Abstract
We report here the characterization of the first agmatine/cadaverine aminopropyl transferase (ACAPT), the enzyme responsible for polyamine biosynthesis from an archaeon. The gene PF0127 encoding ACAPT in the hyperthermophile Pyrococcus furiosus was cloned and expressed in Escherichia coli, and the recombinant protein was purified to homogeneity. P. furiosus ACAPT is a homodimer of 65 kDa. The broad substrate specificity of the enzyme toward the amine acceptors is unique, as agmatine, 1,3-diaminopropane, putrescine, cadaverine, and sym-nor-spermidine all serve as substrates. While maximal catalytic activity was observed with cadaverine, agmatine was the preferred substrate on the basis of the kcat/Km value. P. furiosus ACAPT is thermoactive and thermostable with an apparent melting temperature of 108°C that increases to 112°C in the presence of cadaverine. Limited proteolysis indicated that the only proteolytic cleavage site is localized in the C-terminal region and that the C-terminal peptide is not necessary for the integrity of the active site. The crystal structure of the enzyme determined to 1.8-Å resolution confirmed its dimeric nature and provided insight into the proteolytic analyses as well as into mechanisms of thermal stability. Analysis of the polyamine content of P. furiosus showed that spermidine, cadaverine, and sym-nor-spermidine are the major components, with small amounts of sym-nor-spermine and N-(3-aminopropyl)cadaverine (APC). This is the first report in Archaea of an unusual polyamine APC that is proposed to play a role in stress adaptation.
Propylamine transferases represent a class of enzymes catalyzing the transfer of a propylamine group from S-adenosyl-(5′)-3-methylthiopropylamine (decarboxy-AdoMet) to various amine acceptors (7, 39, 48, 60), according to the following reaction: decarboxy-AdoMet + polyamine substrate → 5′-methylthioadenosine + polyamine product. The reaction involves a nucleophilic attack on the C-3 methylene of the propylamine moiety adjacent to the positively charged sulfur of decarboxy-AdoMet (7, 39, 48, 60).
In 1958, Tabor et al. (49) provided the first evidence that decarboxy-AdoMet is the donor of aminopropyl groups in the biosynthesis of spermidine in cell extracts of Escherichia coli. The enzyme catalyzing this reaction was subsequently purified to homogeneity by Bowman et al. (4). The development of rapid and sensitive methods of polyamine analysis, together with the utilization of affinity chromatography, allowed the purification to homogeneity and a complete characterization of spermine and spermidine synthases from bovine brain (36) and rat ventral prostate (41), respectively. Moreover, a propylamine transferase with unusual molecular and kinetic properties has been purified to homogeneity from Sulfolobus solfataricus (S. solfataricus aminopropyl transferase [APT]), an extreme thermophilic archaeon (8). Recently, recombinant forms of the spermidine synthases of the extremely thermophilic bacterium Thermotoga maritima (T. maritima PAPT) (28) and of the human malaria parasite Plasmodium falciparum (PfSpdS) (18) have been characterized. Crystal structures in the absence and in the presence of the transition state analog S-adenosyl-1,8-diamino-3-thiooctane (AdoDATO) have been obtained for T. maritima PAPT (28).
Putrescine, spermidine, and spermine represent the major components of the polyamine pool of many organisms, where they are involved mainly in important cellular processes mostly related to the biosynthesis of proteins and nucleic acids (31, 37, 47, 48, 52). However, a variety of unusual polyamines, such as longer linear and branched polyamines, have been found in various thermophiles belonging to both Bacteria and Archaea (27, 34). The observation that the cellular content of these unusual polyamines increases with increasing growth temperature (34) suggests that they play a role in maintaining the cellular functions at high temperature.
The polyamine pool of the hyperthermophilic archaeon S. solfataricus is indeed characterized by two unusual polyamines, sym-nor-spermidine and sym-nor-spermine (12, 13). The peculiar carbon chain lengths and charge distributions of these symmetrical polycations could facilitate specific interactions with nucleic acids, resulting in a protection of DNA against thermal denaturation. The observation that sym-nor-spermine increases the melting temperature (Tm) of DNA from Aerobacter aerogenes and from calf thymus (44) supports this hypothesis. Similarly, the thermophilic bacterium Thermus thermophilus produces unusual long-chain linear polyamines, such as caldopentamine and caldohexamine, that are effective in stabilizing DNA and branched polyamines, such as quaternary ammonium compounds, that play a role in stabilizing RNA (51). A novel polyamine biosynthetic pathway is present in T. thermophilus in which an APT utilizes agmatine as a substrate to form N1-aminopropylagmatine. This is hydrolyzed by a ureohydrolase, forming spermidine (33).
The nature of polyamines and their biosynthetic pathways in anaerobic hyperthermophilic Archaea have not been studied previously. Herein the APT from Pyrococcus furiosus, which grows optimally near 100°C, has been characterized. On the basis of its high catalytic activity with agmatine and cadaverine, the enzyme has been named P. furiosus agmatine/cadaverine APT (ACAPT). A detailed kinetic investigation has been performed in order to clarify the substrate specificity of the enzyme. In addition, the polyamine content of P. furiosus has been analyzed and measured by means of high-performance liquid chromatography (HPLC) and mass spectrometry. Finally, the three-dimensional structure of P. furiosus ACAPT has been resolved at 1.8-Å resolution, and this has been compared with the structures of homologs from thermophilic and mesophilic species of the bacterial and archaeal domains.
MATERIALS AND METHODS
Bacterial strains, plasmid, enzymes, and chemicals.
Escherichia coli strain BL21(λDE3) was purchased from Novagen (Darmstadt, Germany). P. furiosus frozen cell paste was purchased from CAMR (Salisbury, United Kingdom) or cells were grown at the University of Georgia. Plasmid pET-22b(+) and the NucleoSpin plasmid kit for plasmid DNA preparation were obtained from Genenco (Duren, Germany). Specifically synthesized oligodeoxyribonucleotides were obtained from MWG-Biotech (Ebersberg, Germany). Restriction endonucleases and DNA-modifying enzymes were obtained from Takara Bio, Inc. (Otsu, Shiga, Japan). Pfu DNA polymerase was purchased from Stratagene (La Jolla, CA). [methyl-14C]AdoMet (50 to 60 mCi/mmol) was supplied by the Radiochemical Centre (Amersham Bioscience, Buckinghamshire, United Kingdom). 5′-Methylthioadenosine (MTA) was prepared from AdoMet (42) and purified by HPLC (11). Agmatine, O-bromoacetyl-N-hydroxysuccinimide, and standard proteins used in molecular mass studies were obtained from Sigma (St. Louis, MO). Isopropyl-β-d-thiogalactopyranoside (IPTG) was from Applichem (Darmstadt, Germany). Sephacryl S-100 and AH-Sepharose 4B were obtained from Amersham Pharmacia Biotech. Polyvinylidene fluoride (PVDF) membranes (0.45-mm pore size) were obtained from Millipore (Bedford, MA). Proline, dansyl chloride, cellulose phosphate, putrescine, spermidine, spermine, 1,3-diaminopropane, 1,5-diaminopentane (cadaverine), 1,10-diaminodecane, and proteinase K were obtained from Sigma-Aldrich (St. Louis, MO). Sym-nor-spermidine and sym-nor-spermine were purchased from Eastman Kodak (Rochester, NY). Decarboxy-AdoMet was kindly supplied by G. Stramentinoli, Bio Research Co., Linate, Italy. All reagents were of the purest commercial grade.
Preparation of [methyl-14C]decarboxy-AdoMet.
[methyl-14C]decarboxy-AdoMet was obtained from [methyl-14C]AdoMet by enzymatic decarboxylation (58). In order to separate the two sulfonium compounds from the contaminants of the incubation mixture, ion-exchange chromatography on a Dowex-50-H+ column was employed (59). As a final purification step, preparative HPLC on an Ultrasil SCX column (Beckman) was employed. The elution was carried out with acetonitrile-50 mM ammonium formate, pH 3.5 (5:95, vol/vol), at a flow rate of 1 ml/min. The retention times of AdoMet and decarboxy-AdoMet standards were 6 min and 15 min, respectively. The eluate was monitored at 254 nm. Decarboxy-AdoMet purified by this procedure was free of AdoMet.
Polyamine extraction and derivatization.
For polyamine extraction, 1 g of P. furiosus cell paste was disrupted by sonication in 10 ml Tris-HCl, pH 7.4. After centrifugation for 1 h at 25,000 × g, the supernatant was treated with perchloric acid to a final concentration of 0.2 N. After extraction for 1 h in an ice bath, samples were centrifuged at 25,000 × g for 15 min. The pellet was washed twice with 5 ml of 0.2 N perchloric acid, and the supernatants were pooled and further extracted for 48 h with mechanical stirring and stored at −80°C until analyzed. Dansylation was performed as described previously (16). Aliquots (200 μl) of the perchloric acid sample were reacted with 4 volumes of dansyl chloride in acetone (5 mg/ml) in the presence of 100 μl of a saturated solution of sodium carbonate. Excess dansyl chloride was neutralized by reaction with proline (1.8 mg/ml). The cyclohexane extract containing the dansyl derivatives was evaporated to dryness, and the residue was dissolved in 100 μl acetonitrile and analyzed by HPLC as described below. Authentic polyamines were used as standards. Yields of polyamines were determined by the addition of 1,10-diaminodecane to cell homogenates as an internal standard. All measurements were done in duplicate.
Enzyme assay.
P. furiosus ACAPT activity was assayed as described by Cacciapuoti et al. (8) by following the formation of [methyl-14C]MTA from [methyl-14C]decarboxy-AdoMet in the presence of appropriate polyamine acceptors. Unless otherwise stated, the standard incubation mixture contained the following: 20 μmol Tris-HCl, pH 7.4, 20 nmol [methyl-14C]decarboxy-AdoMet (2 × 106 cpm/μmol), 1.8 μmol of cadaverine (9 mM final concentration), and the enzyme protein in a final volume of 200 μl. The incubation was performed in sealed glass vials for 10 min at 70°C, except where indicated otherwise. The vials were rapidly cooled in ice, and the reaction was stopped by the addition of 800 μl of HCl at 0.1 N. The mixture was then applied to a cellulose phosphate column (0.6 by 2 cm; Sigma) equilibrated in HCl at 0.1 N. [methyl-14C]MTA produced was eluted with 5 ml of HCl at 0.1 N directly into scintillation vials and evaluated for radioactivity. Control experiments in the absence of the enzyme were performed in order to correct for decarboxy-AdoMet hydrolysis. When the assays were carried out at temperatures above 70°C, the reaction mixture was preincubated for 2 min without the enzyme, which was added immediately before starting the reaction. In all of the kinetic and purification studies, the amounts of protein were adjusted so that no more than 10% of the substrate was converted to product and the reaction rate was strictly linear as a function of time and protein concentration.
To identify the polyamine produced by the reaction, an alternative enzymatic assay was developed. The method involves HPLC analysis of the polyamines derivatized with dansyl chloride. After a 10-min incubation at 70°C, the reaction was stopped by the addition of 100 μl of 0.2 N perchloric acid. After centrifugation, the samples were derivatized and analyzed by HPLC. Separation of dansyl polyamines was carried out by reverse-phase HPLC on a 4.6-by-250-mm Ultrasphere ODS (5-μm-particle-size) column (Beckman) using an Agilent 1100 series chromatograph. The derivatives were eluted by a linear gradient, from 30 to 100% in 20 min, of solvent B (acetonitrile) in solvent A (0.02% aqueous trifluoroacetic acid) at a flow rate of 1 ml/min. Fluorescence intensity (excitation at 320 nm and emission at 500 nm) was detected with a fluorimeter (Agilent Technologies).
Mass spectrometric analysis.
Mass spectrometry analysis of individual HPLC polyamine peaks was performed at CEINGE (Naples, Italy) by matrix-assisted laser desorption ionization mass spectrometry on a Voyager DE-Pro mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a reflectron analyzer and used in delayed extraction mode. Samples were mixed with an equal volume of a saturated solution of α-cyano-4-hydroxycinnaminic acid (10 mg/ml) in acetonitrile-0.2% trifluoroacetic acid, 70:30 (vol/vol), and applied to the metallic sample plate before air drying. Mass calibration was performed by using the standard mixture provided by the manufacturer.
Cloning, expression, and purification of P. furiosus ACAPT.
The putative P. furiosus ACAPT gene (GenBank accession number AE010139) from P. furiosus was cloned into the pET-22b(+) expression vector via two engineered restriction sites (NdeI and BamHI) introduced by PCR with the following primers: 5′-GAAGGTGGCGAGCATATGGAGAGGG-3′ sense and 5′-GACAAAAAGCATGGATCCAGTATTATGG-3′ antisense (the introduced restriction sites are underlined). Genomic DNA from P. furiosus (20 ng) was used as a template. PCR amplification was performed with P. furiosus DNA polymerase and a Minicycler gradient (Eppendorf) programmed for 30 cycles, each cycle consisting of denaturation at 95°C for 1 min, annealing at 58°C for 2 min, and extension at 68°C for 2.5 min plus 5 s/cycle. The amplified gene (25 ng) hydrolyzed by NdeI and BamHI was inserted into pET-22b(+) (150 ng) cut with the same restriction enzymes. The recombinant plasmid was named pET-PfACAPT. The nucleotide sequence of the inserted gene was determined by MWG Biotech (Ebersberg, Germany) to ensure that no mutations were present.
For the expression of recombinant P. furiosus ACAPT, an overnight culture of E. coli BL21(λDE3) transformed with the plasmid pET-PfACAPT was used as a 0.5% inoculum in 1 liter of Luria-Bertani (40) medium containing 100 μg/ml ampicillin at 37°C. When the culture reached an optical density of 1.0, IPTG was added at a 1 mM final concentration, and the induction was prolonged for 3 h. Cells were harvested by centrifugation and lysed (40). The cell debris was removed by centrifugation at 20,000 × g for 60 min at 4°C, and the supernatant was used as a cell extract. Recombinant P. furiosus ACAPT was purified in two steps. The extract from BL21 E. coli cells expressing P. furiosus ACAPT was heated at 100°C for 10 min and centrifuged at 20,000 × g for 60 min. After dialysis overnight against 10 mM Tris-HCl, pH 7.4, the enzyme was applied to an affinity column (2 by 12 cm) of MTA-Sepharose prepared as described by Cacciapuoti et al. (6). The column, equilibrated with 20 mM Tris-HCl, pH 7.4, was washed stepwise with 50 ml of the equilibration buffer and then with the same buffer containing 0.8 M NaCl until the absorbance at 280 nm reached the baseline. P. furiosus ACAPT activity was eluted with 20 mM Tris-HCl, pH 7.4, containing 0.8 M NaCl and 3 mM MTA. Active fractions were pooled, concentrated, and dialyzed extensively against 10 mM Tris-HCl, pH 7.4.
The recombinant form of P. furiosus ACAPT containing an N-terminal hexa-His tag (AHHHHHHGSERA- instead of MERA-) was used for crystallographic analyses. This was produced using the expression plasmid pET24d Bam with E. coli strain BL21 Star DE3 pRIL as the host. The recombinant protein was purified according to the high-throughput protocols established for P. furiosus protein production (45). In brief, cells from a 1-liter culture were heated at 80°C for 30 min to precipitate E. coli proteins, cooled to 4°C, and then clarified by centrifugation (40,000 × g). The supernatant was applied to a column (5 ml) of Histrap Ni affinity resin using an AKTA explorer (GE Healthcare, Piscataway, NJ). After applying the cell extract, the column was washed with 5 column volumes of 20 mM phosphate buffer, pH 7.0, containing 500 mM NaCl, 10 mM imidazole, 5% (vol/vol) glycerol, and 2 mM dithiothreitol. The absorbed protein was eluted with a gradient of 0 to 500 mM imidazole over 20 column volumes. The major protein peak was collected and concentrated to 10 ml by ultrafiltration (Millipore, Bedford, MA), diluted 15-fold in 20 mM Tris buffer, pH 8.0, containing 5% (vol/vol) glycerol and 2 mM dithiothreitol, and then applied to a column (5 ml) of Q Sepharose (GE Healthcare). The column was washed with 5 column volumes of the same buffer, and the bound protein was eluted with a 0 to 1 M NaCl gradient over 20 column volumes. The major protein was concentrated to 5 ml and applied to a 16/60 column of Superdex 75 (GE Healthcare) equilibrated with the same Tris buffer. The major protein peak was collected and concentrated to a volume of 1 ml by ultrafiltration. The protein concentration was estimated by the absorption at 280 nm using a calculated extinction coefficient. The yield from 1 liter of E. coli cells was 15 mg. Protein identity was confirmed by tryptic digestion of the excised band and analysis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Protein mass was determined by electrospray ionization mass spectrometry at the Department of Chemistry, University of Georgia (calculated, 33,241; measured, 33,262).
Crystallography.
Samples of P. furiosus ACAPT were screened against components of nine commercially available screening kits (Crystal Screens 1, 2, Cryo, Membfac, and Peg/Ion [Hampton Research], Wizard Screens 1 and 2 [Decode Genetics], and Classic Screens 1 and 2 [Nextal Biotechnologies]) by sitting drop vapor diffusion as described previously (29). Each 200-nl drop contained equal volumes of protein (12 mg/ml in 20 mM Tris-100 mM NaCl, pH 8.0) and precipitate solution. Crystal hits from the initial screen were optimized using the modified microbatch under oil (70:30 paraffin/silicone oil mixture) method (9). Using this approach, crystals suitable for X-ray diffraction were obtained in 2 weeks from 2-μl drops containing equal volumes of protein solution (as described above) and a precipitate solution containing 100 mM HEPES-NaOH, pH 7.5, in 28% (vol/vol) polyethylene glycol-400. For data collection, a crystal measuring 0.2 by 0.2 by 0.2 mm was harvested, mounted in a 0.3-mm rayon loop (50), and flash frozen in liquid N2. No cryoprotection was necessary. Data were collected under cryogenic conditions (T = 100 K) on a Raxis IV image plate detector using mirror-focused (MSC Blue confocal optics) 5-kW X rays. The data were indexed, integrated, and scaled using HKL 1.9.7 (35). Data collection and processing details are collected in Table 1. The P. furiosus ACAPT structure was solved by molecular replacement (EPMR) (26) using the recently reported structure of the T. maritima homologue (entry 1INL in the Protein Data Bank [2]). The resulting model was refined against data to 1.8-Å resolution using REFMAC V5.1.24 (32) with manual model adjustment using XFIT (30) when needed. Refinement statistics are summarized in Table 1.
TABLE 1.
Statistics from the crystallographic analysis
| Crystale | Name or value |
|---|---|
| Space group | P212121 |
| a | 95.64 |
| b | 110.82 |
| c | 49.26 |
| Z | 8 |
| Data analysis | |
| Wavelength (Å) | 1.5418 |
| Phi step (°) | 1.0 |
| Total rotation (°) | 360 |
| Data processing | HKL 1.9.7 |
| R-syma | 0.048 (0.193) |
| Refinement statistics | |
| Program | REFMAC 5.1.24 |
| Resolution (Å) | 72.55-1.80 |
| Completeness (%)b | 90.7 (47.1) |
| Rcrystb | 0.207 (0.294) |
| Rfreeb | 0.242 (0.332) |
| RMS deviations from ideality | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.008 |
| Ramachandran analysis | |
| Most favored (%)c | 92.0 (99.7) |
| Disallowed (%) | 0.3 |
| Final model | |
| Residues | 3-278 |
| Solvent atoms | 201 |
| Cα deviations (Å)d | 0.354 |
| PDB ID | 1MJF |
Outer-shell (1.86- to 1.80-Å) value in parentheses.
Outer-shell (1.84- to 1.80-Å) value in parentheses.
PROCHECK percentage in all allowed regions shown in parentheses.
RMS deviations (Cα) between A and B chains.
RMS, root mean square; PDB ID, Protein Data Bank identification.
Analytical methods for protein.
Protein concentrations were estimated by the Bradford method (5) using bovine serum albumin as the standard. The molecular mass of the native protein was determined by gel filtration at 20°C on a calibrated Sephacryl S-100 column (2.2 by 95 cm) equilibrated with 10 mM Tris-HCl, pH 7.4, containing 0.3 M NaCl at a flow rate of 4 ml/h. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out at room temperature (56) using 12.5 or 15% acrylamide resolving gel and 5% acrylamide stacking gel. Samples were heated at 100°C for 5 min in 2% SDS and 2% 2-mercaptoethanol prior to electrophoresis. Protein homogeneity was assessed by SDS-PAGE. N-terminal sequence analysis of the purified enzyme was performed by Edman degradation on an Applied Biosystems 473A sequencer according to the manufacturer's instructions. The sample was subjected to SDS-PAGE and electroblotted on a PVDF membrane prior to analysis.
Thermostability studies.
The thermostability of P. furiosus ACAPT was studied by incubating the enzyme (40 μg) in 20 mM Tris-HCl, pH 7.4, in sealed glass vials at temperatures between 70°C and 115°C in an oil bath. Samples (2 μg) were taken at time intervals and residual activity was determined by the standard assay at 70°C. Activity values are expressed as a percentage of zero-time control (100%).
Multiple sequence alignment.
Protein similarity searches were performed using the data from Swiss-Prot and Protein Identification Resource (PIR) data banks. The multiple alignment was constructed using the Clustal method (20) and BioEdit software.
Determination of kinetic constants.
Homogeneous preparation of P. furiosus ACAPT was used for kinetic studies. The purified enzyme gives a linear rate of reaction for at least 15 min at 70°C; thus, an incubation time of 10 min was employed for kinetic experiments. All enzyme reactions were performed in triplicate. Kinetic parameters were determined from Eadie-Hofstee plots of initial velocity data. Km and Vmax values were obtained from linear regression analysis of data fitted to the Eadie-Hofstee equation. Values given are the averages from at least three experiments with standard errors. The kcat values were calculated by dividing Vmax by the total enzyme concentration. Calculations of kcat were based on an enzyme molecular mass of 65 kDa.
Limited proteolysis experiments.
Limited proteolysis experiments were carried out by incubating recombinant P. furiosus ACAPT with proteinase K. Enzymatic digestion was performed in 10 mM Tris-HCl, pH 7.4, at 37°C. The final mass ratio of substrate protein to protease was 10:1. Hydrolysis was stopped by the addition of phenylmethylsulfonyl fluoride (final concentration, 250 μM), and the samples were assayed for ACAPT activity. The degradation of the intact protein over 2 h of incubation was monitored by SDS-PAGE of the digested material followed by staining with Coomassie blue R-250. For the amino acid sequence analysis, samples of the digested recombinant ACAPT from SDS-PAGE analysis were electrophoretically blotted onto a PVDF membrane utilizing a Bio-Rad Mini Trans-Blot transfer cell apparatus (Bio-Rad, CA). Transferred proteins were stained with Coomassie blue (0.1% in 50% methanol) for 5 min, destained in 50% methanol and 10% acetic acid solution for 30 min at room temperature, and allowed to air dry. Stained protein bands were excised from the blot, and their NH2-terminal sequences were determined.
Protein Data Bank accession numbers.
The coordinates for the new structure discovered in this study have been deposited in the Protein Data Bank under the number 1MJF.
RESULTS AND DISCUSSION
Primary sequence comparisons.
In the complete genome sequence of P. furiosus, the gene PF0127 is annotated as a probable spermidine synthase gene. Based on the substrate specificity of the recombinant enzyme determined in this work, it is now designated as P. furiosus ACAPT. From the gene sequence, P. furiosus ACAPT is predicted to contain 281 residues, with a molecular mass of 32,334 Da. N-terminal sequencing of the recombinant protein (see below) showed that the N-terminal methionine is not posttranslationally removed in E. coli. Comparison of its primary structure with those of enzymes present in the GenBank database indicated the highest identity with the hypothetical spermidine synthases from Pyrococcus horikoshi (75%), Pyrococcus abyssi (75%), and Thermococcus kodarensis (67%). Among characterized enzymes, P. furiosus ACAPT shows a moderate degree of identity with the amino acid sequences of Homo sapiens spermidine synthase (HsSpdS) (33%), E. coli spermidine synthase (E. coli SpdS) (35%), T. maritima PAPT (35%), S. solfataricus APT (34%), and PfSpdS (34%) (see Table 2). Like the homologous archaeal propylamine transferase from S. solfataricus, P. furiosus ACAPT does not contain cysteinyl residues. This feature is in contrast to that observed for eukaryal and bacterial enzymes. In fact, 11 cysteinyl residues are present in PfSpdS, 3 in T. maritima PAPT, and 6 in E. coli SpdS. Moreover, 10 cysteine residues have been found in mammalian spermidine synthase, and it has been suggested that the disulfide bond between the two C25 residues in the monomers occurs during formation of the dimer (17).
TABLE 2.
Comparison of structures of P. furiosus ACAPT with homologues
| Source of enzyme | % Sequence identity | Sequence length (amino acids) | PDB IDa | PDB IDb | RMSD (Å)c |
|---|---|---|---|---|---|
| P. furiosus | 100 | 271-271 | 1MJF_A | 1MJF_B | 0.35 |
| C. elegans | 39 | 276-276 | 1MJF | 2B2C | 1.53-1.57 |
| T. maritima | 35 | 264-271 | 1MJF | 1INL | 1.51-1.56 |
| T. maritima | 35 | 264-295 | 1MJF | 1JQ3 | 1.49-1.55 |
| T. thermophilus | 33 | 263-313 | 1MJF | 1UIR | 1.64-1.72 |
| H. sapiens | 33 | 279-286 | 1MJF | 1ZDZ | 1.56-1.62 |
| A. thaliana | 32 | 263-290 | 1MJF | 1XJ5 | 1.47-1.55 |
| B. subtilis | 30 | 259-274 | 1MJF | 1IY9 | 1.42-1.56 |
PDB ID, Protein Data Bank identification.
1JQ3 is a complex between the enzyme and a substrate analogue.
Range based on superposition of equivalent Cα positions for all protein chains in the entry. RMSD, root mean square deviation.
DNA microarray analyses.
An indication of the physiological role of ACAPT comes from transcriptional profiling of P. furiosus grown using different carbon sources and in response to cold shock (95 versus 72°C) using DNA microarrays (43, 57). The results show that PF0127 is expressed at moderate levels in cells grown at 95°C using a disaccharide (maltose) as a carbon source and that this is unaffected compared with cells grown where peptides are the carbon source (at 95°C) or when cells are grown (on maltose) at 72°C. Similarly, there was no change in expression when cells growing at 95°C (on maltose) were transferred to 72°C (at least up to 5 h after the change). Consequently, polyamine production mediated by ACAPT occurs near the optimal growth temperature, but the enzyme does not appear to play a significant role in adaptation to low-temperature growth (72°C). Studies are currently under way to determine if the expression of PF0127 is affected by temperatures higher than 95°C or by oxidative shock of P. furiosus.
Biochemical characterization of recombinant P. furiosus ACAPT.
Recombinant ACAPT was produced in a soluble form in E. coli BL21 cells harboring the plasmid pET-PfACAPT at 37°C in the presence of IPTG. Under the experimental conditions selected for the expression of the recombinant gene, about 10 g (wet weight) of cell paste was obtained from 1 liter of culture. SDS-PAGE analysis of extract from induced cells revealed an additional band of approximately 32 kDa, which corresponded with the calculated molecular mass. This band was absent in extracts of E. coli BL21 carrying the plasmid without the insert (data not shown). The ACAPT activity of recombinant E. coli BL21 cell extract was 9.9 units/mg at 70°C, confirming that the P. furiosus ACAPT gene had been cloned and expressed. Recombinant ACAPT was purified to homogeneity 24.4-fold by a simple two-step purification procedure involving heat treatment and affinity chromatography on MTA-Sepharose. About 3.4 mg of the recombinant enzyme with a 39% yield was obtained from 1 liter of culture. The enzyme had an apparent molecular mass of 65 kDa as estimated by gel filtration on a calibrated Sephacryl S-100 column and a subunit molecular mass of 32.6 kDa by SDS-PAGE. These results indicate that ACAPT is a homodimeric protein, in agreement with the reported structures of ACAPT orthologs from E. coli (4), rat (41), bovine species (36), and human (17). In contrast, the homologous enzymes from hyperthermophilic sources are not dimeric: that from S. solfataricus is a trimer (8) and that from T. maritima is a tetramer in the crystal and in solution (28). The more complex quaternary structures for the latter two hyperthermophilic enzymes could represent a stabilization strategy to enhance interactions among subunits and reduce the solvent-accessible surface, thus increasing thermal stability. However, these preliminary data indicate that this is not the strategy used by the P. furiosus enzyme, and as discussed below, crystallographic analyses confirm this.
Substrate specificity and polyamine determination.
The substrate specificity of P. furiosus ACAPT was determined using various amines as propylamine acceptors. As shown in Table 3, ACAPT was by far the most active (by more than 12-fold) with cadaverine as the substrate. This was surprising, since cadaverine is a very poor substrate for all other APTs characterized so far. To identify the cadaverine derivative formed when ACAPT was incubated with cadaverine and decarboxy-AdoMet, the reaction mixture was subjected to reverse-phase HPLC after derivatization with dansyl chloride. The peak corresponding to the polyamine product was analyzed by mass spectrometry and identified as tridansyl-N-(3-aminopropyl)cadaverine (APC) (858.2 atomic mass units).
TABLE 3.
APT activity in the presence of various amine acceptors
| Substratea | Enzyme activity (nmol/min/mg)b | Relative activity (%) |
|---|---|---|
| Cadaverine | 1,903 ± 10 | 100 |
| Putrescine | 167 ± 8 | 8.7 |
| Agmatine | 93 ± 5 | 4.9 |
| 1,3-Diaminopropane | 56 ± 3 | 2.9 |
| Sym-nor-spermidine | 18 ± 1 | 0.9 |
| Spermidine | 16 ± 1 | 0.8 |
| Sym-nor-spermine | ND | <0.1 |
| Spermine | ND | <0.1 |
Each polyamine substrate was at a saturating concentration.
The enzymatic assay was carried out as described in Materials and Methods. Values are the means of the results (± the standard deviations) of three separate experiments. ND, not detectable.
The highly efficient in vitro synthesis of APC by ACAPT prompted us to analyze the polyamine pool of P. furiosus and to see if the presence of cadaverine and its derivatives could be detected. The method employed for the separation and analysis of polyamines, which couples reversed-phase HPLC and mass spectrometry, allows a rapid and unambiguous determination of these polycations in the cell extract. Table 4 shows the polyamine concentrations in P. furiosus and the retention times and molecular masses of dansyl polyamines. Spermidine, sym-nor-spermidine, and cadaverine are the major components of the polyamine pool of this hyperthermophilic archaeon, followed by sym-nor-spermine and putrescine, while 1,3-diaminopropane and spermine were beyond detection. The presence of APC was also detected in cell extract (Table 4), although in the absence of an authentic polyamine standard, its concentration was estimated using spermidine as a standard.
TABLE 4.
Cellular concentration of polyamines in P. furiosusa
| Polyamine | Retention time (min) | Molecular mass (amu) | Cellular concn (μmol/g [wet wt] cells)b |
|---|---|---|---|
| 1,3-Diaminopropane | 9.3 | 540.1 | ND |
| Putrescine | 10.2 | 554.2 | 0.03 |
| Cadaverine | 10.9 | 568.2 | 0.27 |
| Sym-nor-spermidine | 13.8 | 830.3 | 0.31 |
| Spermidine | 14.4 | 844.3 | 1.12 |
| APC | 12.9 | 858.2 | 0.02 |
| Sym-nor-spermine | 17.3 | 1,120.4 | 0.09 |
| Spermine | 18.1 | 1,134.4 | ND |
The average recovery of 1,10-diaminodecane was 85%.
ND, not detectable.
The overall polyamine content of P. furiosus is of the same order of magnitude as other hyperthermophilic Archaea (19, 23). Moreover, the presence of cadaverine and APC in cell extract of P. furiosus is unusual and indicates that cadaverine can act as a propylamine acceptor from decarboxy-AdoMet in the synthesis of APC. It is interesting to note that APC was identified in a polyamine-requiring mutant of E. coli, grown in the absence of polyamines (14). Higher homologs of cadaverine, such as APC, were also synthesized as a compensatory polyamine by E. coli polyamine-requiring mutants (24). It has also been demonstrated that starvation of the polyamine-dependent Chinese hamster ovary cells resulted in the formation of cadaverine and its aminopropyl derivatives (22). Moreover, formation of cadaverine derivatives has been demonstrated in Saccharomyces cerevisiae exposed to ethanol (54). Finally, the thermotolerant mesophilic fungus Aspergillus fumigatus and S. cerevisiae synthesize higher homologues of cadaverine under conditions of thermal stress (53, 55). The presence of these unusual polyamines under several stress conditions suggests that in P. furiosus, cadaverine and APC, like symmetrical, longer, and branched polyamines in other Archaea, could play a role in the high-temperature adaptation or could represent a molecular response to the oxidative or chemical stress.
Kinetic characterization.
The small amounts of APC and the much higher levels of spermidine measured in P. furiosus cells seem to be in contrast with the activity of ACAPT towards cadaverine that is an order of magnitude higher than that towards putrescine and agmatine observed in vitro (Table 3). Therefore, a detailed kinetic characterization of ACAPT was performed in order to assess whether P. furiosus is able to synthesize spermidine from agmatine using the novel biosynthetic pathway described in the thermophilic bacterium T. thermophilus (33). The Km and Vmax values for 1,3-diaminopropane, putrescine, agmatine, and cadaverine were measured in the presence of saturating concentrations of decarboxy-AdoMet (Fig. 1). The affinity of P. furiosus ACAPT for decarboxy-AdoMet (Km, 20 μM) is similar to that measured for P. falciparum spermidine synthase (Km, 35 μM) (18) but lower than that of corresponding enzymes from S. solfataricus (Km, 7.9 μM) (8), E. coli (Km, 2.2 μM) (4), and rat prostate (Km, 1.1 μM) (41).
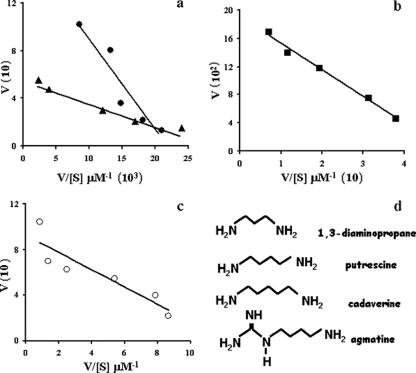
FIG. 1.
Determination of Km values for 1,3-diaminopropane and putrescine (a), cadaverine (b), and agmatine (c). (d) Chemical structures of the polyamines tested. The enzyme was incubated for 10 min at 70°C in the presence of 100 μM decarboxy-AdoMet and various concentrations of 1,3-diaminopropane (▴), putrescine (•), cadaverine (▪), and agmatine (○), as shown. The amount of the polyamine product was determined by radiochemical or HPLC assay, as described in Materials and Methods. Results are shown as a plot of Vmax (V) against the Vmax/[substrate] ratio (V/[S]).
The Vmax values with putrescine, 1,3-diaminopropane (Fig. 1a), and agmatine (Fig. 1c) are comparable and about 20-fold less than that observed with cadaverine (Fig. 1b). However, the enzyme has an extremely high affinity for agmatine (Fig. 1c and Table 5), where the Km value (7.6 μM) is about three orders of magnitude lower than that measured with putrescine (Km = 7,703 μM) or cadaverine (Km = 3,767 μM). The enzyme's higher affinity for agmatine can be explained in part by the PF0127 crystal structure, which shows that agmatine can be easily accommodated in the binding site pocket and that several additional hydrogen bonds (compared to putrescine or cadaverine) can be formed between the agmatine amidine group and residues Glu7, Tyr9, Val51, and Asp162 (data not shown).
TABLE 5.
Kinetic parameters of ACAPT from P. furiosus
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1) (102) |
|---|---|---|---|
| 1,3-Diaminopropane | 1,877 | 0.06 | 0.32 |
| Putrescine | 7,703 | 0.18 | 0.23 |
| Cadaverine | 3,767 | 2.09 | 5.5 |
| Agmatine | 7.6 | 0.10 | 134 |
Consequently, the kcat/Km value for agmatine is about 600-fold higher than that for putrescine, indicating that, in analogy with T. thermophilus, P. furiosus utilizes agmatine, not putrescine, to synthesize spermidine. The presence of putrescine in P. furiosus cells (Table 4) suggests that, at least under the particular growth conditions that were used, this polyamine could function as an aminopropyl acceptor. Moreover, the kcat/Km value for agmatine is also about 20-fold higher than it is for cadaverine (Table 5), in further support of agmatine as a relevant substrate in vivo. We hypothesize that agmatine is the physiological substrate of P. furiosus ACAPT in the constitutive synthesis of spermidine, while cadaverine acts as an aminopropyl acceptor under stress conditions when an active and rapid synthesis of APC could be required.
On the basis of these results, ACAPT shows a unique substrate specificity. It recognizes propylamine acceptor amines with a variable aliphatic spacer between the primary amino groups. Moreover, the ability to utilize both agmatine and cadaverine as substrates makes P. furiosus ACAPT very different from the homologous archaeal propylamine transferase from S. solfataricus (8). The Sulfolobus enzyme is regarded as having a broad substrate specificity, as it can transfer the propylamine group from decarboxy-AdoMet to a primary amino group linked to a trimethylenic as well as to a tetramethylenic chain. However, the S. solfataricus enzyme is almost inactive with cadaverine, in stark contrast to P. furiosus ACAPT.
Thermal properties and limited proteolysis.
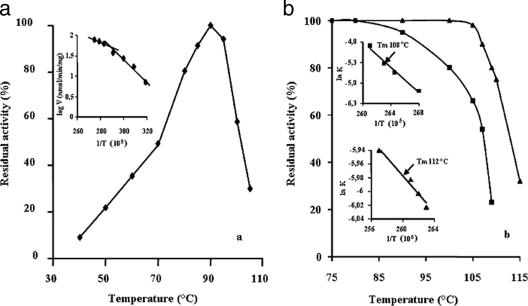
The temperature dependence of the activity of ACAPT in the range from 40°C to 105°C is shown in Fig. 2a. The enzyme is highly thermophilic; its activity increased sharply up to the optimal temperature of 90°C, and a 50% activity was still observable at 103°C. This behavior led to a discontinuity in the Arrhenius plot at about 78°C, with two different activation energies. This result suggests that conformational changes in the protein structure occur near this temperature, resulting in enzymatic forms characterized by different catalytic properties. Two forms were also observed with S. solfataricus APT, whose stability toward denaturation is characterized by a specific temperature dependence (15).
FIG. 2.
The effect of temperature on P. furiosus ACAPT activity and stability. (a) The activity observed at 90°C is expressed as 100%. The assay was performed as described in Materials and Methods. Arrhenius plot is reported in the inset. Temperature (T) is measured in kelvins (K). (b) Residual activity after 10-min incubation at temperatures shown in the absence (▪) or in the presence (▴) of 5 mM cadaverine. Apparent Tm values are reported in the insets.
The stability of ACAPT with respect to reversible denaturation was investigated by carrying out short-time kinetic studies of thermal denaturation. The diagram of the residual activity after 5 min of preincubation as a function of temperature (Fig. 2b) is characterized by a sharp transition. From the corresponding plot (see inset), the transition temperature (apparent Tm) is 108°C. To evaluate the possible stabilizing effect of the substrate, the melting temperature was measured in the presence of 5 mM cadaverine. As shown in Fig. 2b, this polyamine exerts a protection toward temperature inactivation, causing an increase of the apparent Tm to 112°C (see inset). This result indicates that the binding of cadaverine raises the conformational stability of the enzyme, thus reducing its susceptibility to thermal denaturation.
To obtain information about the flexible regions of the protein exposed to the solvent and susceptible to proteolytic attack, ACAPT was subjected to limited proteolysis. The results of the time course analysis for the hydrolysis of the enzyme with proteinase K followed by SDS-PAGE analysis demonstrate that during the proteolytic process, the catalytic activity remains unchanged while a protein band with an apparent molecular mass of about 10 kDa less than that of ACAPT is observable (data not shown). Analysis by Edman degradation of the proteolytic fragment showed that the N terminus was preserved, thus the cleavage site is localized in the C-terminal region, and that the C-terminal peptide is not necessary for the integrity of the active site. When the experiment was carried out in the presence of cadaverine, the proteolytic pattern remained almost unchanged, indicating that this substrate is not able to stabilize the enzyme against the protease.
Figure 3 shows the sequence alignment of P. furiosus ACAPT with the homologous enzymes characterized from various sources. As indicated, the amino acid residues at the active site of T. maritima PAPT, involved in the binding with transition state analog AdoDATO (28), are well conserved in P. furiosus ACAPT with few substitutions. It is interesting to note that Tyr239 and Trp244 at the C terminus of T. maritima PAPT are conserved positions in P. furiosus ACAPT. Nevertheless, the loss of the C-terminal peptide fragment does not affect the catalytic activity of P. furiosus ACAPT, suggesting a different structural environment in the active site of the two enzymes at the level of the polyamine pocket. Moreover, the observation that cadaverine is a very poor substrate for T. maritima PAPT with a maximal rate of 4.6% of that of putrescine (28) and is not a substrate for S. solfataricus APT (9) clearly indicates that the length of the aliphatic spacer between the primary amino groups is critical for polyamine recognition and could prevent precise positioning of the substrate for the reaction.
FIG. 3.
Multiple sequence alignment of APTs from different sources. Sequences: TmPAPT, spermidine synthase from Thermotoga maritima; PfACAPT, ACAPT from Pyrococcus furiosus; SsAPT, APT from Sulfolobus solfataricus; EcSpdS, spermidine synthase from E. coli; HsSpdS, spermidine synthase from Homo sapiens; PFSpdS, spermidine synthase from Plasmodium falciparum. Identical and conserved residues in proteins are highlighted in dark and pale gray, respectively. The amino acid residues of T. maritima PAPT involved in the binding with AdoDATO are indicated (▴).
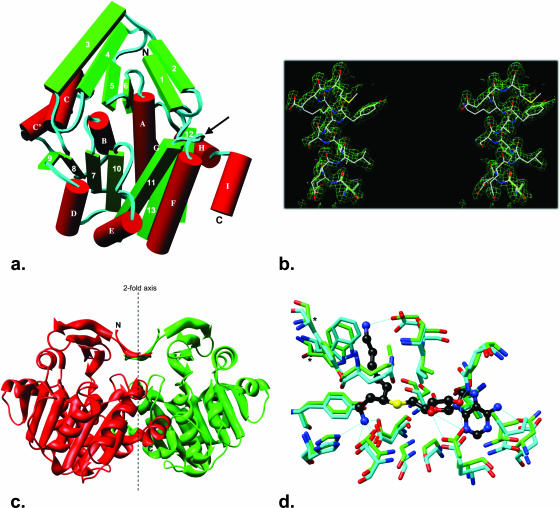
Structure of P. furiosus ACAPT.
The structure of ACAPT was determined at 1.8-Å resolution by molecular replacement. In accordance with the biochemical analyses, the enzyme exists as a homodimer with dimensions of 78 by 47 by 38 Å (Fig. 4a) and a molecular mass of 64.6 kDa. The two molecules in the biologically active dimer exhibit disordered N- and C-terminal regions (residues 1 to 4 and 280 to 281) as well as a five-residue disordered loop (residues 164 to 168). In addition, residues 147 to 148 of the B subunit of the dimer are not observed in the electron density. The refined model (4,527 atoms) includes 201 solvent molecules modeled as water and has an R value of 20.70% (Rfree of 24.20%) and excellent stereochemistry as determined by MOLPROBITY (1, 10). Figure 4b illustrates the quality of the 1.8-Å Fo-Fc omit electron density map (residues 200 to 212 were omitted from the REFMAC refinement).
FIG. 4.
Structural features of ACAPT. (a) A stereo view of the ACAPT dimer perpendicular to the dimer axis. The arrow shows the position of the probable proteolytic cleavage site. (b) A cross-eyed stereo view showing the 1.8-Å (Fo-Fc omit) electron density map contoured at 3σ for residues 200 to 212 (helix αF), which were omitted from the REFMAC refinement. (c) Connectivity diagram (residues in parentheses): β1 (5 to 9), β2 (13 to 16), β3 (19 to 27), β4 (32 to 38), β5 (43 to 47), β6 (50 to 54), αA (58 to 73), β7 (79 to 83), αB (87 to 94), β8 (100 to 105), αC (107 to 118), αC’ (123 to 130), β9 (136 to 140), αD (142 to 150), β10 (153 to 159), αE (172 to 182), β11 (183 to 195), αF (199 to 214), β12 (216 to 222), β13 (231 to 238), αG (247 to 253), αH (260 to 269), and αI (271 to 279). (d) A view of the ACAPT binding pocket (cyan, residues Q31, L47, Q52, Y61, H62, G83, G84, D86, E105, I106, D142, D159, S160, T161, D162, P166, L170, Y227, and W231) with the T. maritima PAPT binding pocket (green, residues Q46, L62, M67, Y76, H77, G98, G99, D101, E121, V122, N152, D170, S171, T172, D173, Q178, L182, Y239, and W244) and bound substrate analogue (black) superimposed. Starred residues Tyr227 and Trp231 are lost upon proteolysis.
The ACAPT monomer (Fig. 4c) shares the two-domain APT architecture observed in related enzyme structures with a mostly β N-terminal domain (residues 3 to 54) and a larger C-terminal catalytic domain (residues 56 to 279) that resembles the Rossmann fold found in a number of nucleotide- and dinucleotide-binding proteins and in S-adenosyl-l-methionine-dependent methyltransferases. The overall ACAPT structure is similar to the structures reported for homologs from the hyperthermophilic bacterium T. maritima (1INL, 1JQ3) (28), the moderately thermophilic bacterium Thermus thermophilus (1UIR), the mesophilic bacterium Bacillus subtilis (1IY9), and the eukaryotes Arabidopsis thaliana (1XJ5) and human (1ZDZ). Superposition (38) of the PF0127 structure (monomer) with the above-mentioned structures gives root mean square deviations (Cα atoms) ranging from 1.42 to 1.72 Å (Table 2). The P. furiosus ACAPT structure was found to contain an additional helix (residues 123 to 130), denoted helix C’ in Fig. 4c, which links helix C to β strand 9. This region showed partial helix structure in the other hyperthermophilic homolog T. maritima PAPT (1INL), possibly suggesting a role in thermal stabilization. The PAPT family signature sequence (residues Val79 to Glu92) (21) spans the end of β strand 7 extending into helix B, similar to that found in related enzymes. As proposed above, the active-site geometry is similar to that observed in the T. maritima PAPT structure (1INL, A chain) used as the model in the molecular replacement calculations, with the “gatekeeper loop” (residues 155 to 164) also adopting a partial open configuration. Superposition of the active-site residues (Gln31, Leu47, Gln52, Tyr61, His62, Gly83, Gly84, Asp86, Glu105, Ile106, Asp142, Asp159, Ser160, Thr161, Asp162, Pro166, Leu170, Tyr227, and Trp231) (Fig. 3) for the two structures gives a root mean square deviation (Cα atoms) of 0.546 Å (Fig. 4d). Consequently, insight into what determines the differences in substrate specificities of T. maritima PAPT (for putrescine) and P. furiosus ACAPT, and the relative activities of ACAPT for its proposed physiological substrates (agmatine and cadaverine), was not apparent. Such information will require structures of ACAPT with substrate analogs, and such studies are planned.
The subunits in the P. furiosus ACAPT dimer are related by a pseudo twofold axis and show similar conformations. Superposition of the A and B chains of the dimer gives a root mean square deviation (Cα atoms) of 0.354 Å. The dimer interface occupies ∼1,741 Å2 (∼14.8%) of the monomer surface and contains 34 solvent molecules. The interface is stabilized by 19 intermolecular hydrogen bonds and 23 bridging water molecules. The intermolecular contacts occur mainly between residues in β2, β12, αH, αI, the loop connecting β11 and αF, and the loop connecting αG with αH.
The structure of P. furiosus ACAPT was analyzed in terms of the results obtained from limited proteolysis of the protein, which showed a loss of a 10-kDa C-terminal fragment. Based on the proteolysis data, the cleavage point should lie in the loop between β strand 11 and helix F (residues Ser195, Val196, Tyr197, Leu198, and Phe199), which is exposed to solvent. In addition, according to the structure in Fig. 4d, the loss of the C-terminal fragment should have only minimal impact on the substrate binding, since only two residues, Tyr227 and Trp231, which do not have direct interaction with the substrate, are lost.
The structure of ACAPT was also used to investigate features of the protein that may contribute to its thermostability. A comparison was made between P. furiosus ACAPT and six related structures from the Protein Data Bank (entries 1INL, 1UIR, 1IY9, 1XJ5, 2B2C, and 1ZDZ) (Table 6). The comparison focused on structural features common to thermophilic proteins (46), such as average loop length, the number of intramolecular hydrogen bonds, the number of intramolecular ionic or polar interactions, the molecular- and solvent-accessible surface area, and the number and size of internal cavities. Since the enzyme exists as a homodimer, additional criteria relating to the dimer interface (size, shape, intersubunit hydrogen bonds and salt bridges, and bridging solvent molecules) were also evaluated (Table 6).
TABLE 6.
Structural parameters of P. furiosus ACAPT and homologs known to affect protein stabilitya
| Parameter | Name or value | ||||||
|---|---|---|---|---|---|---|---|
| PDB ID | 1MJF | 1INL | 1UIR | 1IY9 | 1XJ5 | 2B2C | 1ZDZ |
| Sourceb | P. furiosus | T. maritima | T. thermophilus | B. subtilis | A. thaliana | C. elegans | Human |
| Sequence length | 281 | 296 | 314 | 275 | 334 | 314 | 304 |
| Probable molecular assemblyb | 2 | 4 | 4 | 4 | 4 | 2 | 2 |
| Resolution (Å)c | 1.80 | 1.50 | 2.00 | 2.30 | 2.70 | 2.80 | 2.12 |
| Optimal growth temp (°C) | 100 | 80 | 70 | 35 | 25 | 25 | 37 |
| Protein | |||||||
| No. of loopsc | 21 | 22 | 24 | 20 | 20 | 22 | 21 |
| No. of residues/loop (avg.) | 4.19 | 4.33 | 4.04 | 4.65 | 7.48 | 5.86 | 5.00 |
| No. of hydrogen bondsd | 252 | 277 | 309 | 236 | 265 | 235 | 274 |
| No. of hydrogen bonds/residue | 0.90 | 0.94 | 0.98 | 0.86 | 0.79 | 0.75 | 0.90 |
| No. of salt bridges | 11 | 12 | 16 | 8 | 5 | 11 | 17 |
| No. of CASTp pocketse | 32 | 36 | 32 | 33 | 44 | 41 | 39 |
| Largest CASTp pocket (Å3)e | 637 | 895 | 2596 | 1606 | 1337 | 665 | 1185 |
| Dimer | |||||||
| Total surface area (Å2)f | 18367 | 22292 | 22288 | 18869 | 19932 | 20917 | 19655 |
| Accessible surface area (Å2)f | 19738 | 24246 | 23415 | 20449 | 20715 | 22270 | 20885 |
| Interface accessible surface (Å2)f | 1741 | 1609 | 1787 | 1581 | 1820 | 1634 | 1661 |
| % Interface surface areaf | 14.82 | 11.66 | 13.20 | 13.17 | 13.92 | 12.52 | 13.23 |
| Interface lengthf | 39 | 42 | 40 | 40 | 49 | 45 | 39 |
| Interface breadthf | 35 | 33 | 37 | 33 | 39 | 33 | 39 |
| % Polar atoms at interfacef | 35.26 | 45.91 | 30.00 | 31.21 | 33.63 | 35.17 | 38.68 |
| % Nonpolar atoms at interfacef | 64.70 | 54.00 | 70.00 | 69.70 | 66.30 | 64.80 | 61.30 |
| No. of interface hydrogen bondsf | 19 | 24 | 16 | 12 | 24 | 14 | 20 |
| No. of interface salt bridgesf | 2 | 2 | 0 | 0 | 0 | 0 | |
| No. of bridging HOH moleculesf | 23 | 0 | 15 | 0 | 0 | 6 | 20 |
Comparisons were made between the best-aligning chains for each structure. PDB ID, Protein Data Bank identification.
Protein Data Bank (www.rcsb.org/pdb).
CHIMERA (www.cgl.ucsf.edu/chimera).
CASTp (sts.bioengr.uic.edu/castp).
Protein-Protein Interaction Server (www.biochem.ucl.ac.uk/bsm/PP/server).
Three of the seven structures that were studied were from thermophilic organisms (Pyrococcus furiosus, 1MJF; Thermotoga maritima, 1INL; and Thermus thermophilus, 1UIR), while the others were from bacteria (Bacillus subtilis, 1IY9), plant (Arabidopsis thaliana, 1XJ5), worm (Caenorhabditis elegans, 2B2C), and human (1ZDZ). In general, enzymes from the nonthermophilic organisms had somewhat longer loops, more internal pockets (3), and fewer intermolecular salt bridges (25) stabilizing the dimer interface. The enzymes from the thermophiles had, on average, slightly more intramolecular hydrogen bonds and salt bridges than their mesophilic counterparts. Within the thermophilic enzymes studied, P. furiosus ACAPT should have the most thermostable structure, given that P. furiosus has the highest growth temperature by at least 20°C. Analysis of the data presented in Table 6, however, gives no clear indication as to which of the structural factors surveyed are responsible for its high thermostability, other than the fact that the largest internal pocket (637 Å3) found in ACAPT is significantly smaller than that observed for other thermophilic enzymes and that P. furiosus ACAPT has the smallest solvent-accessible surface area (19,738 Å2) of any of the enzymes. Since the structures used in the study share a common fold (see Table 2), this would suggest that ACAPT has the most compact hydrophobic core of the enzymes studied, which could explain, in part, its high thermostability. In addition, the ACAPT dimer interface, approximately 1,741 Å2, represents the largest interface (in terms of the percentage of total accessible surface area) of the enzymes studied. The P. furiosus ACAPT dimer interface is also stabilized by 19 intermolecular hydrogen bonds, a salt bridge, and 23 bridging water molecules. These factors should also contribute to the thermostability of the dimeric enzyme.
Acknowledgments
This research was supported by grants from Ministero dell'Università e della Ricerca Scientifica PRIN 2004, the National Institutes of Health (GM-60329 to M.W.W.A. and GM-62407 to B.-C.W.), and the Department of Energy (FG05-95ER20175 to M.W.W.A.).
Footnotes
Published ahead of print on 1 June 2007.
REFERENCES
- 1.Arendall, W. B., III, W. Tempel, J. S. Richardson, W. Zhou, S. Wang, I. W. Davis, Z. J. Liu, J. P. Rose, W. M. Carson, M. Luo, D. C. Richardson, and B. C. Wang. 2005. A test of enhancing model accuracy in high-throughput crystallography. J. Struct. Funct. Genomics 6:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, and H. Weissig. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binkowski, T. A., S. Naghibzadeh, and J. Liang. 2003. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 31:3352-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, W. H., C. W. Tabor, and H. Tabor. 1973. Spermidine biosynthesis. Purification and properties of propylamine transferase from Escherichia coli. J. Biol. Chem. 248:2480-2486. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cacciapuoti, G., M. Porcelli, C. Bertoldo, M. De Rosa, and V. Zappia. 1994. Purification and characterization of extremely thermophilic and thermostable 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus. Purine nucleoside phosphorylase activity and evidence for intersubunit disulfide bonds. J. Biol. Chem. 269:24762-24769. [PubMed] [Google Scholar]
- 7.Cacciapuoti, G., M. Porcelli, G. Pontoni, and V. Zappia. 1989. Propylamine transfer reactions in thermophilic archaebacteria: enzymological aspects and comparative biochemistry, p. 47-60. In U. Bacharach and Y. M. Heimer (ed.), The physiology of polyamines, vol. 2. CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 8.Cacciapuoti, G., M. Porcelli, M. Cartenì-Farina, A. Gambacorta, and V. Zappia. 1986. Purification and characterization of propylamine transferase from Sulfolobus solfataricus, an extreme thermophilic archaebacterium. Eur. J. Biochem. 161:263-271. [DOI] [PubMed] [Google Scholar]
- 9.Chayen, N. E., P. D. Shaw Stewart, D. L. Maeder, and D. M. Blow. 1990. An automated system for micro-batch protein crystallization and screening. J. Appl. Crystallogr. 23:297-302. [Google Scholar]
- 10.Davis, I. W., L. W. Murray, J. S. Richardson, and D. C. Richardson. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615-W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Della Ragione, F., M. Cartenì-Farina, M. Porcelli, G. Cacciapuoti, and V. Zappia. 1981. High-performance liquid chromatographic analysis of 5′-methylthioadenosine in rat tissues. J. Chromatogr. 226:243-249. [DOI] [PubMed] [Google Scholar]
- 12.De Rosa, M., S. De Rosa, A. Gambacorta, M. Cartenì-Farina, and V. Zappia. 1976. Occurrence and characterization of new polyamines in the extreme thermophile Caldariella acidophila. Biochem. Biophys. Res. Commun. 69:253-261. [DOI] [PubMed] [Google Scholar]
- 13.De Rosa, M., S. De Rosa, A. Gambacorta, M. Cartenì-Farina, and V. Zappia. 1978. The biosynthetic pathway of new polyamines in Caldariella acidophila. Biochem. J. 176:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dion, A. S., and S. S. Cohen. 1972. Polyamine stimulation of nucleic acid synthesis in an uninfected and phage-infected polyamine auxotroph of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 69:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facchiano, F., R. Ragone, M. Porcelli, G. Cacciapuoti, and G. Colonna. 1992. Effect of temperature on the propylamine transferase from Sulfolobus solfataricus, an extreme thermophilic archaebacterium. 1. Conformational behavior of the oligomeric enzyme in solution. Eur. J. Biochem. 204:473-482. [DOI] [PubMed] [Google Scholar]
- 16.Gaboriau, F., R. Havouis, J. P. Moulinoux, and J. G. Delcros. 2003. Atmospheric pressure chemical ionization-mass spectrometry method to improve the determination of dansylated polyamines. Anal. Biochem. 318:212-220. [DOI] [PubMed] [Google Scholar]
- 17.Goda, H., T. Watanabe, N. Takeda, M. Kobayashi, M. Wada, H. Hosoda, A. Shirahata, and K. Samejima. 2004. Mammalian spermidine synthase. Identification of cysteine residues and investigation of putrescine binding site. Biol. Pharm. Bull. 27:1327-1332. [DOI] [PubMed] [Google Scholar]
- 18.Haider, N., M. L. Eschbach, S. de Souza Dias, T. W. Gilberger, R. D. Walter, and K. Luersen. 2005. The spermidine synthase of the malaria parasite Plasmodium falciparum: molecular and biochemical characterisation of the polyamine synthesis enzyme. Mol. Biochem. Parasitol. 142:224-236. [DOI] [PubMed] [Google Scholar]
- 19.Haman, K., T. Tanaka, R. Hosoya, M. Niitsu, and T. Itih. 2003. Cellular polyamines of the acidophilic, thermophilic and thermoacidophilic archaebacteria. Acidilobus, Ferroplasma, Pyrobaculum, Pyrococcus, Staphylothermus, Thermococcus, Thermodiscus and Vulcanisaeta. J. Gen. Microbiol. 49:287-293. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment of a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtta, E., and P. Pohjanpelto. 1983. Polyamine starvation causes accumulation of cadaverine and its derivatives in a polyamine-dependent strain of Chinese-hamster ovary cells. Biochem. J. 210:945-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoya, R., K. Hamana, M. Niitsu, and T. Itoh. 2004. Polyamine analysis for chemotaxonomy of thermophilic eubacteria: polyamine distribution profiles within the orders Aquificales, Thermotogales, Thermodesulfobacteriales, Thermales, Thermoanaerobacteriales, Clostridiales and Bacillales. J. Gen. Appl. Microbiol. 50:271-287. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi, K., K. Kashiwagi, H. Hamasaki, A. Miura, T. Kakegawa, S. Hirose, and S. Matsuzaki. 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J. Bacteriol. 166:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kissinger, C. R., D. K. Gehlhaar, and D. B. Fogel. 1999. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55:484-491. [DOI] [PubMed] [Google Scholar]
- 27.Kneifel, H., K. O. Stetter, J. R. Andreesen, J. Wviegel, H. Konig, and S. M. Schoberth. 1986. Distribution of polyamines in representative species of archaebacteria. Syst. Appl. Microbiol. 7:241-245. [Google Scholar]
- 28.Korolev, S., Y. Ikeguchi, T. Skarina, S. Beasley, C. Arrowsmith, A. Edwards, A. Joachimiak, A. E. Pegg, and A. Savchenko. 2002. The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat. Struct. Biol. 9:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Z. J., W. Tempel, J. D. Ng, D. Lin, A. K. Shah, L. Chen, P. S. Horanyi, J. E. Habel, I. A. Kataeva, H. Xu, H. Yang, J. C. Chang, L. Huang, S. H. Chang, W. Zhou, D. Lee, J. L. Praissman, H. Zhang, M. G. Newton, J. P. Rose, J. S. Richardson, D. C. Richardson, and B. C. Wang. 2005. The high-throughput protein-to-structure pipeline at SECSG. Acta Crystallogr. D 61:679-684. [DOI] [PubMed] [Google Scholar]
- 30.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 31.Moinard, C., L. Cynober, and J. P. deBanth. 2005. Polyamines: metabolism and implications in human diseases. Clin. Nutr. 24:184-197. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov, G. N. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 33.Ohnuma, M., Y. Terui, M. Tamakoshi, H. Mitome, M. Niitsu, K. Samejima, E. Kawashima, and T. Oshima. 2005. N1-aminopropyl agmatine: a new polyamine produced as a key intermediate in polyamine biosynthesis of an extreme thermophile Thermus thermophilus. J. Biol. Chem. 280:30073-30082. [DOI] [PubMed] [Google Scholar]
- 34.Oshima, T. 1989. Polyamines in thermophiles, p. 35-46. In U. Bacharach and Y. M. Heimer (ed.), The physiology of polyamines, vol. 2. CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 35.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 36.Pajula, R. L., A. Raina, and T. Eloranta. 1979. Polyamine synthesis in mammalian tissues. Isolation and characterization of spermine synthase from bovine brain. Eur. J. Biochem. 101:619-626. [DOI] [PubMed] [Google Scholar]
- 37.Pegg, A. E. 1986. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 234:249-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 39.Raina, A., T. Eloranta, T. Hyvonen, and R. L. Pajula. 1983. Mammalian propylamine transferases, p. 245-253. In U. Bachrach, A. Kaye, and R. Chayen (ed.), Advances in polyamine research, vol. 4. Raven Press, New York, NY. [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Samejima, K., and B. Yamanoha. 1982. Purification of spermidine synthase from rat ventral prostate by affinity chromatography on immobilized S- adenosyl(5′)-3-thiopropylamine. Arch. Biochem. Biophys. 216:213-222. [DOI] [PubMed] [Google Scholar]
- 42.Schlenk, F., and D. J. Ehninger. 1964. Observations on the metabolism of 5′-methylthioadenosine. Arch. Biochem. Biophys. 106:95-100. [DOI] [PubMed] [Google Scholar]
- 43.Schut, G. J., S. D. Brehm, S. Datta, and M. W. W. Adams. 2003. Whole genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 183:3935-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens, L. 1967. Studies on the interaction of homologues of spermine with deoxyribonucleic acid and with bacterial protoplasts. Biochem. J. 103:811-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugar, F. J., F. E. Jenney, F. L. Poole, P. S. Brereton, M. Izumi, C. Shah, and M. W. W. Adams. 2005. Comparison of small- and large-scale expression of selected Pyrococcus furiosus genes as an aid to high-throughput protein production. J. Struct. Funct. Genomics 6:149-158. [DOI] [PubMed] [Google Scholar]
- 46.Szilagyi, A., and P. Zavodszky. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8:493-504. [DOI] [PubMed] [Google Scholar]
- 47.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53:749-790. [DOI] [PubMed] [Google Scholar]
- 48.Tabor, H., and C. W. Tabor. 1976. 1,4-Diaminobutane (putrescine), spermidine and spermine. Annu. Rev. Biochem. 45:285-306. [DOI] [PubMed] [Google Scholar]
- 49.Tabor, H., S. M. Rosenthal, and C. W. Tabor. 1958. The biosynthesis of spermidine and spermine from putrescine and methionine. J. Biol. Chem. 233:907-914. [PubMed] [Google Scholar]
- 50.Teng, T. Y. 1990. Mounting of crystals for macromolecular crystallography in a freestanding thin-film. J. Appl. Crystallogr. 23:387-391. [Google Scholar]
- 51.Terui, Y., M. Ohnuma, K. Hiraga, E. Kawashima, and T. Oshima. 2005. Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile Thermus thermophilus. Biochem. J. 388:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace, H. M., A. D. Fraser, and A. Hughes. 2003. A perspective of polyamine metabolism. Biochem. J. 376:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walters, D. R., and T. Cowley. 1996. Formation of cadaverine derivatives in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 145:255-259. [DOI] [PubMed] [Google Scholar]
- 54.Walters, D. R., and T. Cowley. 1998. Polyamine metabolism in Saccharomyces cerevisiae exposed to ethanol. Microbiol. Res. 153:179-184. [DOI] [PubMed] [Google Scholar]
- 55.Walters, D. R., T. Cowley, and A. McPherson. 1997. Polyamine metabolism in the thermotolerant mesophilic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 153:433-437. [DOI] [PubMed] [Google Scholar]
- 56.Weber, K., J. R. Pringle, and M. Osborn. 1972. Measurement of molecular weight by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 260:3-27. [DOI] [PubMed] [Google Scholar]
- 57.Weinberg, M. V., G. J. Schut, S. Brehm, S. Datta, and M. W. W. Adams. 2005. Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J. Bacteriol. 187:336-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickner, R. B., C. W. Tabor, and H. Tabor. 1970. Purification of adenosylmethionine decarboxylase from E. coli W: evidence for covalently-bound pyruvate. J. Biol. Chem. 245:2132-2139. [PubMed] [Google Scholar]
- 59.Wiest, L., and A. E. Pegg. 1997. Assay of spermidine and spermine synthases, p. 51-57. In D. Morgan (ed.), Methods in molecular biology, vol. 79. Polyamine protocols. Humana Press, Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 60.Williams-Ashman, H. G., and A. E. Pegg. 1981. Amino propyl group transfers, p. 43-54. In D. R. Morris and L. J. Marton (ed.), Polyamines in biology and medicine. Dekker, New York, NY.