resolution (Å) | 30.00-2.54 | 30.00-1.90 |
| No. of reflections (work/test) | 11,245/1,239 | 24,629/2,652 |
| Rc/Rfree | 0.21/0.25 | 0.195/0.23 |
| No. of atoms | ||
| Protein | 1,885 | 969 |
| Ligand/ion | 0 | 8 |
| Water | 42 | 109 |
| B factors (Å2) | ||
| Protein | 44.14 | 22.015 |
| Main chain | 0.60 | 0.90 |
| RMSDd (main chain) | 25.53 | |
| Side chain | 45.21 | 1.80 |
| RMSD (side chain) | 1.65 | |
| Ligand/ion | - | 25.45 |
| Water | 48.02 | 36.42 |
| RMSD from ideal stereochemistry | ||
| Bond lengths (Å) | 0.019 | 0.004 |
| Bond angles (°) | 1.83 | 1.28 |
The values for the highest-resolution bins used for refinement are shown in parentheses.
Rsym = Σ I − <I>/ΣI, where I is the observed integrated intensity and <I> is the average integrated intensity obtained from multiple measurements.
R = Σ‖Fo − Fc‖, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree is equivalent to R but is calculated using a 10% disjoint set of reflections set aside prior to refinement.
RMSD, root mean square deviation.
Data collection and structure determination of unactivated PhoPN.
Data were collected from a single selenomethionine-derivatized crystal of unactivated PhoPN at 1.07217-Å wavelength, and the data were processed and scaled with Denzo and Scalepack (34). The crystals belong to space group P65 with unit cell dimensions as follows: a = b = 102.64 Å and c = 62.64 Å, corresponding to two molecules of unactivated PhoPN per asymmetric unit (Table 1). The structure was solved using data from 15 to 3 Å by molecular replacement with Phaser (45) integrated in the CCP4i (39) interface, using the structure of activated PhoPN as the search model. A step of rigid body refinement was performed in CNS to optimize the positions and orientations of the two molecules. At this point, all the side chains and loops were removed and the model was rebuilt by iterative cycles of simulated annealing, positional refinement, temperature factor refinement, and manual rebuilding using XtalView, CNS version 1.1, and Refmac 5.2.0005 (32) until convergence by inspecting the SigmaA-weighted 2Fo − Fc and Fo −Fc electron density maps after every cycle. Noncrystallographic symmetry restraints (residues 2 to 75 in chains A and B) were used for refinement using Refmac in the initial cycles of refinement until the Rcryst dropped to ∼27%, after which the two protein chains were built individually. The final model was refined to 2.54 Å and had R/Rfree values of 0.21/0.25. The final model for the two protomers of PhoPN in the asymmetric unit contains 239 amino acids and 42 water molecules (Table 1). The data quality and structure stereochemistry were validated using the programs SFCHECK (49), PROCHECK (24), and MolProbity (26). From PROCHECK analysis, 95% of the residues appear in the most favorable regions and 5% occur in allowed regions.
Analytical size exclusion chromatography.
Analytical size exclusion chromatography was used to probe the oligomeric states of PhoPN and PhoP. The chromatography for PhoPN was performed using a TosoHaas G2000SW (7.5 mm by 300 mm; particle size, 10 μm) HPLC column (Supelco, PA) and for PhoP using a TosoHaas G3000SWXL (7.8 mm by 300 mm; particle size, 5 μm) HPLC column. Experiments were carried out at room temperature at a flow rate of 0.5 ml/min using 50 mM MES (morpholineethanesulfonic acid), pH 6.5, 0.1 M NaCl for PhoPN and 50 mM Tris-HCl, pH 8.0, 0.1 M NaCl for intact PhoP. Aliquots of 20-μl to 30-μl volumes were injected at protein concentrations of ∼1 mg/ml. All samples were filtered through 0.2-μm filters prior to injection. A standard curve was generated using bovine serum albumin (66 kDa), chicken ovalbumin (44 kDa), soybean trypsin inhibitor (21 kDa), and S. enterica CheY (13.8 kDa).
Protein Data Bank accession codes.
The coordinates and structure factors for the unactivated and activated PhoPN structures have been deposited in the Protein Data Bank with accession codes 2PKX and 2PL1, respectively.
RESULTS AND DISCUSSION
BeF3−-activated PhoPN crystallizes as a twofold symmetric dimer.
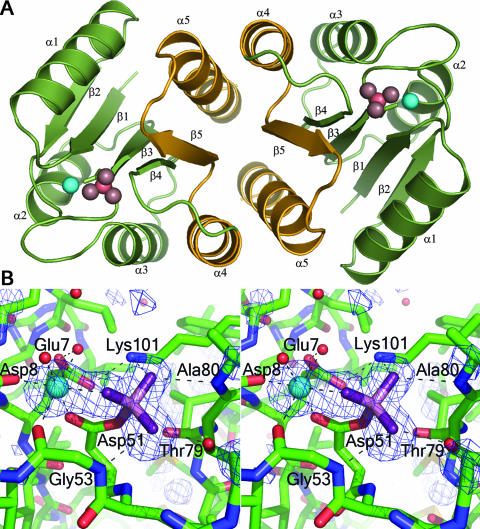
BeF3−-activated PhoPN crystallized in the space group P64 with a solvent content of 64%. The asymmetric unit contains one molecule that forms a twofold symmetric dimer with a symmetry-related molecule using the α4-β5-α5 interface (Fig. 1A).
FIG. 1.
BeF3−-activated PhoPN. (A) The regulatory-domain dimer. Activated PhoPN dimerizes with twofold rotational symmetry, forming an α4-β5-α5 interface. A ribbon depiction of the dimer is shown in green with the α4-β5-α5 dimer interface highlighted in gold and repetitive secondary-structure elements labeled. The noncovalent BeF3− complex and Mg2+ within the active site are shown as pink and cyan spheres, respectively. (B) Stereo view of the active site. A difference electron density map (Fo − Fc) calculated at 1.9 Å with omission of the Mg2+ and BeF3− complex from the model is shown in blue, contoured at 3σ. Carbons, nitrogens, and oxygens of the protein are shown as green, blue, and red sticks, respectively. The noncovalent BeF3− complex is shown in stick format with beryllium and fluorides in light and dark purple, respectively. Water molecules and Mg2+ are shown as red and cyan spheres, respectively. The network of hydrogen bonds that coordinates BeF3− and Mg2+ is shown as dashed lines. All molecular images were generated using Pymol (http://www.pymol.org).
The conformations of conserved active-site residues in active PhoPN are similar to those of equivalent residues in other activated receiver domain structures. The densities for the Mg2+ and the noncovalent BeF3− complex were clear in the experimental maps obtained after SAD phasing and density modification (Fig. 1B). The octahedral coordination of Mg2+ is satisfied by F-1 from BeF3−, the side chain carboxyl oxygens of Asp8 and Asp51, the main-chain carbonyl oxygen of Gly53, and two water molecules that form hydrogen bonds to Glu7 and Asp8. The conserved active-site lysine, Lys101, is involved in salt bridges with Glu7, F-3 of BeF3−, and a water molecule. The conserved switch residue, Thr79, involved in the activation mechanisms of all response regulators, is in an inward orientation, forming a hydrogen bond with F-2 of BeF3−. F-2 also forms a hydrogen bond with the backbone nitrogen of Gly53. As has been observed in other active receiver domains, the inward orientation of Thr79 is correlated with a specific positioning of the backbone nitrogen of the adjacent residue, Ala80, allowing formation of a hydrogen bond with F-3.
The α4-β5-α5 dimer interface of activated PhoPN buries a surface area of ∼1,010 Å2 per protomer. The buried surface area for all nonhydrogen atoms was calculated using the GETAREA 1.1 web server (http://pauli.utmb.edu/cgi-bin/get_a_form.tcl) (10) with a probe radius of 1.4 Å. The interacting surfaces of the activated protomers form a small hydrophobic core surrounded by an extensive network of hydrogen bonds and salt bridges. The interface is very similar to those observed in other activated regulatory-domain structures (1, 47, 48), and the same conserved residues comprise the hydrophobic patch (Val88, Leu91, and Ala110) and participate in salt bridges (Lys87-Glu107, Asp96-Arg118, and Asp97-Arg111). The salt bridge corresponding to Glu96-Lys117 in PhoB (1) and Glu94-Arg115 in ArcA (47), which involves residues that are highly conserved in the primary sequence of response regulators in the OmpR/PhoB family, is not observed in PhoPN. Instead, the corresponding residues, Ser92 and Gln113, interact through hydrogen bonds mediated by a water molecule.
The formation of a rotationally symmetric dimer by the activated regulatory domains and the occurrence of direct-repeat DNA-binding sites for PhoP (21) together imply the lack of a unique interdomain interface in the active protein. This study provides further evidence that members of the OmpR/PhoB family share a common active, DNA-bound state with flexible linkers connecting the regulatory and effector domains, as has been previously proposed for other members of this family (1, 47). While most studies of OmpR/PhoB response regulators to date have indicated a tandem mode of DNA binding, the lack of interdomain constraints would allow versatility in the orientation of DNA-binding domains bound to DNA. The mode of binding would presumably be dictated by protein-protein interactions between effector domains and/or by the DNA sequences. It should be noted that the oligomeric states of active response regulators vary among different members of the OmpR/PhoB family, although most members transition to a higher oligomeric state upon phosphorylation (7, 9, 17, 27, 38). In cases where the oligomeric state is greater than a dimer, we envision that the proposed active transcription factor dimer would be a subunit of the larger complex and that other system-specific interactions among the regulatory domains, effector domains, and/or DNA sequences would dictate the overall quaternary structure of the active transcription complex. It is notable that the binding sites for response regulators with multiple adjacent sites mostly occur in multiples of two (i.e., two, four, six, or eight half-sites), consistent with a dimer being part of a larger assembly (15, 19, 53).
Unactivated PhoPN crystallizes in an “active-like” form in the absence of activating agents.
Unactivated PhoPN was crystallized under the same conditions used for crystallization of BeF3−-activated PhoPN, except that activating agents were excluded. The crystals belong to the space group P65 with two molecules (chain A and chain B) in the asymmetric unit compared to one molecule per asymmetric unit for activated PhoPN. The positions and orientations of the molecules within the unit cell and the unit cell dimensions, except for the doubling of the c axis, are similar to those obtained for activated PhoPN. The doubling of the asymmetric unit results from minor differences between the two protomers of the dimer of unactivated PhoPN, which converts the twofold crystallographic symmetry of activated PhoPN to pseudo-twofold noncrystallographic symmetry. The two molecules in the asymmetric unit of unactivated PhoPN dimerize in the same manner as activated PhoPN, using the same α4-β5-α5 interface.
The occurrence of an “active-like” dimer even in the absence of activating agents may be the result of the high protein concentrations employed for crystallization or it may be the only form allowed by the lattice packing under these crystallization conditions. The unphosphorylated regulatory domain of the response regulator NtrC has been shown to exist in equilibrium between inactive and active states, and phosphorylation shifts the equilibrium toward the active state by stabilizing the active form (50). Similar equilibriums are postulated for all receiver domains. At the high protein concentrations required for crystal formation, a few nucleation events could drive the equilibrium toward the active conformation simply by mass action. This appears to be the most likely explanation for crystallization of an “active-like” dimer of unphosphorylated PhoPN. Several other OmpR/PhoB family members have been observed to crystallize in “active-like” dimers in the absence of activating agents (2, 47, 48). In contrast, the regulatory domain of PhoB, which in its unphosphorylated state exists in solution in fast equilibrium between a monomer and a dimer that is distinctly different from the phosphorylated dimer, was found to crystallize in distinct inactive and active dimer conformations despite similar crystallization conditions other than the absence or presence of a phosphoryl analog (1).
Comparison of the unactivated and activated dimers of PhoPN.
Although the “active-like” dimer observed in the crystal structure of unactivated PhoPN has an overall conformation similar to that of the dimer observed in the crystal structure of activated PhoPN, phosphorylation appears to significantly stabilize the protein. While the two protomers in the dimer of the activated protein are identical, the two protomers in the dimer of the unactivated protein have minor differences, particularly in the α4-β5-α5 region. These differences are correlated with an expansion of the asymmetric unit to accommodate both molecules of the dimer, resulting in a doubling of the c axis of the unit cell in the lattice of unactivated PhoPN crystals relative to that of activated PhoPN crystals.
Activated PhoPN superimposes on chains A and B of unactivated PhoPN with root mean square (RMS) deviations of 0.72 Å and 0.77 Å, respectively, for all Cα atoms (residues 2 to 119). The dimer of activated PhoPN formed by symmetry operations superimposes on the dimer of unactivated PhoPN with an RMS deviation of 0.91 Å for Cα atoms of residues 2 to 119.
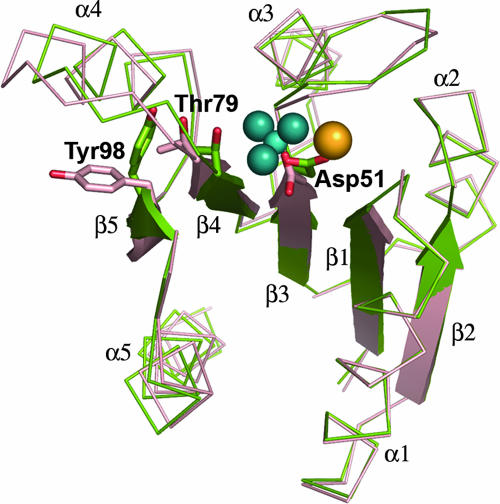
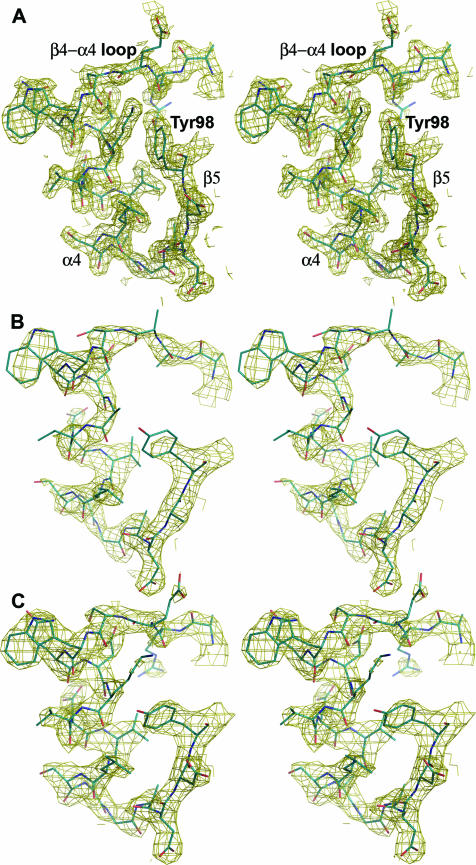
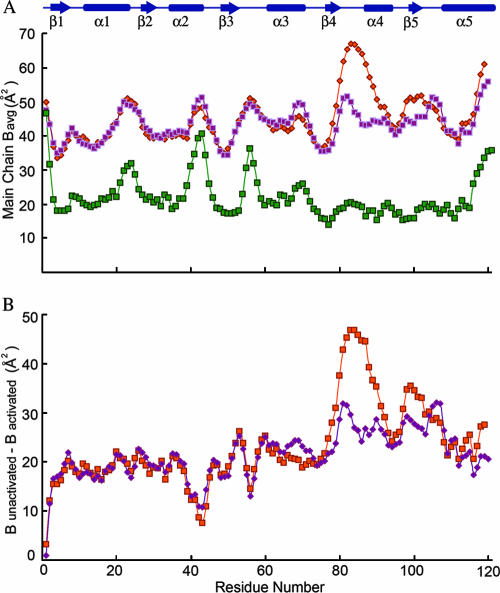
There are minor differences in the conformations of the backbone in the structures of the activated and unactivated proteins (Fig. 2). The greatest differences are observed in the β4-α4 loop region and the N terminus of helix α4. In the activated protein, the β4-α4 loop is positioned toward the active site and the conserved switch residue Thr79 adopts an inward orientation, forming a hydrogen bond with one of the fluorides of the BeF3− complex. This positioning of the loop is also correlated with a slight inward shift of the N terminus of helix α4. In the unactivated domain, Thr79 exists in an outward position, and this is correlated with an outward orientation of the switch residue Tyr98, representing displacements of the side chain hydroxyl groups of 2.31 Å and 7.97 Å (chain A) and 2.36 Å and 8.20 Å (chain B), respectively, relative to their positions in BeF3−-activated PhoPN when the chains are superimposed using residues 2 to 70. Unlike the densities for other regions of the protein, the densities for residues in the β4-α4 loop and helix α4 and for the side chain of Tyr98 are weaker for chain A than for chain B, suggesting that these regions are more mobile than the rest of the molecule in the inactive protein (Fig. 3). Average B factors for all atoms in the β4-α4 loop and helix α4 are 58.1 Å2 and 46.3 Å2 for chains A and B of the unactivated protein and 20.9 Å2 for the active protein. It is clear from the plot of main-chain B factors versus residue numbers in Fig. 4 that the C-terminal half of PhoPN has relatively greater mobility than the rest of the molecule in the unactivated protein compared to the activated protein. As the crystal packing is the same for both unactivated and activated PhoPN proteins, lattice contacts cannot account for the differences in mobility in the two proteins. Greater structural rigidity of the β4-α4 loop and the helix α4 region upon activation has been observed in other regulatory domains (22). These features presumably reflect the equilibrium between inactive and active conformations of the regulatory domain that exist in the absence of phosphorylation.
FIG. 2.
Differences in the backbone conformations of activated versus unactivated PhoPN. The conformational changes of the switch residues Thr79 and Tyr98, when BeF3− is bound at the active site Asp51, are correlated with minor differences in the overall positioning of secondary-structural elements relative to the unactivated protein. The unactivated protomer (chain A) is shown in red, and the activated protein is shown in green. The BeF3− complex is shown in blue, and Mg2+ is shown in yellow. Tyr98 has an outward orientation in the unactivated molecule and an inward orientation in the activated molecule.
FIG. 3.
Electron density in the β4-α4-β5 region of activated and unactivated PhoPN proteins. Stereo views of the σA-weighted 2Fo − Fc electron density maps contoured at 2.5σ and calculated at 1.9 Å for activated PhoPN (A) and at 2.54 Å for (B) chain A and (C) chain B of unactivated PhoPN. The protein model is shown in stick representation (carbon, blue; nitrogen, dark blue; oxygen, red), and the relevant secondary-structural elements and tyrosine 98 are labeled in panel A for reference.
FIG. 4.
Comparison of B factors of unactivated and activated PhoPN. (A) Average main-chain B factors for unactivated PhoPN chain A (pink), unactivated PhoPN chain B (orange), and activated PhoPN (green). (B) Differences between B factors of activated PhoPN and unactivated PhoPN chain A (pink) and unactivated PhoPN chain B (orange). The secondary-structural elements are shown in blue at the top.
Most isolated regulatory domains of OmpR/PhoB family members crystallized in the absence of phosphoryl analogs have been found to exist as “active-like” dimers in the crystal lattice. These include the regulatory domains of E. coli ArcA (47), KdpE, and TorR (48); Thermotoga maritima DrrB and DrrD (V. L. Robinson and A. M. Stock, unpublished results); and Streptococcus pneumoniae MicAN (2). Notably, the two exceptions to this are E. coli PhoB (1, 44) and its ortholog, B. subtilis PhoP (4), both of which form distinct alternative dimers in their inactive states. No isolated regulatory domain of any OmpR/PhoB response regulator has yet been crystallized as a monomer in its inactive state. However, monomeric regulatory domains have been observed in the context of full-length inactive OmpR/PhoB response regulators (6, 11, 33, 38).
In “active-like” dimers of unactivated regulatory domains, the orientation of the conserved tyrosine residue that switches conformations in inactive and active states varies among different dimers and even within the protomers of a single dimer. While these tyrosines are in the outward position in both protomers of E. coli PhoP (this study), in ArcAN, both tyrosines point inward (47), and in TorRN, the tyrosine points outward in one protomer and inward in the other (48). The differences in the conformations of the switch tyrosine residues in different unactivated dimers contrasts with their invariant inward orientation in active dimers, providing further evidence of the flexibility of the α4-β5-α5 region in the absence of phosphorylation.
Solution studies of inactive and active PhoP.
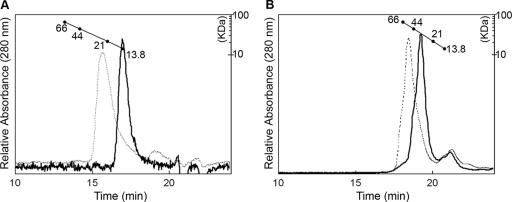
Analytical gel filtration was performed with PhoPN and full-length PhoP to characterize their oligomeric states in solution. The regulatory domain and the intact protein both exhibited elution consistent with a monomer in the inactive state and a dimer in the active state at micromolar concentrations (Fig. 5). Analytical ultracentrifugation sedimentation velocity runs, which indicate the distribution between different forms at a particular concentration, performed at 10 μM, 20 μM, and 40 μM concentrations of unactivated PhoP showed a predominant species consistent with a monomer (T. Mack, unpublished results). Thus, at low micromolar concentrations (<40 μM), such as those used for gel filtration and analytical ultracentrifugation experiments, the inactive protein exists primarily as a monomer. The intracellular concentration of PhoP is ∼2 to 15 μM, depending on the presence or absence of Mg2+ in the growth medium (25). Intracellular concentrations are highly regulated, and autoinduction of PhoP by a positive feedback loop has been shown to be essential for virulence (43). Inside a wild-type cell, the unphosphorylated protein most likely exists as a monomer, which strengthens the assumption that the dimer observed in the crystal represents the active form, induced by the high concentration (∼2 mM) used during crystallization.
FIG. 5.
Size exclusion chromatography of unphosphorylated and phosphorylated PhoPN and PhoP. (A) Elution profiles of PhoPN (solid line) and phosphorylated PhoPN (dotted line) on a TosoHaas G2000SW column. (B) Elution profiles of full-length PhoP (solid line) and phosphorylated PhoP (dotted line) on a TosoHaas G3000SWXL column. The conditions for chromatography and the protein standards used for calibration of the column are described in Materials and Methods.
Interestingly, it has been shown that overexpression of S. enterica PhoP (93% identical to E. coli PhoP) above 30 μM is sufficient for transcriptional activation of one of its target genes, pcgF, independent of histidine kinase PhoQ or phosphorylation of PhoP (25). Furthermore, it was shown that the amount of dimer formed by the overexpressed inactive protein increased with the concentration, and phosphorylation enhanced dimerization of PhoP. Similar results have also been reported for the response regulator UhpA, for which it was shown that overexpression of the protein could activate transcription in the absence of phosphorylation (51). Even though the concentrations of unphosphorylated protein required for dimerization and transcriptional activation in these overexpression studies are higher than physiological levels, it is possible that the “active-like” form adopted by the unphosphorylated protein may have a physiological role, such as binding to high-affinity promoters to provide a low basal level of transcription in a noninducing environment. Consistent with this hypothesis, plasmid-encoded PhoP, expressed at four times its physiological level, under uninduced (high-Mg2+) conditions binds to representative target promoters at 10 to 25% of the levels observed under induced (low-Mg2+) conditions (42).
Our gel filtration analyses of PhoP and PhoPN indicate a monomer-to-dimer transition upon phosphorylation and differ from the results reported in a recent study of S. enterica PhoP (36). Perron-Savard et al. reported that the dimerization and DNA-binding activity of S. enterica PhoP were unaffected by phosphorylation. These results are inconsistent with our study and that of Lejona et al. (25), leading us to conclude that the observations of Perron-Savard et al. were specific to their experimental conditions.
In summary, the structure of activated PhoPN provides further evidence for a common active DNA-bound state for OmpR/PhoB transcription factors. Although all activated receiver domains of OmpR/PhoB family members crystallize in the same configuration, inactive OmpR/PhoB receiver domains have been observed in a variety of configurations. The unactivated regulatory domains of E. coli PhoB and its ortholog, B. subtilis PhoP, crystallize as unique dimers, not mediated by the α4-β5-α5 interface characteristic of the active dimer. Inactive proteins from the OmpR/PhoB family have also been crystallized in the context of full-length proteins. T. maritima DrrD lacks a substantial interdomain interface, and the recognition helix is exposed to the solvent and sterically unhindered (6). T. maritima DrrB contains an extensive interface, involving the α4-β5-α5 face of the regulatory domain and the β-platform of the DNA-binding domain (38). Mycobacterium tuberculosis PrrA also contains an extensive interdomain interface, but it involves a domain arrangement distinct from that of DrrB and buries the recognition helix at the interface (33).
Differences in the modes of regulation employed by different members of the OmpR/PhoB family may arise from variations in their inactive states and are likely to be system dependent. Specifically, different regulatory-domain interactions in inactive response regulators are likely to pose different barriers to the transition between inactive and active states. The observation that high intracellular concentrations of PhoP can produce transcriptional activation in the absence of phosphorylation (25) suggests that the barrier to conversion between inactive and active states of PhoP is relatively low. The “active-like” dimer observed in crystals provides a structural rationale for phosphorylation-independent transcription activity of PhoP.
Acknowledgments
We thank Timothy Mack for analytical ultracentrifugation analysis of PhoP, Eileen Fox and Ti Wu for assistance with molecular biology and protein purification, and Jayita Guhaniyogi for synthesis of phosphoramidate. We thank the staff at beamline X4A at the National Synchrotron Light Source at Brookhaven National Laboratory for technical assistance.
This work was supported by grant R37GM047958 from the U.S. National Institutes of Health. A.M.S. is an investigator of the Howard Hughes Medical Institute.
We have no competing financial interests.
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Bachhawat, P., G. V. Swapna, G. T. Montelione, and A. M. Stock. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bent, C. J., N. W. Isaacs, T. J. Mitchell, and A. Riboldi-Tunnicliffe. 2004. Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states. J. Bacteriol. 186:2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. [DOI] [PubMed] [Google Scholar]
- 4.Birck, C., Y. Chen, F. M. Hulett, and J. P. Samama. 2003. The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface. J. Bacteriol. 185:254-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Buckler, D. R., Y. Zhou, and A. M. Stock. 2002. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure 10:153-164. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., C. Birck, J. P. Samama, and F. M. Hulett. 2003. Residue R113 is essential for PhoP dimerization and function: a residue buried in the asymmetric PhoP dimer interface determined in the PhoPN three-dimensional crystal structure. J. Bacteriol. 185:262-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty, A. J., S. R. Ashford, J. A. Brannigan, and D. B. Wigley. 1995. A superior host strain for the over-expression of cloned genes using the T7 promoter based vectors. Nucleic Acids Res. 23:2074-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 14:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraczkiewicz, R., and W. Braun. 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 19:319-333. [Google Scholar]
- 11.Friedland, N., T. R. Mack, M. Yu, L.-W. Hung, T. C. Terwilliger, G. S. Waldo, and A. M. Stock. 2007. Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry 46:6733-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 15.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 16.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon, Y., Y. S. Lee, J. S. Han, J. B. Kim, and D. S. Hwang. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276:40873-40879. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, C. R., J. Newcombe, S. Thorne, H. A. Borde, L. J. Eales-Reynolds, A. R. Gorringe, S. G. Funnell, and J. J. McFadden. 2001. Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39:1345-1355. [DOI] [PubMed] [Google Scholar]
- 19.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, A., and E. A. Groisman. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 18:2302-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern, D., B. F. Volkman, P. Luginbuhl, M. J. Nohaile, S. Kustu, and D. E. Wemmer. 1999. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 40:894-898. [DOI] [PubMed] [Google Scholar]
- 23.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., M. W. McArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:282-291. [Google Scholar]
- 25.Lejona, S., M. E. Castelli, M. L. Cabeza, L. J. Kenney, E. Garcia Vescovi, and F. C. Soncini. 2004. PhoP can activate its target genes in a PhoQ-independent manner. J. Bacteriol. 186:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 27.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 28.McRee, D. E. 1999. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 29.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (PhoP/ PhoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss, J. E., P. E. Fisher, B. Vick, E. A. Groisman, and A. Zychlinsky. 2000. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol. 2:443-452. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 33.Nowak, E., S. Panjikar, P. Konarev, D. I. Svergun, and P. A. Tucker. 2006. The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis. J. Biol. Chem. 281:9659-9666. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 35.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perron-Savard, P., G. De Crescenzo, and H. Le Moual. 2005. Dimerization and DNA binding of the Salmonella enterica PhoP response regulator are phosphorylation independent. Microbiology 151:3979-3987. [DOI] [PubMed] [Google Scholar]
- 37.Robinson, V. L., J. Hwang, E. Fox, M. Inouye, and A. M. Stock. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10:1649-1658. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, V. L., T. Wu, and A. M. Stock. 2003. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J. Bacteriol. 185:4186-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SERC (UK) Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan, R. C., J. F. McCullough, and Z. T. Wakefield. 1971. Phosphoramidic acid and its salts. Inorg. Synth. 13:23-26. [Google Scholar]
- 41.Shi, Y., M. J. Cromie, F. F. Hsu, J. Turk, and E. A. Groisman. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229-241. [DOI] [PubMed] [Google Scholar]
- 42.Shin, D., and E. A. Groisman. 2005. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem. 280:4089-4094. [DOI] [PubMed] [Google Scholar]
- 43.Shin, D., E. J. Lee, H. Huang, and E. A. Groisman. 2006. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science 314:1607-1609. [DOI] [PubMed] [Google Scholar]
- 44.Solà, M., F. X. Gomis-Rüth, L. Serrano, A. González, and M. Coll. 1999. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 285:675-687. [DOI] [PubMed] [Google Scholar]
- 45.Storoni, L. C., A. J. McCoy, and R. J. Read. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60:432-438. [DOI] [PubMed] [Google Scholar]
- 46.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 84:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toro-Roman, A., T. R. Mack, and A. M. Stock. 2005. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the α4-β5-α5 face. J. Mol. Biol. 349:11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toro-Roman, A., T. Wu, and A. M. Stock. 2005. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 14:3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaguine, A. A., J. Richelle, and S. J. Wodak. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D 55:191-205. [DOI] [PubMed] [Google Scholar]
- 50.Volkman, B. F., D. Lipson, D. E. Wemmer, and D. Kern. 2001. Two-state allosteric behavior in a single domain signaling protein. Science 291:2429-2433. [DOI] [PubMed] [Google Scholar]
- 51.Webber, C. A., and R. J. Kadner. 1997. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol. Microbiol. 24:1039-1048. [DOI] [PubMed] [Google Scholar]
- 52.Wemmer, D. E., and D. Kern. 2005. Beryllofluoride binding mimics phosphorylation of aspartate in response regulators. J. Bacteriol. 187:8229-8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, T., L. Qin, L. A. Egger, and M. Inouye. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114-17123. [DOI] [PubMed] [Google Scholar]
- 54.Zwir, I., D. Shin, A. Kato, K. Nishino, T. Latifi, F. Solomon, J. M. Hare, H. Huang, and E. A. Groisman. 2005. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 102:2862-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]