Abstract
The genes encoding malate synthase (glcB) and isocitrate lyase (aceA) and a 240-bp open reading frame (SMc00767) located downstream of aceA were isolated and functionally characterized in Sinorhizobium meliloti. Independent and double interposon mutants of each gene were constructed, and the corresponding phenotypes were analyzed. aceA mutants failed to grow on acetate, and mutants deficient in SMc00767 were also affected in acetate utilization. In contrast, mutants deficient in glcB grew on acetate similar to wild-type strain Rm5000. Complementation experiments showed that aceA and SMc00767 gene constructs were able to restore the growth on acetate in the corresponding single mutants. aceA-glcB, aceA-SMc00767, and glcB-SMc00767 double knockouts were also unable to grow on acetate, but this ability was recovered when the wild-type aceA-glcB or aceA-SMc00767 loci were introduced into the double mutants. These data confirm the functional role of aceA and SMc00767 and show that glcB, in the absence of SMc00767, is required for acetate metabolism. Isocitrate lyase and malate synthase activities were measured in strain Rm5000, the mutant derivatives, and complemented strains. aceA and glcB were able to complement the enzymatic activity lacking in the corresponding single mutants. The enzymatic activities also showed that SMc00767 represses the activity of isocitrate lyase in cells grown on acetate. Gene fusions confirmed the repressor role of SMc00767, which regulates aceA expression at the transcriptional level. Comparison of the transcriptional profiles of the SMc00767 mutant and wild-type strain Rm5000 showed that SMc00767 represses the expression of a moderate number of open reading frames, including aceA; thus, we propose that SMc00767 is a novel repressor involved in acetate metabolism in S. meliloti. Genetic and functional analyses indicated that aceA and SMc00767 constitute a functional two-gene operon, which is conserved in other α-proteobacteria. Alfalfa plants infected with the aceA and glcB mutants were not impaired in nodulation or nitrogen fixation, and so the glyoxylate cycle is not required in the Rhizobium-legume symbiosis.
Bacteria of the genera Rhizobium, Sinorhizobium, Mesorhizobium, and Bradyrhizobium fix nitrogen within nodules that they form in symbiotic association with legumes. Sinorhizobium meliloti, the bacterium that interacts with alfalfa plants, has been used as a model to study the Rhizobium plant-microbe interaction. An aspect that has been investigated in S. meliloti is carbon metabolism, both in free-living cells and in the symbiotic state. Several rhizobial enzymes, such as citrate synthase (29), isocitrate dehydrogenase (37), succinate dehydrogenase (24), and malate dehydrogenase (17), are essential for N2 fixation, indicating that a functional tricarboxylic acid (TCA) cycle is important in symbiosis (16). Anaplerotic pathways, such as the glyoxylate shunt, are essential for growth on C2 substrates, such as acetate, allowing bacterial cells to replenish the pool of TCA cycle intermediates necessary for supporting gluconeogenesis and other biosynthetic processes (9, 10, 30). This bypass is widespread in prokaryotes and plants (2, 18) and is encoded by two principal genes, aceA (encoding isocitrate lyase [ICL]) and glcB (encoding malate synthase [MS]). ICL cleaves isocitrate to glyoxylate and succinate, and MS condenses glyoxylate with acetyl coenzyme A (acetyl-CoA) to produce malate. Two isoenzymes of MS have been described in Escherichia coli; MSA encoded by the aceB gene is part of the ace operon (91 min), which is required for growth on acetate, and MSG encoded by the glcB gene mapped in the glc locus (64.5 min), which is inducible by glycolate (9). The presence of large amounts of acetate and fatty acids in soybean nodules encouraged early studies on the glyoxylate cycle (30). The role of this pathway in symbiosis was also supported by radiorespirometric studies of Bradyrhizobium japonicum bacteroids which indicated that as much as 50% of the acetyl-CoA entering the TCA cycle is metabolized via MS (48). It was also shown that acetate can be used by isolated B. japonicum bacteroids to support ex planta nitrogen fixation (42, 43). The existence of the glyoxylate cycle in bacteroids is in doubt because the activity of ICL has not been detected in bacteroids isolated from soybean, pea, alfalfa, and clover nodules (27, 30). However ICL activity has been detected in bacteroids from senesced nodules formed by B. japonicum (53). S. meliloti and B. japonicum cells grown on acetate contain ICL activity as well (15, 27, 36). In contrast, MS activity was found in bacteroids isolated from pea, alfalfa, and clover nodules, and substantially higher activities have been detected in bacteroids isolated from bean, cowpea, and soybean nodules (25, 30). The MS enzymatic activity seems to be constitutively expressed, since it has been detected in extracts of cells growing on acetate, glucose, arabinose, pyruvate, and malate (15, 27, 36). A better understanding of the role of the glyoxylate cycle in acetate metabolism and in the rhizobium-legume interaction could be achieved through analysis of mutants. Although putative aceA and glcB genes were annotated SMc00768 and SMc02581 in the complete genome sequence of S. meliloti, the role of these genes in C2 utilization has not been evaluated. This paper reports the functional roles of aceA, glcB, and an open reading frame (ORF) encoding a 79-amino-acid protein (SMc00767) in acetate metabolism and in the S. meliloti- legume symbiosis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and mating.
The bacterial strains and plasmids used are listed in Table 1. S. meliloti was grown in LB (10 g tryptone per liter, 5 g yeast extract per liter, 10 g NaCl per liter) (45), in PY medium (5 g tryptone per liter, 3 g yeast extract per liter) (3), or in M9 (45) minimal medium supplemented with thiamine (1 μg/ml), pantothenic acid (2 μg/ml), biotin (0.1 μg/ml), and potassium acetate (2 mM or 5 mM) as a carbon source. To determine growth rates on acetate, S. meliloti cells were grown to saturation in PY medium and then transferred to minimal medium with acetate. When required, the following antibiotics were added: rifampin (50 μg/ml), spectinomycin (25 μg/ml), tetracycline (2 μg/ml), streptomycin (25 μg/ml), and gentamicin (30 μg/ml). E. coli strains were grown in LB supplemented with 25 μg/ml spectinomycin, 10 μg/ml tetracycline, 20 μg/ml gentamicin, and 100 μg/ml ampicillin when needed. E. coli and rhizobia were grown at 30°C. Conjugation experiments were performed in the presence of the helper strain E. coli HB101 containing pRK2013 (20) as previously described (6).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| Rm5000 | SU47 rif-5 | 21 |
| RH190 | Rm5000 aceA::ΩSp/Sm | This study |
| RH198 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA | This study |
| RH218 | Rm5000 glcB::ΩTc | This study |
| RH222 | Rm5000 aceA::ΩSp/Sm glcB::ΩTc | This study |
| RH302 | Rm5000 glcB::ΩTc, pBBR1MCS-5 glcB | This study |
| RH312 | Rm5000 aceA::ΩSp/Sm glcB::ΩTc, pBBR1MCS-5 aceA-glcB | This study |
| RH326 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 | This study |
| RH327 | Rm5000 aceA::ΩSp/Sm, glcB::ΩTc, pBBR1MCS-5, aceA-SMc00767 | This study |
| RH419 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm | This study |
| RH421 | Rm5000 SMc00767::ΩSp/Sm | This study |
| RH429 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm, pBBR1MCS-5 glcB | This study |
| RH435 | Rm5000 SMc00767::ΩSp/Sm, pBBR1MCS-5 SMc00767 | This study |
| RH436 | Rm5000 glcB::ΩTc, SMc00767::ΩSp/Sm, pBBR1MCS-5 SMc00767 | This study |
| RH442 | Rm5000 SMc00767::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 with an internal 700-bp deletion in aceA that leaves wild-type SMc00767 and the intact aceA promoter region | This study |
| RH443 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 with an internal 700-bp deletion in aceA that leaves wild-type SMc00767 and the intact aceA promoter region | This study |
| RH462 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA-glcB | This study |
| RH465 | Rm5000 containing an aceA::gusA fusion in pBBR1MCS-5 | This study |
| RH467 | RH421 containing an aceA::gusA fusion in pBBR1MCS-5 | This study |
| E. coli strains | ||
| DH5α | recA1 φ80dlacZΔM15 gyrA96 | Gibco BRL |
| HB101 | recA13 rspL20 (Smr) | Gibco BRL |
| Plasmids | ||
| pCR2.1 | TA cloning vector for PCR products, Ampr Kmr | Invitrogen |
| pJQ200mp18 | Suicide vector for gene replacement, GmrsacB Mob | 44 |
| pRK2013 | ColE1 replicon with RK2 transfer region, Kmr Nmr | 20 |
| pHP45 | ΩSpr/Smr, vector Apr | 19 |
| pHP45 | ΩTcr, vector Apr | 19 |
| pBBRMCS-5 | Broad-host-range cloning vector, Mob InsP Gmr | 34 |
| pWM6 | ΩuidA2-aph Kmr/Nmr, vector Apr | 38 |
| pHL76 | pCR2.1 aceA | This study |
| pHL79 | pCR2.1 aceA::ΩSp/Sm | This study |
| pHL85 | pBBR1MCS-5 aceA | This study |
| pHL86 | pCR2.1 glcB | This study |
| pHL87 | pBBR1MCS-5 glcB | This study |
| pHL88 | pBBR1MCS-5 aceA-glcB | This study |
| pHL89 | pBBR1MCS-5 aceA-SMc00767 | This study |
| pHL90 | pBBR1MCS-5 aceA-SMc00767 | This study |
| pHL91 | pBBR1MCS-5 SMc00767 | This study |
| pHL92 | pBBR1MCS-5 aceA-SMc00767 with an internal 720-bp deletion in aceA that leaves wild-type SMc00767 and the intact aceA promoter region | This study |
| pHL93 | aceA::gusA fusion in pBBR1MCS-5 | This study |
| pHL95 | pJQ200mp18 aceA::ΩSp/Sm | This study |
| pHL96 | pCR2.1 with a 1,154-bp glcB fragment | This study |
| pHL97 | pCR2.1 with a 1,154-bp glcB fragment::ΩTc | This study |
| pHL98 | pJQ200mp18 with a 1,154-bp glcB fragment::ΩTc | This study |
| pHL99 | pJQ200mp18 aceA-SMc00767 | This study |
| pHL100 | pJQ200mp18 aceA-SMc00767::ΩSp/Sm | This study |
DNA manipulations.
Plasmid purification and genomic DNA extraction were performed according to published protocols (45). For hybridization, DNA was digested with EcoRI or PstI and then transferred from agarose gels to nylon membranes. Probes were labeled with 32P by polymerase extension using random primers, and hybridization was carried out under high-stringency conditions (47). For sequencing, double-stranded DNA was purified with a High Pure plasmid isolation kit (Boehringer Manheim, Germany), and sequencing was performed with an automatic Perkin-Elmer/Applied Biosystems 377-18 system.
Sequence analysis, primers, and PCR amplification.
The S. meliloti ORFs designated SMc00768 and SMc02581 were annotated in the genome as the glyoxylate cycle genes aceA and glcB, respectively. For SMc00767, which encodes a small hypothetical protein, no functional homologs were identified. Sequence analysis was carried out with the GCG programs from the Genetics Computer Group program suite (12). For DNA manipulations the aceA, glcB, and SMc00767 genes were amplified by PCR using genomic DNA from wild-type strain Rm5000. The following primers were used: for aceA, aceA3 (5′-GAGATTCAAATAGGAAGGAG-3′) and aceA8 (5′-ACAGTCATCGGAGTGCT-3′); and for glcB, glcB210 (5′-CAAGGACGGCTCGGGACA-3′), glcB3973 (5′-GCTCACAGACCACGACCACG-3′), msg387 (5′-CAATGCCCGCTGGGGCTCGCT-3′), and ms139 (5′-ATCGCCCACATGCCCTTG-3′). To amplify aceA-SMc00767, primers aceA3 and sm3 (5′-AATTCGGCATGAGCCTCCAG-3′) were used. PCR amplifications were performed in a 9700 thermocycler (Perkin-Elmer) with the following conditions: initial denaturation at 94°C for 3 min, followed by 34 cycles of denaturation (94°C, 2 min), annealing (55°C, 2 min), and extension (72°C, 3 min) and a final extension at 72°C for 5 min. PCR samples were electrophoresed through 0.8 to 1% agarose gels in Tris-acetate-EDTA buffer and stained with ethidium bromide.
Generation of mutants and complementation experiments.
To generate recombinant plasmids for mutagenesis, 1,755-bp, 1,154-bp, and 2,162-bp PCR products corresponding to aceA, glcB, and aceA-SMc00767 were cloned in pCR2.1 (Invitrogen) to obtain plasmids pHL76, pHL96, and pHL89, respectively (see Fig. S1A.1, S1A.9, and S1B.1 in the supplemental material). The antibiotic resistance cassettes pHP45Ω Sp/Sm and pHP45Ω Tc (19) were inserted into the aceA and glcB genes to generate plasmids pHL79 and pHL97 (see Fig. S1A.3 and S1A.10 in the supplemental material), and the interrupted genes were subcloned into the pJQ200mp18 vector (44), generating plasmids pHL95 and pHL98 (see Fig. S1A.4 and S1A.11 in the supplemental material). To generate a construct for SMc00767 mutagenesis, the aceA and SMc00767 genes from plasmid pHL89 were subcloned into pJQ200mp18, generating plasmid pHL99 (see Fig. S1B.5 in the supplemental material). This plasmid was digested with MluI and filled in with the Klenow fragment, and then the antibiotic resistance cassette (Sp/St) previously digested with SmaI was inserted into the SMc00767 gene to generate plasmid pHL100 (see Fig. S1B.6 in the supplemental material).
Recombinant plasmids harboring the interrupted genes were introduced into Rm5000, and mutants generated by double crossover were selected for each gene. To generate the aceA-glcB double knockout, the interrupted glcB gene was transferred into the aceA-deficient mutant. To generate the glcB-SMc00767 mutant, the interrupted SMc00767 gene was transferred into the glcB-deficient strain, and then mutants generated by double recombination events were isolated. To obtain knockouts, we used the sacRB selection system (44) and the appropriate antibiotics. Gene replacement was confirmed by PCR using appropriate primers and Southern blot hybridization.
To complement the mutant phenotypes, a 3,783-nucleotide PCR fragment corresponding to glcB and flanking sequences was cloned in pCR2.1, generating plasmid pHL86 (see Fig. S1A.7 in the supplemental material). The aceA, glcB, and aceA-SMc00767 genes from plasmids pHL76, pHL86, and pHL89 were then subcloned into pBBR1MCS-5, generating plasmids pHL85, pHL87, and pHL90, respectively (see Fig. S1A.2, S1A.8, and S1B.2 in the supplemental material). To complement the aceA-glcB double mutant, plasmid pHL86 was digested with XbaI and SpeI, and the liberated glcB gene was cloned into plasmid pHL85, generating plasmid pHL88, which contains aceA and glcB (see Fig. S1A.6 in the supplemental material). Additionally, the aceA-glcB mutant was complemented with plasmid pHL90 carrying the aceA and SMc00767 genes (see Fig. S1B.2 in the supplemental material). To complement the SMc00767 mutant, the SMc00767 gene was excised from plasmid pHL89 with StuI and XbaI and then ligated into pBBR1MCS-5, generating plasmid pHL91 (see Fig. S1B.4 in the supplemental material). Alternatively, this mutant was complemented with plasmid pHL92 (see Fig. S1B.3 in the supplemental material). To obtain this plasmid, pHL90 was digested with StuI and NruI and relegated to obtain a 720-bp internal deletion of the aceA gene.
Growth curves.
Bacterial strains were grown in PY medium overnight, and 4 ml was transferred to 50 ml of M9 medium with 2 mM acetate and cultivated overnight at 30°C at 200 rpm. Cells from the 2 mM acetate culture were used to inoculate 50 ml of M9 medium containing 5 mM acetate to an initial optical density at 595 nm (OD595) of 0.05. The cultures were incubated at 30°C at 200 rpm, and growth was followed by measuring the OD595 every 24 h.
Preparation of cell extracts.
To determine ICL activities, cells were grown to an OD595 of 0.7, harvested by centrifugation, washed with a saline solution, and resuspended in breaking buffer [20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (pH 7.0), 100 mM NaCl, 5 mM MgCl2, 0.4 mM EDTA, 1.5 mM dithiothreitol, 2% (wt/vol) glycerol] (27). The cells were sonicated on ice five times for 45 s with 45-s rest periods using a Soniprep 150 (MSE). The homogenate was centrifuged, and the supernatant was used for activity measurement. To determine MS activities, cells were grown to an OD595 of 0.7, harvested by centrifugation, washed, and resuspended in 100 mM Tris-HCl (pH 7. 5). The cells were sonicated on ice three times for 15 s with 1-min rest periods (23). The lysate was centrifuged to remove cell debris, and the supernatant was used for MS activity measurement. The protein concentrations in cell extracts were determined by the Bradford method (4), using bovine serum albumin as the standard.
Enzyme assays.
ICL activity was measured as described by Dixon and Kornberg (14). The assay mixtures (1 ml) contained 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.3), 5 mM MgCl2, 1 mM EDTA, 4 mM phenylhydrazine HCl, and S. meliloti extract. Isocitric acid was added to a final concentration of 12.5 mM to initiate the reaction. The increase in the level of the phenylhydrazone derivative of glyoxylate was measured at 324 nm. Negative controls without isocitrate were included in each experiment. MS activity was monitored by determining the glyoxylate-dependent release of free CoA from acetyl-CoA (36). The assay mixtures (0.5 ml) contained 100 mM Tris HCl (pH 7.5), 10 mM MgCl2, 2.5 mM glyoxylic acid, and S. meliloti extract. The reaction was initiated by addition of acetyl-CoA to a final concentration of 0.43 mM. After incubation for 5 min at room temperature, the reaction was stopped with 1 ml of 6 M urea. Color was developed by addition of 5,5′-dithiobis(2-nitrobenzoic acid) to a final concentration of 10 mM, and absorbance was determined at 412 nm. Negative controls without glyoxylate were included in each experiment. Enzymatic assays of ICL and MS activities were repeated four times.
Construction of transcriptional aceA-gusA reporter fusions.
An S. meliloti aceA-gusA transcriptional gene fusion was constructed as follows. Plasmid pHL85 (see Fig. S1A.2 in the supplemental material) harboring the aceA regulatory region as well as the entire aceA gene was digested with StuI (the restriction site was located 490 bp downstream of the aceA start codon), and plasmid pWM6 (38) was digested with SmaI to obtain a 3,727-bp fragment containing the gusA reporter gene. The 3,727-bp fragment was inserted into the StuI restriction site of plasmid pHL85, generating plasmid pHL93 (see Fig. S1A.5 in the supplemental material). The pHL93 plasmid containing the aceA::gusA fusion was introduced into wild-type strain Rm5000, as well as into the SMc00767 mutant RH421, generating strains RH465 and RH467, respectively.
β-Glucuronidase activity measurement.
To measure β-glucuronidase activity, separate cultures were grown in M9 containing 5 mM acetate to OD595s of 0.35, 0.7, and 1. A 1-ml aliquot of culture was centrifuged and resuspended in a salt wash solution supplemented with chloramphenicol (100 μg ml−1). Quantitative β-glucuronidase assays were performed with the p-nitrophenyl glucuronide substrate as described previously (52). Data were normalized to the total cell protein concentration by the Bradford method (4). The results presented below are the means of three independent experiments.
RNA isolation, synthesis of labeled cDNA, and microarray hybridization.
Sinorhizobium strains were grown at 30°C in M9 medium containing acetate to an OD595 of 0.7. Bacterial cells (100 ml) were collected, and total RNA was isolated by acid hot-phenol extraction as described previously (13). The concentration of RNA was determined by measuring the absorbance at 260 nm. The integrity of RNA was determined by running a 1.5% agarose gel. Ten micrograms of RNA was labeled differentially with Cy3-dCTP and Cy5-dCTP using a CyScribe First-Strand cDNA labeling kit (Amersham Biosciences). Pairs of Cy3- and Cy5-labeled cDNA samples were mixed and hybridized to the array as described by Hegde et al. (28). After washing, the arrays were scanned using a pixel size of 10 μm with a Scan Array Lite microarray scanner (Perkin-Elmer, Boston, MA). The S. meliloti 6205 70-mer oligonucleotide set was acquired from QIAGEN (Hilden, Germany) (https://www.operon.com/arrays/oligosets_sinorhizobium.php). The oligonucleotide set was resuspended and spotted in duplicate on SuperAmine-coated slides (25 by 75 mm; TeleChem International, Inc.) by a high-speed robot at the microarray facility at the Cellular Physiology Institute (Universidad Nacional Autónoma de México).
DNA microarray analysis.
Spot detection, mean signals, mean local background intensities, image segmentation, and signal quantification were determined for the microarray images using the Array-Pro Analyzer 4.0 software (Media Cybernetics, L.P). Microarray data analysis was performed with genArise software, developed in the Computing Unit of the Cellular Physiology Institute at Universidad Nacional Autónoma de México (http://www.ifc.unam.mx/genarise/). This software identifies differentially expressed genes by calculating an intensity-dependent z-score. It uses a sliding window algorithm to calculate the mean and standard deviation within a window surrounding each data point and defines a z-score where z measures the number of standard deviations that a data point is from the mean: zi = [Ri·mean(R)]/sd(R), where zi is the z-score for each element, mean(R) is the mean log ratio, Ri is the log ratio for each element, and sd(R) is the standard deviation of the log ratio. With this criterion, the elements in all experiments with a z-score of >2 standard deviations were considered significantly differentially expressed genes. DNA microarray experiments were performed three times with RNA isolated from independent cultures.
Plant nodulation experiments.
Seeds of Medicago sativa were surface sterilized for 15 min in sulfuric acid and for 5 min in 1.5% sodium hypochlorite and washed in sterile distilled water. They were germinated for 48 h on 0.75% agar at 30°C in the dark. Fourteen seedlings were transplanted into a pot containing vermiculite. For nodulation experiments, each plant was inoculated with 1 × 105 bacteria. After inoculation, the plants were transferred to a growth chamber and incubated at 21°C with a photoperiod consisting of 16 h of light and 8 h of darkness. After 35 days, nitrogen fixation was determined by examining acetylene reduction by using a gas chromatograph (5). Nodules were surface sterilized and then crushed in a sterile saline solution and plated on PY medium. One hundred colonies were replica plated with the appropriate antibiotic to ensure that cross-contamination had not taken place.
RESULTS AND DISCUSSION
Growth rates of aceA, SMc00767, and glcB mutants on acetate.
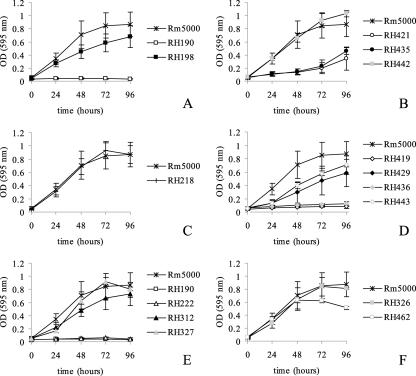
Mutants with mutations in the glyoxylate cycle genes were constructed and analyzed in liquid minimal medium supplemented with 5 mM potassium acetate as the carbon source (Fig. 1). The aceA mutant RH190 failed to grow on acetate, in contrast to wild-type strain Rm5000 and the complemented aceA mutant RH198, which contains the entire aceA gene (Fig. 1A). This result supports the essential role of aceA in the glyoxylate shunt. Downstream of and contiguous with aceA is a gene encoding a small hypothetical protein (79 amino acids) annotated SMc00767 (22) which appears to be part of a transcriptional unit with aceA. In order to evaluate if SMc00767 has a functional role in acetate metabolism, an SMc00767 mutant (RH421) was constructed. Growth profiles of mutant RH421 show that it was able to grow on acetate, albeit at a very reduced rate (Fig. 1B), indicating that the SMc00767 gene has a role in acetate metabolism. Complementation experiments with SMc00767 mutant RH421 were performed. The intergenic region between aceA and SMc00767 is 111 bp long. Based on this organization, we complemented SMc00767 mutant RH421 with plasmid pHL91, which contains the entire SMc00767 ORF and sequences 800 bp upstream and 200 bp downstream (see Fig. S1B.4 in the supplemental material). The complemented SMc00767 mutant strain, designated RH435, had the same growth rate as the parental SMc00767 mutant RH421 strain on acetate (Fig. 1B). This result suggested that SMc00767 transcription may require the aceA promoter. To validate this assertion, pHL92, a derivative plasmid of pHL90, was constructed. pHL92 has an internal deletion (720 bp) in the aceA structural gene and contains 485 bp of the aceA putative promoter, 570 bp of the aceA structural gene, and the entire intergenic aceA-SMc00767 region, as well as the complete SMc00767 gene (see Fig. S1B.3 in the supplemental material). Conjugal transfer of pHL92 into the SMc00767 mutant RH421 generated complemented strain RH442. The growth rate of the complemented SMc00767 mutant RH442 on acetate was the same as that of the wild-type strain (Fig. 1B). This finding indicates that expression of SMc00767 requires the promoter region of aceA and that these two genes are organized in an operon. In contrast to the aceA- and SMc00767-deficient strains, the glcB mutant RH218 grew similar to the Rm5000 wild-type strain on acetate (Fig. 1C), indicating that single mutations in glcB do not affect acetate utilization.
FIG. 1.
Functional role of aceA, SMc00767, and glcB in acetate utilization by S. meliloti. Bacteria were grown in M9 minimal medium supplemented with 5 mM potassium acetate. The OD595 was measured every 24 h over a 96-h period. The bacterial strains used were wild-type strain Rm5000, aceA mutant RH190, SMc00767 mutant RH421, glcB mutant RH218, an aceA mutant complemented with the wild-type aceA gene (RH198), an SMc00767 mutant complemented with the SMc00786 gene (RH435), an SMc00767 mutant complemented with the wild-type SMc00767 gene and the intact aceA promoter region (RH442, glcB-SMc00767 double mutant RH419, a glcB-SMc00767 double mutant complemented with glcB (RH429), a glcB-SMc00767 double mutant complemented with SMc00767 (RH436), a glcB-SMc00767 double mutant complemented with the aceA promoter region and SMc00767 (RH443), aceA-glcB double mutant RH222, an aceA-glcB double mutant complemented with aceA-glcB (RH312), an aceA-glcB double mutant complemented with aceA-SMc00767 (RH327), an aceA mutant complemented with aceA-SMc00767 (RH326), and an aceA mutant complemented with aceA-glcB (RH462). Growth kinetics were determined at least four times, and the graphs show the averages of all experiments.
Our results showed that the aceA mutant was unable to growth on acetate, while the growth rate of the SMc00767 mutant was reduced 65% in comparison to the growth rate of the wild- type strain. This observation indicated that another gene besides SMc00767 was involved in acetate utilization. Other data supporting the presence of an additional component for acetate utilization include the fact that the mutation in aceA had a polar effect on SMc00767, and so a second genetic component must be involved in the growth of the complemented aceA strain RH198 on acetate. In order to identify the additional genetic locus involved in acetate metabolism, an SMc00767-glcB double mutant, RH419, was constructed. glcB was chosen because it is involved in acetate metabolism in many bacterial species, although we have shown that the glcB single mutant was not affected in acetate utilization (Fig. 1C). The growth profiles of the SMc00767-glcB double mutant (Fig. 1D) show that this strain is unable to grow on acetate, indicating that glcB has a functional role in the absence of SMc00767. This result indicates that aceA itself is unable to restore growth on acetate and demonstrates the necessity of SMc00767 or glcB for growth on acetate. To support this hypothesis, we transferred plasmids pHL92 (harboring the aceA promoter and the SMc00767 structural gene), pHL87 (containing the glcB gene), and pHL91 (harboring the SMc00767 gene without the aceA promoter) (see Fig. S1B.3, S1A.8, and S1B.4 in the supplemental material) independently to the SMc00767-glcB double mutant RH419, obtaining the complemented strains RH443, RH429, and RH436, respectively. The growth of the SMc00767-glcB double mutant complemented with the aceA promoter and the SMc00767 structural gene (RH443) and the growth of the strain with glcB (RH429) were partially reestablished on acetate. In contrast, the SMc00767-glcB mutant complemented with SMc00767 without the aceA promoter was unable to restore growth (Fig. 1D). The results of these experiments support the hypothesis that SMc00767 and glcB have a functional role in acetate metabolism and provide further genetic evidence of the aceA-SMc00767 operon organization. To ascertain the requirement for SMc00767 and glcB for growth on acetate, an aceA-glcB double mutant (RH222) was constructed. The aceA-SMc00767 mutant strain RH190 (see above) and the aceA-glcB mutant strain RH222 were evaluated for growth on acetate. Neither mutant was able to grow on acetate as a carbon source (Fig. 1E). Complementation experiments with the aceA-glcB RH222 mutant strain were performed. Plasmid pHL88 (see Fig. S1A.6 in the supplemental material), which harbors the aceA and glcB genes, was transferred to the aceA-glcB RH222 mutant, generating the RH312 derivative. The complemented aceA-glcB mutant strain RH312 exhibited growth similar to that of the wild-type strain on acetate (Fig. 1E). In addition we generated the complemented strain RH327 by introducing plasmid pHL90 (see Fig. S1B.2 in the supplemental material), which contains aceA as well as SMc00767, into aceA-glcB mutant RH222. As shown in Fig. 1E, the aceA-glcB mutant, complemented with the wild-type aceA and SMc00767 genes, also reestablished growth on acetate. The aceA-SMc00767 double mutant RH190 was also complemented with plasmid pHL88, which contains aceA and glcB, and plasmid pHL90, which contains aceA and SMc00767 (see Fig. S1A.6 and Fig. S1B.2 in the supplemental material) (strains RH462 and RH326, respectively). The growth profiles on acetate (Fig. 1F) of the aceA-SMc00767 mutant complemented with aceA-glcB or aceA and SMc00767 are similar to those of wild-type strain Rm5000 (Fig. 1F). These results show that aceA and SMc00767, as well as glcB, are involved in acetate metabolism in S. meliloti. In order to determine if aceA, SMc00767, and glcB are functional with other carbon sources, growth rate experiments were performed with different carbon compounds. Previously, we reported (23) that glcB was induced on minimal medium supplemented with arabinose as the carbon source, and it has also been reported that GlcB activity was detectable in minimal medium supplemented with succinate, arabinose, or malate (15, 27). To evaluate if glcB has a functional role in the utilization of these carbon compounds, we performed growth rate experiments with the glcB mutant RH218 and the complemented glcB strain RH302 in minimal medium supplemented with arabinose, succinate, or glucose and in PY medium. With the different carbon sources tested, the growth of the glcB mutant and the complemented RH302 strain was identical to the growth of the wild-type Rm5000 strain (data not shown). This indicates that glcB is not essential for arabinose, succinate, glucose, or PY medium utilization. Growth rate experiments were also performed with the aceA mutant RH190 and the SMc00767 mutant RH421 in minimal medium supplemented with glucose and in PY medium. We found that the growth of the aceA mutant RH190 and the growth of the SMc00767 mutant RH421 were similar to the growth of the Rm5000 wild-type strain (data not shown), indicating that the aceA and SMc00767 loci are not involved in glucose or PY medium utilization.
Activity of glyoxylate cycle enzymes.
A series of enzymatic assays were carried out with cell extracts of mutants and complemented strains growing on acetate as the carbon source. Table 2 shows the loss of ICL activity in the aceA mutant RH190. The complemented aceA mutant RH198, harboring the single aceA gene, had eightfold more ICL activity than the wild-type strain (Table 2). The glcB mutant RH218 lacked MS activity, while the glcB mutant complemented with glcB (RH302) exhibited a level of MS activity similar to that of the wild-type strain (Table 2). These results indicate that the inability of the aceA mutant to grow on acetate was due to the aceA mutation (Fig. 1A). The null activity of the glcB mutant and the restored activity of the complemented strain showed that a single MS gene was present in S. meliloti. The activities of the glyoxylate cycle enzymes ICL and MS were also measured in the aceA-glcB double mutant RH222. The double mutant was unable to grow on acetate (Fig. 1E) and had no ICL and MS activities (Table 2). The aceA-glcB mutant complemented with aceA and glcB (strain RH312) exhibited growth on acetate (Fig. 1E), as well as ICL and MS activities, and in the case of ICL, it had sevenfold more activity that the wild-type Rm5000 strain. The aceA-glcB double mutant complemented with aceA-SMc00767 (strain RH327) also exhibited ICL activity, but it showed only a threefold increase in ICL activity compared to the wild type (Table 2).
TABLE 2.
ICL and MS activities in cell extracts of S. meliloti strainsa
| Strain | Genotype | Enzyme sp act (nmol min−1 mg protein−1)
|
|
|---|---|---|---|
| ICL | MS | ||
| Rm5000 | Wild type | 138 ± 21 | 179 ± 27 |
| RH190 | Rm5000 aceA::ΩSp/Sm | 6.7 | 43 |
| RH198 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA | 1,080 ± 190 | 172 ± 21 |
| RH218 | Rm5000 glcB::ΩTc | 297 ± 26 | ND |
| RH222 | Rm5000 aceA::ΩSp/Sm glcB::ΩTc | ND | ND |
| RH302 | Rm5000 glcB::ΩTc, pBBR1MCS-5 glcB | 190 ± 18 | 230 ± 25 |
| RH312 | Rm5000 aceA::ΩSp/Sm glcB::ΩTc, pBBR1MCS-5 aceA-glcB | 1,030 ± 209 | 251 ± 26 |
| RH326 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 | 341 ± 22 | 115 ± 24 |
| RH327 | Rm5000 aceA::ΩSp/Sm glcB::ΩTc, pBBR1MCS-5 aceA-SMc00767 | 443 ± 50 | ND |
| RH419 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm | 12 ± 10 | ND |
| RH421 | Rm5000 SMc00767::ΩSp/Sm | 685 ± 39 | 132 ± 19 |
| RH429 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm, pBBR1MCS-5 glcB | 731 ± 37 | 907 ± 220 |
| RH435 | Rm5000 SMc00767::ΩSp/Sm, pBBR1MCS-5 SMc00767 | 657 ± 36 | 141 ± 2 |
| RH442 | Rm5000 SMc00767::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 with an internal 700-bp deletion in aceA that leaves wild-type SMc00767 and the intact aceA promoter region | 96 ± 18 | 116 ± 6 |
| RH443 | Rm5000 glcB::ΩTc SMc00767::ΩSp/Sm, pBBR1MCS-5 aceA-SMc00767 with an internal 700-bp deletion in aceA that leaves wild-type SMc00767 and the intact aceA promoter region | 216 ± 31 | ND |
| RH462 | Rm5000 aceA::ΩSp/Sm, pBBR1MCS-5 aceA-glcB | 832 ± 194 | 1,343 ± 303 |
Bacteria were grown in M9 medium supplemented with 5 mM potassium acetate as the carbon source. In most cases the values are the averages of at least four separate experiments; the exception is the values for strain RH190, for which only one experiment was done. ND, not detected.
These results show that in the absence of SMc00767 ICL overexpression occurs, suggesting that SMc00767 is a repressor of the aceA gene. To validate this suggestion, ICL activities were measured. The aceA-SMc00767 mutant RH190, as mentioned above, has no ICL activity, but when it was complemented with the aceA and SMc00767 genes (strain RH326), it exhibited a twofold increase in ICL activity compared with the wild type (Table 2). In contrast, mutant RH421, lacking SMc00767, had five times as much in ICL activity as the wild type (Table 2). The SMc00767 mutant complemented with the SMc00767 gene without the aceA promoter (strain RH435) showed no reduction in ICL activity, while the SMc00767 mutant complemented with the aceA promoter and the SMc00767 gene (strain RH442) had a reduced level of ICL activity similar to the level of wild-type strain Rm5000. In addition, when glcB-SMc00767 mutant RH419, which showed no growth on acetate (Fig. 1D), was complemented with glcB (strain RH429), it showed a fivefold increase in ICL activity, while the glcB-SMc00767 mutant complemented with the aceA promoter and the SMc00767 gene (strain RH443) had ICL activity similar to that of the wild type. Together, these data indicate that SMc00767 repressed aceA expression. To determine if the glyoxylate cycle activities were present during grown on other carbon sources, ICL and MS enzymatic assays were performed. In PY medium, ICL activity was not detected in wild-type strain Rm5000 or in the SMc00767 mutant RH421 (which overexpressed ICL activity in the presence of acetate), supporting the specificity of ICL activity in acetate metabolism. To evaluate if MS activity was present during grown on other carbon sources, wild-type strain Rm5000 and the glcB mutant RH218 were grown with glucose, arabinose, or succinate as the sole carbon source. The glcB mutant RH218, as expected, had no MS activity on any carbon source tested. In contrast, on arabinose, succinate, and glucose, wild-type strain Rm5000 had MS activities that were 20, 15, and 17% of the activity on acetate. Together, the growth rate experiments and the MS activities suggest that this enzyme is not required for arabinose, succinate, or glucose metabolism.
SMc00767 regulates aceA expression on acetate.
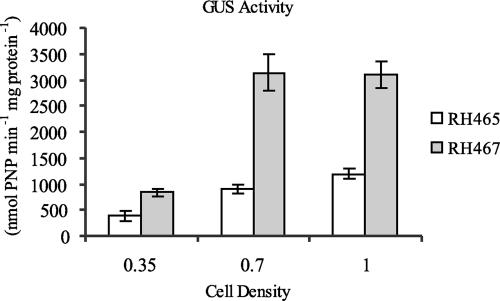
The enzymatic data indicate that SMc00767 represses aceA expression. To evaluate if this repression occurs at the transcriptional level, gene fusions were constructed in which the gusA reporter gene was inserted downstream of the aceA promoter of plasmid pHL85 to obtain plasmid pHL93 (see Fig. S1A.5 in the supplemental material). The resulting aceA promoter-gusA fusion plasmid was introduced into wild-type strain Rm5000 and the SMc00767 mutant to generate strains RH465 and RH467, respectively. β-Glucuronidase activity was determined for both strains in M9 minimal medium supplemented with 5 mM acetate. The transcriptional assays revealed that at three different culture OD595s (0.35, 0.7, and 1), the strain with the aceA promoter in the absence of SMc00767 had significantly greater β-glucuronidase activity than wild-type strain Rm5000 harboring the aceA promoter-gusA fusion plasmid (Fig. 2). Thus, we demonstrated, by using expression assays of the aceA promoter as well as by examining ICL enzymatic activity, that SMc00767 is a repressor of the aceA gene. To determine if SMc00767 repressed aceA expression in the presence of other carbon sources, β-glucuronidase assays were performed with these strains using succinate-, arabinose-, and glucose-grown cultures. At three different OD595s (0.35, 0.7, and 1), aceA expression was not detected with arabinose, glucose, or succinate in RH465 and RH467 (data not shown), supporting the hypothesis that aceA and SMc00767 have a specific role in acetate metabolism.
FIG. 2.
β-Glucuronidase activities of RH465 (wild-type strain Rm5000 containing the aceA::gusA fusion) and RH467 (SMc00767 mutant RH421 harboring the aceA::gusA fusion) grown in M9 medium containing acetate. The cultures were collected at OD595s of 0.35, 0.7, and 1. The values are the means of three independent experiments performed in duplicate. PNP, p-nitrophenyl; GUS, β-glucuronidase.
Transcriptional profiling of the S. meliloti wild-type Rm5000 and SMc00767 mutant strains grown on acetate.
The enzymatic and transcriptional results presented above indicate that SMc00767 is a novel regulator involved in acetate metabolism in rhizobia. To investigate the role of the SMc00767 gene in the global regulation of acetate metabolism and to identify further putative target genes of SMc00767, transcriptional profiles of wild-type strain Rm5000 and the SMc00767 mutant were compared. These strains were grown on 5 mM acetate to an OD595 of 0.7, the bacteria were collected, and the total RNA was isolated, labeled, and hybridized with slides that contained the genome oligonucleotide set acquired from QIAGEN. We found that in the SMc00767 mutant seven genes were overexpressed with a z-score of >2 standard deviations (Table 3). These genes correspond to four hypothetical proteins, dnaB, potH, and aceA. These genes are dispersed in the S. meliloti genome: SMb21456 and SMb21463 are encoded on pSymB, SMa2071 is encoded on pSymA, and aceA, SMc00769, potH, and dnaB are encoded on the chromosome. Interestingly, one of the genes encoding a hypothetical protein (SMc00769) and potH are located downstream of aceA. In S. meliloti, SMc00767 is clustered with aceA, SMc00769 potF, potG potH, and potI, and the microarray data show that three of these genes are overexpressed in the SMc00767 mutant, suggesting that this cluster of genes is involved in acetate utilization. The reason why the microarray experiments failed to detect all the genes of this cluster could be RNA degradation, RNA instability, or low mRNA levels. However, with the exception of aceA, the genes overexpressed in the microarray experiments represent novel genes for acetate metabolism in S. meliloti. Additional work is necessary to assign specific roles to these genes. The moderate number of overexpressed genes obtained with the microarray experiments shows that SMc00767 is a local repressor of acetate metabolism in S. meliloti. In agreement with the enzymatic data and expression analysis of the aceA gene, the microarray experiment also shows that SMc00767 represses aceA transcription; thus, we considered aceA a good internal control to validate the effect of the SMc00767 gene on the expression of other S. meliloti genes.
TABLE 3.
Genes significantly induced in the SMc00767 mutant as detected by microarray analysis
| ORF | Description and/or gene | Fold change in SMc00767 mutant vs wild type |
|---|---|---|
| SMa2071 | Hypothetical protein | 2.6 |
| SMb21456 | Hypothetical protein | 2.4 |
| SMb21473 | Conserved hypothetical protein | 2.9 |
| SMc00561 | Probable replicative DNA helicase protein, dnaB | 3.5 |
| SMc00768 | ICL protein, aceA | 4.6 |
| SMc00769 | Conserved hypothetical protein | 2.6 |
| SMc00772 | Probable putrescine transport system permease protein, potH | 2.0 |
SMc00767 regulates the expression of a conserved cluster of genes in rhizobia.
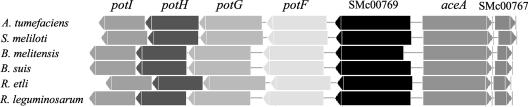
The enzymatic data and the transcriptional results show that SMc00767 regulates the expression of aceA, and the microarray experiments indicate that SMc00767 also regulates the expression of two genes located downstream of aceA. To determine if the genes repressed by SMc00767 are conserved in rhizobia, genomes of several α-proteobacteria were analyzed. The alignment in Fig. 3 shows that the transcriptional repressor SMc00767, as well as aceA, SMc00769, potF, potG, potH, and potI, are contiguous in Agrobacterium tumefaciens (26, 54), Brucella melitensis (11), Brucella suis (41), Rhizobium etli (25), Rhizobium leguminosarum (55), and S. meliloti (22). This finding, in light of the S. meliloti microarray data, suggests that these genes have a role in acetate metabolism in α-proteobacteria.
FIG. 3.
Genome context of the SMc00767, aceA, SMc00769, potF, potG, potH, and potI genes in different members of the α-proteobacteria. In all cases, SMc00767 is clustered in this group of genes, suggesting that they are involved in a common process. Genes and intergenic distances are drawn in proportion. The figure was generated using GeCont (7).
Symbiotic phenotypes of the S. meliloti aceA and glcB mutants.
Recently, studies with Candida albicans (35), Magnaphorte grisea (51), Mycobacterium tuberculosis (39), Rhodococcus equi (50), Rhodococcus fascians (49), and Stagonospora nodorum (46) showed that the glyoxylate bypass is essential for virulence in both animal and plant pathogens. However, the functional role of this pathway in the Rhizobium-legume symbiosis has not been explored. To determine if the glyoxylate cycle has a role in symbiosis, plant nodulation experiments were performed. Fourteen alfalfa plants were inoculated independently with the aceA mutant, the glcB mutant, and the aceA-glcB double mutant and analyzed to determine the number of nodules, acetylene reduction, and dry weight 42 days postinoculation. The results show that the wild-type Rm5000 strain produced 18 nodules per plant and the aceA, glcB, and aceA-glcB mutants formed 19, 17, and 15 nodules per plant, respectively. Wild-type strain Rm5000, the aceA and glcB mutants, and the aceA-glcB double mutant had nitrogenase specific activities of 0.063, 0.096, 0.087, and 0.078 μmol of acetylene·plant−1·h−1, respectively. The dry weights of the 14 plants inoculated with wild-type strain Rm5000 and the aceA, glcB, and aceA-glcB mutants were 17.5, 19.5, 22, and 23.6 mg, respectively. Together, these data indicate that the glyoxylate cycle genes in S. meliloti are not involved in nodulation and nitrogen fixation.
The glyoxylate bypass has been established in several rhizobia (23, 27, 30, 36), but little research has focused on the functionality of this pathway in these organisms. The enzymatic activities of ICL and MS in several Rhizobium species indicated that the glyoxylate shunt was functional (15, 27, 30, 36). However, systematic studies of mutants with mutations in each of these genes have not been done until now. In this report we show that aceA and a 240-bp ORF designated SMc00767 are the principal genes for acetate metabolism in S. meliloti. The results obtained indicate that in rhizobia, as well as in other bacterial species, aceA is required in the glyoxylate cycle, since mutations in this gene completely abolish growth on acetate. A remarkable and interesting finding of this work was the identification of SMc00767, which is present only in symbionts, plant and animal pathogens such as S. meliloti, R. etli, R. leguminosarum, A. tumefaciens, and different Brucella spp. In all the organisms analyzed, SMc00767 is downstream of aceA in an operon, indicating that in rhizobia these genes operate in the same metabolic process. Our results support this hypothesis, since we show that the presence of SMc00767 is essential for optimal growth on acetate and that it regulates aceA transcription. Our results indicate that the C2 metabolism in rhizobia is completely different from that in other bacteria, such as E. coli (31, 40). This assertion is also supported by the fact that S. meliloti glcB mutants are able to grow on acetate; this is an important result since until now most studies of acetate metabolism have shown that glcB mutants are unable to grow on acetate, while in S. meliloti glcB has a secondary role in the utilization of C2 compounds. This result is similar to that obtained by Cornah et al. (8), who reported that Arabidopsis mutants lacking MS are capable of gluconeogenesis from acetate. These authors suggested that a new metabolic pathway to metabolize acetate to sugars in the absence of MS is present in Arabidopsis seedlings, and recent studies with Rhodobacter sphaeroides (1) and Methylobacterium extorquens (32, 33) provided evidence of alternative acetate assimilation pathways. Thus, the possibility of new metabolic pathways for C2 compounds in rhizobia exists. In the case of S. meliloti we believe that the primary route for acetate utilization depends on aceA and SMc00767, but in the absence of SMc00767, aceA and glcB are able to support growth on acetate, indicating that S. meliloti is able to utilize acetate in these two ways.
The symbiotic performance of the S. meliloti aceA and glcB mutants shows that these genes are not involved in the interaction with alfalfa plants. Recently, we isolated and sequenced two aceA genes from Rhizobium tropici, one located on the chromosome and the other encoded on the symbiotic plasmid. An R. tropici aceA double mutant was constructed, and analysis of the symbiotic performance on bean plants (Phaseolus vulgaris) showed that this mutant was not affected in nodulation or nitrogen fixation (data not shown). Thus, aceA does not appear to be involved in the S. meliloti-alfalfa and R. tropici-P. vulgaris symbioses.
Previous reports showed that MS activity is present in bacteroids from pea, alfalfa, and clover, and substantially higher activities were detected in bacteroids from bean, cowpea, and soybean. However, our results show that MS is not involved in nodulation or nitrogen fixation in the interaction of S. meliloti with alfalfa plants, and we previously reported that pea plants inoculated with an R. leguminosarum glcB mutant showed no significant differences in nitrogen fixation (23). Thus, we have demonstrated that glcB is dispensable in plant-microbe interactions in two symbiotic systems.
Supplementary Material
Acknowledgments
We thank G. Uribe-Figueroa, S. I. Fuentes, F. J. Santana, M. Fernández-Mora, A. Vazquez, J. Miranda-Ríos, H. Salgado, P. Mavingui, R. Oropeza, and J. Caballero-Mellado for technical help and useful scientific comments to improve the manuscript.
This research was supported by grants to I.H.-L. from DGAPA/UNAM (IN206802 and IN206705). J.A.R.-T. is a recipient of a Ph.D. studentship from the Consejo Nacional de Ciencia y Tecnología (Mexico).
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 62:297-309. [DOI] [PubMed] [Google Scholar]
- 2.Beeckmans, S. April 2001, posting date. Glyoxylate cycle. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom. http://www.els.net/.
- 3.Beringer, J. E. 1974. R-factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burris, R. H. 1972. Nitrogen fixation assay—methods and techniques. Methods Enzymol. 24:415-431. [DOI] [PubMed] [Google Scholar]
- 6.Charles, T. C., and T. M. Finan. 1990. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47B. J. Bacteriol. 172:2469-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciria, R., C. Abreu-Goodger, E. Morett, and E. Merino. 2004. GeConT: gene context analysis. Bioinformatics 20:2307-2308. [DOI] [PubMed] [Google Scholar]
- 8.Cornah, J. E., V. Germain, J. L. Ward, M. H. Beale, and S. M. Smith. 2004. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279:42916-42923. [DOI] [PubMed] [Google Scholar]
- 9.Cozzone, A. J. 1998. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu. Rev. Microbiol. 52:127-164. [DOI] [PubMed] [Google Scholar]
- 10.Cronan, J. E., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 201-206. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 11.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereaux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vries, S., H. Hoge, and T. Bisseling. 1988. Isolation of total and polysomal RNA from plant tissues, p. 1-13. In S. B. Gelvin, R. A. Schilperoort, and D. P. S. Verma (ed.), Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 14.Dixon, G. H., and H. L. Kornberg. 1959. Assay methods for key enzymes of the glyoxylate cycle. Biochem. J. 72:3P. [Google Scholar]
- 15.Duncan, M. J., and D. G. Fraenkel. 1979. Alpha-ketoglutarate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 137:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, M. F. 1998. Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol. Rev. 22:105-123. [DOI] [PubMed] [Google Scholar]
- 17.Dymov, I. S., D. J. J. Meek, B. Steven, and B. T. Driscoll. 2004. Insertion of transposon Tn5tac1 in the Sinorhizobium meliloti malate dehydrogenase (mdh) gene results in conditional polar effects on downstream TCA cycle genes. Mol. Plant-Microbe Interact. 17:1318-1327. [DOI] [PubMed] [Google Scholar]
- 18.Escher, C. L., and F. Widmer. 1997. Lipid mobilization and gluconeogenesis in plants: do glyoxylate cycle enzyme activities constitute a real cycle? A hypothesis. Biol. Chem. 378:803-813. [PubMed] [Google Scholar]
- 19.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 20.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finan, T. M., E. Hartwieg, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, A. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, M. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F. J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K. Chen Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-de los Santos, A., A. Morales, L. Baldoma, S. R. Clark, S. Brom, C. K. Yost, I. Hernandez-Lucas, J. Aguilar, and M. F. Hynes. 2002. The glcB locus of Rhizobium leguminosarum VF39 encodes an arabinose-inducible malate synthase. Can. J. Microbiol. 48:922-932. [DOI] [PubMed] [Google Scholar]
- 24.Gardiol, A., A. Arias, C. Cerveñansky, and G. Martínez-Drets. 1982. Succinate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 151:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 103:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 27.Green, L. S., D. B. Karr, and D. W. Emerich. 1998. Isocitrate dehydrogenase and glyoxylate cycle enzyme activities in Bradyrhizobium japonicum under various growth conditions. Arch. Microbiol. 169:445-451. [DOI] [PubMed] [Google Scholar]
- 28.Hegde, P., R. Qi, R. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 29.Hernández-Lucas, I., M. A. Pardo, L. Segovia, J. Miranda, and E. Martínez. 1995. Rhizobium tropici chromosomal citrate synthase gene. Appl. Environ. Microbiol. 61:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, G. V., H. J. Evans, and T. Ching. 1966. Enzymes of glyoxylate cycle in rhizobia and nodules of legumes. Plant Physiol. 41:1330-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornberg, H. L. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 99:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korotkova, N., L. Chistoserdova, V. Kuksa, and M. E. Lidstrom. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korotkova, N., M. E. Lidstrom, and L. Chistoserdova. 2005. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 187:1523-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 35.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 41:83-86. [DOI] [PubMed] [Google Scholar]
- 36.Mandal, N. C., and P. K. Chakrabartty. 1992. Regulation of enzymes of glyoxylate pathway in root-nodule bacteria. J. Gen. Appl. Microbiol. 38:417-427. [Google Scholar]
- 37.McDermott, T. R., and M. Kahn. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 39.Muñoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ornston, L. N., and M. K. Ornston. 1969. Regulation of glyoxylate metabolism in Escherichia coli K-12. J. Bacteriol. 98:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson, J. B., and T. A. LaRue. 1981. Utilization of aldehydes and alcohols in soybean bacteroids. Plant Physiol. 68:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson, J. B., and T. A. LaRue. 1982. Soluble aldehyde dehydrogenase and metabolism of aldehydes by soybean bacteroids. J. Bacteriol. 151:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch., and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Solomon, P. S., R. C. Lee, T. J. Wilson, and R. P. Oliver. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53:1065-1073. [DOI] [PubMed] [Google Scholar]
- 47.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 48.Stovall, I., and M. Cole. 1978. Organic acid metabolism by isolated Rhizobium japonicum bacteroids. Plant Physiol. 61:787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vereecke, D., K. Cornelis, W. Temmerman, M. Jaziri, M. Van Montagu, M. Holsters, and K. Goethals. 2002. Chromosomal locus that affects pathogenicity of Rhodococcus fascians. J. Bacteriol. 184:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wall, D. M., P. S. Duffy, C. Dupont, J. F. Prescott, and W. G. Meijer. 2005. Isocitrate lyase activity is required for virulence of the intracellular pathogen Rhodococcus equi. Infect. Immun. 73:6736-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Z. Y., C. R. Thornton, M. J. Kershaw, L. Debao, and N. J. Talbot. 2003. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 47:1601-1612. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, K. J., S. G. Huges, and R. A. Jefferson. 1992. The Escherichia coli gus operon, induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria, p. 7-23. In S. R. Gallagher (ed.), Gus protocols, using the gus gene as a reporter of gene expression, vol. 1. Academic Press, San Diego, CA. [Google Scholar]
- 53.Wong, P. P., and H. J. Evans. 1971. Poly-β-hydroxybutyrate utilization by soybean (Glycine max Merr) nodules and assessment of its role in maintenance of nitrogenase activity. Plant Physiol. 47:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 55.Young, J. P., L. C. Crossman, A. W. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.