Abstract
The cold shock response of Escherichia coli is elicited by downshift of temperature from 37°C to 15°C and is characterized by induction of several cold shock proteins, including CsdA, during the acclimation phase. CsdA, a DEAD-box protein, has been proposed to participate in a variety of processes, such as ribosome biogenesis, mRNA decay, translation initiation, and gene regulation. It is not clear which of the functions of CsdA play a role in its essential cold shock function or whether all do, and so far no protein has been shown to complement its function in vivo. Our screening of an E. coli genomic library for an in vivo counterpart of CsdA that can compensate for its absence at low temperature revealed only one protein, RhlE, another DEAD-box RNA helicase. We also observed that although not detected in our genetic screening, two cold shock-inducible proteins, namely, CspA, an RNA chaperone, and RNase R, an exonuclease, can also complement the cold shock function of CsdA. Interestingly, the absence of CsdA and RNase R leads to increased sensitivity of the cells to even moderate temperature downshifts. The correlation between the helicase activity of CsdA and the stability of mRNAs of cold-inducible genes was shown using cspA mRNA, which was significantly stabilized in the ΔcsdA cells, an effect counteracted by overexpression of wild-type CsdA or RNase R but not by that of the helicase-deficient mutant of CsdA. These results suggest that the primary role of CsdA in cold acclimation of cells is in mRNA decay and that its helicase activity is pivotal for promoting degradation of mRNAs stabilized at low temperature.
Cold shock response is elicited when exponentially growing cells of Escherichia coli are shifted from 37°C to 15°C (for a review, see reference 23). This response is characterized by a transient arrest of cell growth termed the acclimation phase, during which a number of genes are induced, in contrast to a severe inhibition of general protein synthesis. The highly induced cold shock proteins include CspA (8) and its homologues, such as CspB (15), CspG (22), and CspI (35), transcription factor NusA (7), polynucleotide phosphorylase (6), initiation factor IF2 (9), RecA (34), histone-like protein H-NS (5), DNA gyrase (12), ribosome-binding factor RbfA (4), and RNA helicase CsdA (31).
CsdA is a DEAD-box protein which belongs to the large family of putative RNA helicases conserved from bacteria to humans (17). Along with the related DEXD/H-box proteins, they play important roles in many cellular processes, such as processing, transport, or degradation of RNA or ribosome biogenesis (for a review, see reference 10). E. coli contains five DEAD-box genes, csdA (formerly called deaD), dbpA, rhlB, rhlE, and srmB. Despite their importance in RNA metabolism, the exact function of these proteins has not been elucidated. Except for DbpA, these proteins do not exhibit RNA specificity in vitro. Among these, CsdA has been identified as a multifunctional protein. It is essential at low temperature, and deletion of its gene impairs growth upon cold shock (3, 13). On the other hand, it is dispensable at 37°C. The deaD/csdA gene was originally identified as a multicopy suppressor of a mutation in the rpsB gene that encodes r-protein (31). This suggested that CsdA may play a role in the biogenesis of the small ribosomal subunit, and later on it was reported that overexpression of CsdA in an S2 mutant restores the incorporation of r-proteins S2 and S1 into the 30S ribosomal subunits (21). It has also been suggested that CsdA assists in translation by promoting translation initiation of structured mRNAs (18). This suggestion is supported by the observation that CsdA shows 54% identity and 87% sequence similarity to the eukaryotic initiation factor eIF4A that catalyzes the ATP-dependent unwinding of RNA duplexes and stimulates translational initiation in eukaryotic cells (18).
CsdA has been implicated in the stabilization and degradation of mRNAs (14, 37). It accumulates during the early stages of cold acclimatization and assembles into degradosomes with RNase E synthesized in cold-adapted cultures (27). It was also reported that CsdA may be involved in the efficient and selective degradation of Csp mRNAs by unwinding the mRNA secondary structure that impedes the processive activity of polynucleotide phosphorylase (PNPase) (37). Recently, it was shown that CsdA is involved in the biogenesis of the 50S rather than the 30S ribosomal subunits and that deletion of the csdA gene leads to a deficit in free 50S subunits and accumulation of a 40S-like particle. The authors also showed that CsdA associates with 50S precursors at low temperature and can complement the ribosome defect of an srmB deletion strain (3). Altogether, these data suggest that CsdA has multiple overlapping functions in several important physiological processes. It is not clear which of the activities of CsdA play a role in its essential cold shock function or whether they all do, and so far no protein has been shown to complement its function in vivo. To elucidate the cold shock function of CsdA, we screened for genes which when expressed from multicopy plasmids can complement the cold-sensitive phenotype of the csdA deletion cells at 15°C. We observed that another DEAD-box RNA helicase, RhlE, can complement CsdA function at low temperature. This finding emphasized the importance of helicase activity for the acclimation of cells to cold temperatures. We thus carried out mutational analysis of DEAD box of CsdA and show that the first two amino acids of the DEAD box are essential for the cold-acclimation activity of CsdA. We also observed that although not detected in our genetic screening, two cold shock-inducible proteins, CspA, an RNA chaperone, and RNase R, an exonuclease, can also complement the cold shock function of CsdA. We also observed that the absence of CsdA and RNase R results in increased sensitivity of the cells to moderate temperature downshifts. We tested the correlation between the helicase activity of CsdA and stability of mRNAs of cold-inducible genes by use of cspA mRNA, which was significantly stabilized in the ΔcsdA cells. This stabilization was reversed by the overexpression of wild-type CsdA or RNase R but not by the overexpression of a helicase-deficient mutant of CsdA. Based on the results presented here, we conclude that involvement of CsdA in low-temperature mRNA decay may be its primary role and that its helicase activity is crucial for promoting degradation of mRNAs stabilized at low temperature.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains MC4100 [F− araD139Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR] (19) and JM83 [F− araΔ(lac-proAB) rpsL (Strr)] (38) were used as wild-type strains. The ΔcsdA-MC4100 deletion strain was described previously (37). The Δrnr-JW5741 strain and ΔrhlE-JW0781 strain were obtained from the Keio Collection, Japan. The ΔcsdA, Δrnr, and ΔrhlE single, double, and triple deletions were created in the JM83 strain by P1 transduction. The bacterial cultures were grown in Luria-Bertani broth (LB). Antibiotics such as ampicillin (50 μg ml−1) and chloramphenicol (Cm) (50 μg ml−1) were supplemented as required.
The plasmids pACYC-duet-csdA, pACYC-duet-rhlB, pACYC-duet-rhlE, pACYC-duet-rnr, and pACYC-duet-cspA were constructed by cloning the coding regions corresponding to csdA, rhlB, rhlE, rnr, and cspA into pACYC-duet vector, respectively, by use of NdeI and KpnI. The leaky expression of the inserted genes in this vector system was used; IPTG (isopropyl-β-d-thiogalactopyranoside) was not added to the cells. The plasmids pINIII-rnr, pINIII-csdA, pINIII-rnb, and pINIII-pnp were constructed by cloning the regions corresponding to the rnr, csdA, rnb, and pnp, respectively, by use of NdeI and BamHI. The leaky expression of the inserted genes in this vector system was used; IPTG was not added to the cells.
Site-directed mutagenesis.
The point mutations of the three acidic amino acids of the DEAD box were introduced through site-directed mutagenesis using PCR by the methods described by Lerner et al. and Spee et al. (16, 30). The resultant DNA fragments were cloned in pACYC-duet vectors to create plasmids expressing CsdAD173A, CsdAE174A, or CsdAD176A by use of NdeI and KpnI. The plasmids were sequenced to confirm the mutant sequences.
RNA isolation.
E. coli cells grown overnight in LB medium at 37°C were diluted into fresh medium. The wild-type and the ΔcsdA cells carrying pACYC-duet vector alone and the ΔcsdA cells expressing wild-type CsdA, CsdA(DAAD), or RNase R proteins were grown at 37°C to exponential phase (optical density at 600 nm [OD600] of 0.5) and then were transferred to 15°C for 1 h; transcription was stopped by adding rifampin (200 μg/ml). The cells were then harvested for RNA isolation at two time points, 0 min and 40 min. Total RNA was extracted by the hot-phenol method described previously (29). It was quantified by measuring absorbance at 260 nm. The purity of the RNA was confirmed by agarose gel electrophoresis.
Primer extension.
The primer extension and the deoxyoligonucleotide used for detection of cspA were described previously (26). The primer was labeled with [γ-32P]ATP (DuPont-New England Nuclear) by using T4 polynucleotide kinase (Gibco BRL). Primer extension was carried out with 5 μg of RNA at 42°C for 1 h in a final reaction volume of 10 μl with 50 mM Tris-HCl (pH 8.5)-8 mM MgCl2-30 mM KCl-1 mM dithiothreitol-0.4 pmol of 32P-labeled primer-0.5 mM (each) deoxynucleoside triphosphates-10 U RNase inhibitor (Boehringer Mannheim)-6.25 U of reverse transcriptase (Boehringer Mannheim). The products were analyzed on a 6% polyacrylamide gel under denaturing conditions.
RESULTS AND DISCUSSION
Genetic screening for protein counterparts of CsdA.
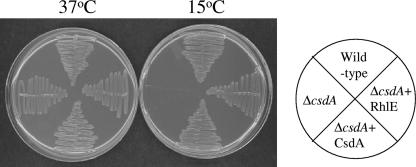
Several functions have been attributed to CsdA. In order to elucidate which of these functions is important for the role of CsdA in cold acclimation of cells, we explored genes that can complement the cold-sensitive phenotype of the ΔcsdA cells at 15°C. A genomic library was created by partial digestion of the E. coli chromosomal DNA with Sau3AI. The partially digested DNA fragments were cloned into the pACYC184 plasmid by use of BamHI. The ΔcsdA cells were transformed with the genomic library. Transformants were isolated for their ability to grow at 15°C. Plasmid DNAs were isolated from the colonies that showed growth at 15°C, purified, and retransformed into the ΔcsdA cells to confirm their ability to support growth at low temperature. The majority of the selected transformants showed the presence of the csdA gene as expected. A significant number (10% [CsdA, 27 of 30; RhlE, 3 of 30]) of the transformants showed the presence of a gene that encodes another RNA helicase, RhlE. To confirm whether RhlE indeed complements the cold-sensitive phenotype of the ΔcsdA cells, the wild-type or ΔcsdA cells harboring the pACYC-duet plasmid as controls and the ΔcsdA cells expressing plasmid-encoded CsdA or RhlE were streaked on LB-Cm plates and incubated at 37°C and 15°C. As expected, all the cells were able to grow at 37°C. The wild-type cells were able to grow at 15°C, while the ΔcsdA cells showed the cold-sensitive phenotype. The growth of ΔcsdA cells was restored at 15°C by the presence of either CsdA or RhlE on a plasmid (Fig. 1). This result confirms that RhlE can complement the CsdA function at low temperature. No role has been so far attributed to RhlE; however, it is interesting that in similarity to CsdA, it shows broader phylogenetic distribution (10) and that multiple RhlE homologues are present in some organisms. RhlE can unwind blunt duplexes whereas CsdA requires 3′ or 5′ single-stranded extensions, and neither of these shows RNA specificity in vitro. However, the lack of strong complementation within the family of DEAD-box helicases suggests that they may possess in vivo specificity, possibly as a result of interacting with some other factors (10).
FIG. 1.
RhlE can complement the cold-sensitive phenotype of ΔcsdA cells at 15°C. E. coli wild-type cells were transformed with pACYC-duet plasmid as a control, and ΔcsdA cells were transformed with pACYC-duet plasmid alone as a control or containing csdA or rhlE. The cells were streaked on LB plates containing chloramphenicol (50 μg ml−1) and incubated at 37°C and 15°C. The results obtained with the plates incubated at 37°C for 24 h and at 15°C for 72 h are presented.
Analysis of the helicase activity of CsdA.
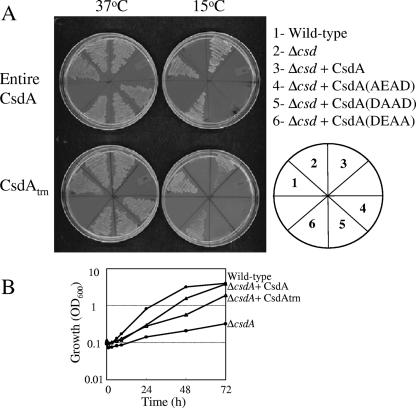
The results of our genetic screening emphasized the importance of helicase activity of CsdA in its cold shock acclimation function, as the only protein that can complement its function is another DEAD-box RNA helicase. This determination is supported by a recent paper by Turner et al. (32). Those authors mutated the second amino acid of the DEAD box and showed that it is critical for the unwinding activity of CsdA. Also, using in vitro studies, Bizebard et al. (2) have shown that the C-terminal segment of CsdA is not essential for its unwinding activity. To test our hypothesis that the helicase activity of CsdA plays a crucial role in its cold-acclimation function, we decided to analyze mutations in the DEAD box of CsdA. We created a truncated CsdA construct (444 amino acids) in which the C-terminal domain (185 amino acids) is deleted and also created mutations in all three acidic residues (D156, E157, and D159) of the DEAD box in the entire as well as in the C-terminal-truncated CsdA. The three mutant proteins created were CsdA(AEAD), CsdA(DAAD), and CsdA(DEAA). The plasmids harboring their respective examples of mutant csdA were transformed into the ΔcsdA cells, and growth was monitored at 37°C and 15°C. The wild-type and ΔcsdA cells harboring the pACYC-duet plasmid were also streaked on the plates as controls. As seen in Fig. 2A, all the cells were able to grow at 37°C. The ΔcsdA cells harboring plasmids with either entire or truncated CsdA carrying the D159A mutation, i.e., CsdA(DEAA), were able to grow at 15°C. Two conclusions can be drawn from this observation: (i) the last Asp residue in the DEAD box is not important for its helicase activity, or else CsdA with this mutation may still retain some helicase activity which is enough to suppress the ΔcsdA cold-sensitive phenotype, and (ii) the C-terminal region is not essential for the cold-complementation function of CsdA. The latter observation is consistent with the observation by Bizebard et al. (2) that the C-terminal region is not essential for the unwinding activity of CsdA. A similar result was seen with the liquid culture, wherein wild-type cells, ΔcsdA cells, and the ΔcsdA cells expressing wild-type or C-terminal-truncated CsdA were grown at 37°C until an OD600 of 0.5 was reached and were then transferred to 15°C. C-terminal-truncated CsdA was able to complement the cold-sensitive growth of the ΔcsdA cells (Fig. 2B), although this strain grew slower than the strain expressing wild-type CsdA. Turner et al. observed complementation of the ΔcsdA cells by the C-terminal-truncated CsdA only at 25°C (32). They also observed that it exerts an inhibitory effect on the ΔcsdA cells at 15°C. These different observations may be due to the different levels of protein expression in the two systems used. The expression of CsdA in our system occurred in the form of leaky expression from the pACYC-duet plasmid, and this low level of expression did not inhibit the cell growth. Interestingly, this level-dependent inhibitory effect was seen only for the C-terminal-truncated CsdA and not for the intact CsdA.
FIG. 2.
Analysis of the role of acidic amino acids of the DEAD box in the helicase activity of CsdA. (A) E. coli wild-type cells were transformed with pACYC-duet plasmid as a control, and ΔcsdA cells were transformed with pACYC-duet plasmid alone as a control or expressing entire CsdA or C-terminal-truncated CsdA (CsdAtrn) carrying DEAD-box amino acid mutation D156A, E157A, or D159A; the corresponding mutant proteins are designated CsdA(AEAD), CsdA(DAAD) and CsdA(DEAA), respectively. The cells were streaked on LB plates containing chloramphenicol (50 μg ml−1) and incubated at 37°C and 15°C. The results of the plate incubation at 37°C for 24 h and at 15°C for 72 h are presented. (B) Growth curves of wild-type cells with pACYC-duet vector alone, ΔcsdA cells, and ΔcsdA cells expressing wild-type CsdA (ΔcsdA+CsdA) or C-terminally truncated CsdA (ΔcsdA+CsdAtrn) grown at 15°C.
On the other hand, D156A or E157A mutation of both the entire and the C-terminal-truncated CsdA resulted in failure of growth at 15°C. The substitution of the E157 residue of CsdA has been shown to lead to loss of its helicase activity (32), and here we also observed that the CsdA(DAAD) protein fails to help the cold acclimation of the ΔcsdA cells. Therefore, it is reasonable to presume that the lack of cold-acclimation activity of the CsdA(AEAD) mutant protein may also be due to its lack of helicase activity. Based on these observations, we conclude that the helicase activity of CsdA is essential for its cold-acclimation function and that the first two residues of the DEAD box are essential for this activity.
Cold sensitivity of ΔcsdA cells can be complemented by proteins other than helicases.
Recently, Iost and Dreyfuss reviewed DbpA, RhlB, RhlE, SrmB, and CsdA, the five E. coli DEAD-box RNA helicases (10). They created double-deletion mutants of each of the 10 combinations of these helicases and observed that in none of these cases was the growth of the double mutants slower than that of single mutants. It has been also shown that RhlB, RhlE, and CsdA are interchangeable for the degradation of structured mRNA fragment in vitro (14, 27); however, the in vivo data regarding this interchangeability are not known. In their review article, Iost and Dreyfuss mentioned their unpublished data showing that RhlE may complement the in vivo function of CsdA (10). Our genetic screening showed that this is indeed the case, and RhlE was the sole candidate that emerged in our in vivo screening as a protein that can complement the cold-sensitive phenotype of the ΔcsdA cells. Since none of the other E. coli DEAD-box RNA helicases showed complementation of function of CsdA in vivo, we considered whether cold-inducible proteins other than RNA helicases can complement CsdA function at low temperature. We considered two types of proteins, known cold shock-inducible RNA chaperones and cold shock-inducible RNases. The role of CsdA in mRNA decay was demonstrated by its association with the cold shock degradosome (27) and its involvement in degradation of certain mRNAs at low temperature (37). RNA structures are stabilized upon temperature downshift; therefore, the RNA chaperone (helicase) activity of CsdA may be important for unwinding these mRNAs for degradation by RNases. SrmB and CsdA are the only DEAD-box RNA helicases in E. coli whose deletion leads to cold sensitivity, the growth defect being more severe in the case of the ΔcsdA strain. Interestingly, however, the defect in ribosome biogenesis is milder in the ΔcsdA strain at low temperature (3). This suggests that the role of CsdA in ribosome biogenesis may not be its primary function. Thus, the essentiality of CsdA upon cold shock may be due to its role in mRNA decay, in which its helicase activity plays a crucial role. On the basis of this consideration, we tested two proteins as CsdA counterparts at low temperature: CspA, the major cold shock protein and extensively studied RNA chaperone (1, 11, 24), and RNase R, a cold shock-inducible exonuclease that has recently been shown to be associated with RhlE, the DEAD-box RNA helicase, in the cold-adapted bacterium Pseudomonas syringae (28).
E. coli has nine CspA homologues, some of which are cold shock inducible. These proteins act as RNA chaperones by destabilizing secondary structures in nucleic acids, thus facilitating transcription and translation (1, 11). By virtue of their nucleic acid melting activity these act as transcription antiterminators, an activity that has been shown to be critical for the cold acclimation of cells (24). A csp quadruple-deletion mutant is thus cold sensitive (36). In similarity to CsdA, Csps cannot act on blunt-ended duplexes and require 3′ or 5′ single-stranded extensions in the form of an overhang or loop at least four bases in length (25). The Csp-mediated nucleic acid melting is relatively independent of the sequence of target nucleic acid and does not require ATP. RNase R is a processive, 3′-to-5′ hydrolytic exoribonuclease that, together with polynucleotide phosphorylase, is considered to play an important role in the degradation of structured RNAs. However, RNase R differs from other exoribonucleases in that it can by itself degrade RNAs with extensive secondary structure provided that a single-stranded 3′ overhang is present (33).
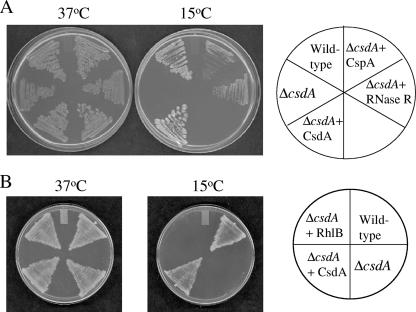
The ΔcsdA cells were transformed with the vector pACYC-duet alone or with the vector expressing CsdA, CspA, or RNase R. The cells were grown on LB-Cm plates at 37°C and at 15°C. Wild-type cells containing the vector were included as a control. As seen in Fig. 3A, all of the cells were able to grow at 37°C. The wild-type cells and ΔcsdA cells expressing either plasmid-encoded CsdA form show growth at 15°C. The ΔcsdA cells expressing CspA or RNase R also show growth at 15°C, albeit complementation by these two proteins is weaker than that by CsdA. This may be the reason why these two proteins were not chosen in our genetic screening. We also tested whether another DEAD-box helicase, RhlB, can complement the cold-sensitive phenotype of the ΔcsdA cells, but consistent with our results in the genetic screening, RhlB did not complement the cold-sensitive phenotype of these cells, as seen in Fig. 3B.
FIG. 3.
(A) Complementation of the cold-sensitive phenotype of the ΔcsdA cells by CspA and RNase R. E. coli wild-type cells were transformed with pACYC-duet plasmid as a control, and ΔcsdA cells were transformed with pACYC-duet plasmid alone as a control or expressing CsdA, CspA, or RNase R. The cells were streaked on LB plates containing chloramphenicol (50 μg ml−1) and incubated at 37°C and 15°C. The results for the plates incubated at 37°C for 24 h and at 15°C for 72 h are presented. (B) DEAD-box RNA helicase: RhlB does not complement the cold-sensitive phenotype of the ΔcsdA cells. E. coli wild-type cells were transformed with pACYC-duet plasmid as a control, and ΔcsdA cells were transformed with pACYC-duet plasmid alone as a control or expressing CsdA or RhlB. The cells were streaked on LB plates containing chloramphenicol (50 μg ml−1) and incubated at 37°C and 15°C. The results for the plates incubated at 37°C for 24 h and at 15°C for 120 h are presented.
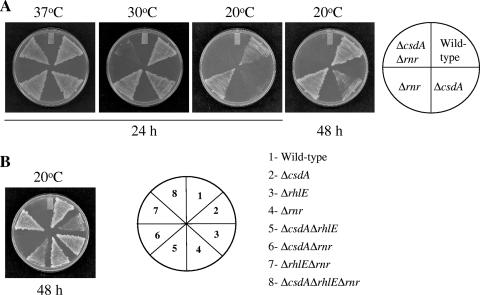
Next, we further explored the functional relationship between CsdA and RNase R by analyzing the growth patterns of the Δrnr and the ΔcsdAΔrnr strains. The wild-type, ΔcsdA, Δrnr, and ΔcsdAΔrnr strains were grown on LB plates at 37, 30, and 20°C. As seen in Fig. 4A, all the strains were able to grow at 37°C. The wild-type and Δrnr and ΔcsdA strains were able to grow at 30°C. Interestingly, the double-deletion mutant of rnr and csdA showed significantly slower growth even at 30°C. This suggests that the presence of either CsdA or RNase R is required for growth when cells experience even a very modest temperature downshift. This observation is especially interesting in light of the fact that RNase R, although cold shock inducible, is not essential at low temperature and that CsdA is essential only at low temperature. This result is consistent with the result presented in Fig. 3 showing that there may be a functional overlap between CsdA and RNase R and that the presence of at least one of these is essential for cell growth even after a modest temperature downshift. Upon 48 h of incubation, the ΔcsdAΔrnr strain showed growth at 30°C but not at 20°C, while the ΔcsdA strain showed growth after 48 h of incubation at 20°C. As expected, ΔcsdA or ΔcsdAΔrnr strains did not show growth at 15°C (data not shown). Interestingly, the absence of RhlE does not lead to increased sensitivity to temperature either singly or in combination with the absence of CsdA or RNase R, as the ΔrhlE, ΔcsdA ΔrhlE, and ΔrhlEΔrnr strains were able to grow at 20°C (Fig. 4B).
FIG. 4.
Double-deletion strain of csdA and rnr has increased temperature sensitivity. E. coli wild-type and ΔcsdA, Δrnr, and ΔcsdAΔrnr cells were streaked on LB plates and incubated at 37°C, 30°C, and 20°C for the time intervals indicated (A), and E. coli wild-type, ΔcsdA, ΔrhlE, Δrnr, ΔcsdAΔrhlE, ΔcsdAΔrnr, ΔrhlEΔrnr, and ΔcsdAΔrhlEΔrnr cells were streaked on LB plates and incubated at 20°C for 48 h (B).
Complementation of the temperature sensitivity of the ΔcsdA Δrnr strain by CsdA and RNase R.
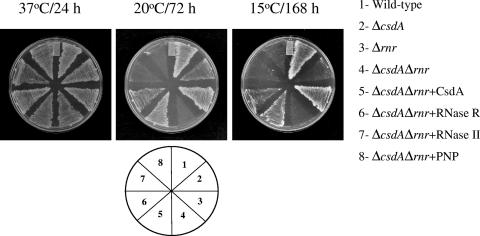
We next tested whether CsdA or RNase R can complement the growth defect of ΔcsdAΔrnr cells at 20°C. We included wild-type, ΔcsdA, Δrnr and ΔcsdAΔrnr cells harboring pINIII vector alone as a control. The ΔcsdAΔrnr cells were transformed with pINIII vector containing csdA or rnr. The cells were grown on LB-Cm plates at 37°C, 20°C, and 15°C. As seen in Fig. 5, the ΔcsdAΔrnr cells showed cold sensitivity at both temperatures which can be complemented by both CsdA and RNase R. CsdA showed the apparently stronger complementation, as the ΔcsdAΔrnr cells with a plasmid carrying csdA showed growth within 48 h, while those with rnr showed growth after 72 h of incubation at 20°C. At 15°C, cells expressing plasmid-encoded CsdA showed growth after 72 h, while those expressing RNase R required around 1 week. These experimental results indicate that there is a functional overlap between CsdA and RNase R and that at least one of the two proteins is essential for cell growth even after a modest temperature downshift. In the light of the observation that CsdA is associated with the cold shock degradosome (27), we also tested whether PNPase, a component of degradosome, can complement the cold sensitivity of the ΔcsdAΔrnr strain along with another RNase, RNase II. As seen in Fig. 5, both of these proteins failed to complement the cold shock acclimation function of CsdA/RNase R.
FIG. 5.
CsdA and RNase R, but not RNase II or PNPase, can complement the cold-sensitive phenotype of the ΔcsdAΔrnr cells. E. coli wild-type, ΔcsdA, Δrnr, and ΔcsdAΔrnr cells were transformed with pINIII plasmid as a control, and ΔcsdAΔrnr cells were transformed with pINIII plasmid alone as a control or with pINII-csdA, pINIII-rnr, pINIII-rnb, or pINIII-pnp expressing CsdA, RNase R, RNase II, or PNPase, respectively. The cells were streaked on LB plates containing ampicillin (50 mg ml−1). The plates were incubated at 37°C, 20°C, and 15°C for the time intervals indicated.
Helicase activity of CsdA is important for its role in mRNA stabilization.
Published data show that (i) CsdA and SrmB are the only cold shock-inducible DEAD-box RNA helicases of E. coli, (ii) CsdA associates with 50S precursors at low temperature and can complement the ribosome defect of the srmB deletion strain, (iii) overexpression of SrmB does not suppress the cold sensitivity of the ΔcsdA strain and vice versa, and (iv) although deletion of both SrmB and CsdA leads to cold sensitivity, the growth defect is more severe in case of the ΔcsdA strain. Interestingly, however, the defect in ribosome biogenesis is milder in the ΔcsdA strain at low temperature (3). Our present results show that RNase R can complement the cold shock function of CsdA. These observations suggest that the role of CsdA in ribosome biogenesis may not be its primary function. Thus, the essentiality of CsdA upon cold shock may be due to its role in mRNA decay, in which its helicase activity plays a crucial role.
We tested this hypothesis by analyzing (i) whether an mRNA of a model cold shock-inducible gene is stabilized in the ΔcsdA strain at 15°C, (ii) whether this effect can be counteracted by introduction of CsdA into the ΔcsdA cells, (iii) whether a helicase-deficient mutant of CsdA will fail to reverse the stabilization of this mRNA, and (iv) whether RNase R will exhibit an effect on the mRNA stabilization similar to that of the wild-type CsdA. We chose CspA as a model gene for this experiment, as it is the major cold shock protein of E. coli and the stability of its mRNA has been reported to be affected by CsdA at 15°C (37). The exponentially growing wild-type and ΔcsdA cells harboring vector alone and the ΔcsdA cells expressing wild type, CsdA(DAAD), or RNase R were cold shocked at 15°C for 1 h, the transcription was stopped by the addition of rifampin, and total RNAs were isolated at 0 min and 40 min time points. We chose the DAAD mutant of CsdA for this experiment, as our present results and the results reported by Turner et al. (32) show that this CsdA mutant cannot complement the cold-sensitive phenotype of the ΔcsdA cells and is helicase deficient. Equal amounts of RNA samples were used for primer extension with deoxyoligonucleotide corresponding to the cspA gene. The results of the primer extension are shown in Fig. 6. As seen in lane 2 in the figure, at 40 min after the addition of rifampin, the cspA mRNA amount is greatly diminished. This is consistent with a previous report showing the half-life of cspA mRNA at 15°C to be ∼20 min (20). In the ΔcsdA cells, the cspA mRNA was significantly stabilized and most of the mRNA was apparent even 40 min after transcription was stopped (lane 4). In the ΔcsdA cells expressing plasmid-encoded CsdA, this stabilization is lost and the level of cspA mRNA at the 40-min time point resembles that seen with the wild-type cells (compare lanes 2 and 6). Interestingly, in the ΔcsdA cells expressing the plasmid-borne, helicase-deficient CsdA(DAAD), cspA mRNA remains stabilized (lane 8). In the ΔcsdA cells expressing plasmid-borne RNase R, the stabilization of cspA mRNA is diminished (lane 10), albeit this effect is distinctly less than that seen with wild-type CsdA (lane 6). This result suggests that CsdA is involved in mRNA decay at low temperature and that its helicase activity is critical for this function. The data also suggest that RNase R can substitute for CsdA at low temperature, as it can act on complex secondary structures present in RNAs to facilitate their degradation by other RNases. However, it has less effect on cspA mRNA stability than CsdA. This may be the reason for the weaker complementation of the cold-sensitive phenotype of the ΔcsdA cells by RNase R and why it was not detected in our genetic screening. In addition, we also observed that higher expression of RNase R is somewhat toxic to the cell growth (data not shown).
FIG. 6.
Helicase activity of CsdA is essential for its role in mRNA degradation at low temperature. Cells from which total RNAs were isolated by the hot-phenol method are indicated above the respective lanes. The exponentially growing cells were cold shocked at 15°C for 1 h, and transcription was stopped by adding rifampin. Equal amounts of RNA samples were used for primer extension (0 min and 40 min time points for each type of cells) with deoxyoligonucleotide corresponding to the cspA gene.
Conclusion.
Based on the observations that the helicase activity of CsdA is essential for its cold-acclimation function and that it can be complemented by the presence of cold shock-inducible RNase R, we conclude that the primary role of CsdA in cold acclimation of cells is in mRNA decay and that its helicase activity is pivotal for promoting degradation of mRNAs stabilized at low temperature. It is really interesting that CspA and RNase R can complement CsdA function at low temperature, as all three of these proteins share a common feature in that they can destabilize mRNAs in spite of the fact that CsdA and RNase R have different enzymatic activities and that CspA has no enzymatic activity. We propose that all these three cold-inducible proteins synergistically work together and may play a role in mRNA decay which is crucial for cell growth at low temperature.
Acknowledgments
We thank Hirotada Mori from Keio Collection, Japan, for the gift of rnr and rhlE deletion strains.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bizebard, T., I. Ferlenghi, I. Iost, and M. Dreyfus. 2004. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry 43:7857-7866. [DOI] [PubMed] [Google Scholar]
- 3.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dammel, C. S., and H. F. Noller. 1995. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9:626-637. [DOI] [PubMed] [Google Scholar]
- 5.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245:255-259. [DOI] [PubMed] [Google Scholar]
- 6.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, D. I., E. R. Olson, C. Georgopoulos, K. Tilly, I. Herskowitz, and F. Banuett. 1984. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol. Rev. 48:299-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881-5889. [DOI] [PubMed] [Google Scholar]
- 10.Iost, I., and M. Dreyfus. 2006. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 34:4189-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 12.Jones, P. G., R. Krah, S. R. Tafuri, and A. P. Wolffe. 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174:5798-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, P. G., M. Mitta, Y. Kim, W. Jiang, and M. Inouye. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khemici, V., I. Toesca, L. Poljak, N. F. Vanzo, and A. J. Carpousis. 2004. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54:1422-1430. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. J., A. Xie, W. Jiang, J. P. Etchegaray, P. G. Jones, and M. Inouye. 1994. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol. Microbiol. 11:833-839. [DOI] [PubMed] [Google Scholar]
- 16.Lerner, C. G., T. Kobayashi, and M. Inouye. 1990. Isolation of subtilisin pro-sequence mutations that affect formation of active protease by localized random polymerase chain reaction mutagenesis. J. Biol. Chem. 265:20085-20086. [PubMed] [Google Scholar]
- 17.Linder, P., P. F. Lasko, M. Ashburner, P. Leroy, P. J. Nielsen, K. Nishi, J. Schnier, and P. P. Slonimski. 1989. Birth of the D-E-A-D box. Nature 337:121-122. [DOI] [PubMed] [Google Scholar]
- 18.Lu, J., H. Aoki, and M. C. Ganoza. 1999. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int. J. Biochem. Cell Biol. 31:215-229. [DOI] [PubMed] [Google Scholar]
- 19.McClelland, M., M. Nelson, and E. Raschke. 1994. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22:3640-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitta, M., L. Fang, and M. Inouye. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 26:321-335. [DOI] [PubMed] [Google Scholar]
- 21.Moll, I., S. Grill, A. Grundling, and U. Blasi. 2002. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol. 44:1387-1396. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima, K., K. Kanamaru, T. Mizuno, and K. Horikoshi. 1996. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J. Bacteriol. 178:2994-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125-136. [PubMed] [Google Scholar]
- 24.Phadtare, S., M. Inouye, and K. Severinov. 2002. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277:7239-7245. [DOI] [PubMed] [Google Scholar]
- 25.Phadtare, S., and K. Severinov. 2005. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 33:5583-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phadtare, S., V. Tadigotla, W. H. Shin, K. Sengupta, and K. Severinov. 2006. Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J. Bacteriol. 188:2521-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prud'homme-Genereux, A., R. K. Beran, I. Iost, C. S. Ramey, G. A. Mackie, and R. W. Simons. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome.’ Mol. Microbiol. 54:1409-1421. [DOI] [PubMed] [Google Scholar]
- 28.Purusharth, R. I., F. Klein, S. Sulthana, S. Jager, M. V. Jagannadham, E. Evguenieva-Hackenberg, M. K. Ray, and G. Klug. 2005. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 280:14572-14578. [DOI] [PubMed] [Google Scholar]
- 29.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 30.Spee, J. H., W. M. de Vos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toone, W. M., K. E. Rudd, and J. D. Friesen. 1991. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 173:3291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, A. M., C. F. Love, R. W. Alexander, and P. G. Jones. 2007. Mutational analysis of Escherichia coli DEAD-box protein CsdA. J. Bacteriol. 189:2769-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent, H. A., and M. P. Deutscher. 2006. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 281:29769-29775. [DOI] [PubMed] [Google Scholar]
- 34.Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48:60-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka, K., and M. Inouye. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183:2808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]