Abstract
Malic enzymes (MEs) catalyze the oxidative decarboxylation of malate in the presence of a divalent metal ion. In eukaryotes, well-conserved cytoplasmic, mitochondrial, and plastidic MEs have been characterized. On the other hand, distinct groups can be detected among prokaryotic MEs, which are more diverse in structure and less well characterized than their eukaryotic counterparts. In Escherichia coli, two genes with a high degree of homology to ME can be detected: sfcA and maeB. MaeB possesses a multimodular structure: the N-terminal extension shows homology to ME, while the C-terminal extension shows homology to phosphotransacetylases (PTAs). In the present work, a detailed characterization of the products of E. coli sfcA and maeB was performed. The results indicate that the two MEs exhibit relevant kinetic, regulatory, and structural differences. SfcA is a NAD(P) ME, while MaeB is a NADP-specific ME highly regulated by key metabolites. Characterization of truncated versions of MaeB indicated that the PTA domain is not essential for the ME reaction. Nevertheless, truncated MaeB without the PTA domain loses most of its metabolic ME modulation and its native oligomeric state. Thus, the association of the two structural domains in MaeB seems to facilitate metabolic control of the enzyme. Although the PTA domain in MaeB is highly similar to the domains of proteins with PTA activity, MaeB and its PTA domain do not exhibit PTA activity. Determination of the distinct properties of recombinant products of sfcA and maeB performed in the present work will help to clarify the roles of MEs in prokaryotic metabolism.
Malic enzymes (MEs) catalyze the oxidative decarboxylation of malate coupled with the reduction of NAD+ or NADP+ to produce pyruvate and CO2 (Fig. 1A) (3, 8). Based on their coenzyme specificities and their abilities to decarboxylate oxaloacetate (OAA), MEs are divided into three categories: EC 1.1.1.38 (NAD dependent; decarboxylates added OAA), EC 1.1.1.39 (NAD dependent; does not decarboxylate added OAA), and EC 1.1.1.40 (NADP dependent; decarboxylates added OAA) (International Union of Biochemistry and Molecular Biology nomenclature home page [http://www.chem.qmul.ac.uk/iubmb/]). MEs have been found in organisms representative of all the major biological divisions, due to the extensive participation of the substrates and products in different metabolic pathways. In eukaryotes, cytoplasmic, mitochondrial, and plastidic ME isoforms have been found and the crystal structures of some typical MEs have been resolved. According to the results, crystallized MEs are homotetramers composed of monomers with molecular masses between 62 and 68 kDa (3, 7).
FIG. 1.
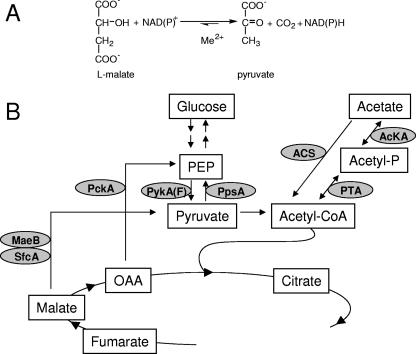
(A) ME reaction. ME catalyzes the oxidative decarboxylation of the C4 dicarboxylic acid malate to the C3 acid pyruvate and CO2. Depending on the type of ME, NAD or NADP is used. A metal ion cofactor is essential for the reaction. (B) The PEP-pyruvate-OAA node and its connection with glycolysis/gluconeogenesis and acetate activation. ME (SfcA and/or MaeB) in combination with PEP synthetase (PpsA) allows the formation of PEP, the direct precursor for gluconeogenesis, from TCA intermediates. Alternatively, PCK (PckA) can also direct C4 intermediates to PEP. The conversion of PEP to pyruvate, an essential step in glycolysis, is catalyzed by pyruvate kinase (PykA or PykF). Also shown is the activation of acetate, which involves two possible pathways. One is catalyzed by the AMP-forming acetyl-CoA synthetase (ACS), and an alternative pathway uses acetate kinase (AcKA) in combination with PTA.
Prokaryotic MEs are more diverse in structure and less well characterized than their eukaryotic counterparts. For example, MEs with large subunits (approximately 83 kDa each), characterized by the presence of an extra carboxyl-terminal region with homology to phosphotransacetylases (PTA) (EC 2.3.1.8), have been found in Sinorhizobium meliloti (25, 38). These chimerical NAD or NADP MEs can also be identified in other bacteria by sequence homology. On the other hand, MEs with smaller subunits were found in several gram-positive bacteria, such as ME from Bacillus stearothermophilus, a tetramer of 50-kDa subunits (17). Smaller MEs of approximately 40 kDa per subunit were characterized from Streptococcus bovis [NAD(P) ME] (16), Corynebacterium glutamicum (NADP ME) (11), and Lactococcus lactis (34). The last enzyme was found to catalyze the decarboxylation of OAA only. Finally, MEs with subunit molecular mass values more closely related to the MEs from eukaryotic organisms (approximately 60 kDa) can also be found, such as SfcA from Escherichia coli (36). Interestingly, the analysis of the complete genome sequence of Bacillus subtilis revealed the presence of four paralogous genes encoding MEs with predicted molecular masses between 45 and 60 kDa (6). The protein products of the four B. subtilis ME genes exhibited ME activity, showing that YtsJ has an important role in efficient growth on malate that could not be fulfilled by any of the other MEs (19).
In bacteria, MEs are part of the phosphoenolpyruvate (PEP)-pyruvate-OAA node, which links glycolysis/gluconeogenesis to the tricarboxylic acid (TCA) cycle (31). Under gluconeogenic conditions, PEP carboxykinase (PCK) and/or ME, in combination with PEP synthetase, is used for directing C4 intermediates from the TCA cycle to PEP, the direct precursor for gluconeogenesis (Fig. 1B). Thus, PCK or MEs are essential enzymes for growth on TCA cycle intermediates or on substrates that enter central metabolism via acetyl-coenzyme A (CoA), as is the case for acetate (Fig. 1B).
In E. coli, two genes possess homology to ME: sfcA (or maeA) and maeB (or ypfF). The product of sfcA, characterized as a NAD ME, has been used in metabolic engineering (36) and recently to restore wild-type N2 fixation to an S. meliloti NAD ME mutant (26). The product of maeB is a putative ME with a multimodular structure, which has not been characterized at all, although it was recently used to complement a B. subtilis mutant (19). Earlier, in the 1970s, several attempts were made to characterize E. coli MEs by obtaining purified preparations with NAD and/or NADP ME activity (29, 30, 35). However, the subunit molecular masses and amino acid compositions of the purified NAD and NADP MEs failed to match the predicted molecular masses and amino acid compositions of the two MEs deduced from the E. coli genome sequence.
In the present work, a detailed characterization of the products of sfcA and maeB, as well as of truncated versions of the multimodular protein MaeB, was performed. The genes were amplified from E. coli K-12, and the products were expressed and purified from E. coli BL21(DE3). Determination of the kinetic properties of SfcA and MaeB is expected to help in the elucidation of the distinct roles of the two MEs in E. coli metabolism, as well as in the future manipulation of the enzyme in order to obtain modified versions well suited for metabolic engineering.
MATERIALS AND METHODS
Bacterial strains and growth media.
E. coli DH5α was used as a general cloning host. E. coli strain K-12 was used for cloning the open reading frames of sfcA and maeB. Strains were routinely cultured aerobically in Luria-Bertani (LB) broth with appropriate antibiotics. For expression and purification of E. coli MEs, E. coli strain BL21(DE3) was used.
Cloning, expression, and purification of the products of sfcA and maeB.
The sfcA and maeB open reading frames were amplified by colony PCR of E. coli K-12 using the following primer sets: 5′-CCATGGATATTCAAAAAGAGTGAGTG-3′ in combination with 5′-CTCGAGTTAGATGGAGGTACGGCGG-3′ for sfcA and 5′-CCATGGATGACCAGTTAAAACAAAGT-3′ in combination with 5′-CTCGAGAATTACAGCGGTTGGGTT-3′ for maeB. The primers were designed based on the published sequence of each gene (GenBank gi:90111281 for sfcA and gi:16130388 for maeB), and unique NcoI and XhoI restriction sites were introduced at the 5′ and 3′ terminals of each gene for further cloning. The amplified products were cloned into pGEM T-Easy (Promega, Madison, WI), generating plasmids pGEMT-SfcA and pGEMT-MaeB, and their identities were confirmed by sequencing. The NcoI-XhoI fragments were subcloned into pET-32 (Novagen; Merck, Darmstadt, Germany), and the resulting plasmids were named pET-SfcA and pET-MaeB. The expression products were designed to contain an N-terminal hexahistidine tag for purification on Ni-agarose.
For expression, BL21(DE3) cells transformed with pET-SfcA or pET-MaeB were grown to an A600 of 0.6 in LB medium containing the appropriate antibiotic (ampicillin). The cultures were then induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside. After 12 h of aerobic growth at 15°C, the cells were harvested by centrifugation for 5 min at 4,000 × g; resuspended in buffer A (0.5 M NaCl, 20 mM Tris-HCl, pH 7.9) containing 5 mM imidazole, 0.01 μg/μl leupeptin, and 2 mM phenylmethylsulfonyl fluoride; sonicated; and centrifuged for 10 min at 7,000 × g at 4°C. The supernatant (soluble fraction) was loaded onto a 5-ml Niquel-NTA (QIAGEN, Hilden, Germany) column according to the protocol provided by the pET plasmid's manufacturer but using 200 mM imidazole for elution. The His-tagged enzymes were then concentrated on Centricon YM-50 (Amicon) and desalted using buffer B (100 mM Tris-HCl, pH 7.5, 10% [vol/vol] glycerol, and 20 mM β-mercaptoethanol). Purified SfcA was then incubated with enterokinase (1:10) in buffer B at 16°C for 2 h, while MaeB was incubated with thrombin (1:10) under the same conditions, in order to remove the N terminus encoded by the expression vector. The proteins were further purified using an Affi Gel Blue affinity column (Bio-Rad, Hercules, CA), followed by a second Niquel-NTA column. The purified SfcA and MaeB proteins were concentrated, supplemented with 50% (vol/vol) glycerol, and stored at −80°C for further characterization.
The protein concentration was determined by the method of Sedmak and Grossberg (33) using bovine serum albumin as a standard.
Construction of truncated MaeB proteins and the PTA fragment for expression.
Three truncated versions of MaeB were constructed by PCR using the plasmid pGEMT-MaeB as a template. Three lower primers were designed to contain a stop codon and a restriction site (XhoI) for further cloning. These primers were used in combination with the upper primer used for cloning the open reading frame of maeB. In this way, the primer sp1stop (5′-CTCGAGAAATCAGCAATCGGACGAG-3′) generated a PCR product of 1,246 bp and thus a truncated MaeB protein, called Sp1, consisting of the first 413 amino acids of MaeB. Primer sp2stop (5′-CTCGAGTTAGCGCGGATCAAACGGT-3′) generated a PCR product of 1,172 bp and a truncated MaeB (Sp2) of 388 amino acids, while sp3stop (5′-CTCGAGTTACAGTCCCAGCGTTACCAGT-3′) generated a product of 1,412 bp and a truncated MaeB protein (Sp3) of 468 amino acids (Fig. 2). Finally, the carboxyl-terminal part of MaeB (MaeB-PTA, the last 351 amino acids, homologous to PTA) (Fig. 2) was cloned using the plasmid pGEMT-MaeB as a template. The lower primer used was the same as the lower primer used for cloning the open reading frame of maeB in combination with the primer MaeBPTAstart (5′-CCATGGCTGATTTCGACGTCTACAT-3′), which introduced a restriction site (NcoI) for cloning. The PCR fidelity was checked by sequencing in all cases. The generated PCR products were cloned in pGEM T-Easy (Promega), and NcoI-XhoI fragments were subcloned into pET-32 (Novagen). The plasmids were named pET-Sp1, pET-Sp2, pET-Sp3, and pET-MaeBPTA and were introduced into BL21(DE3) cells for expression of the fused His-tagged proteins. The purification and digestion conditions used for each protein were the same as for the complete version of MaeB described above.
FIG. 2.
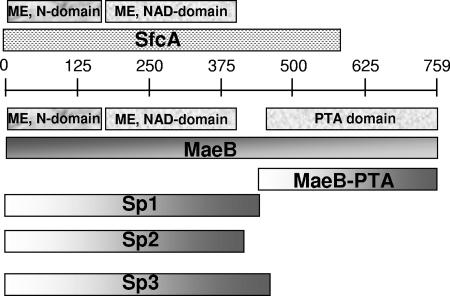
Recombinant E. coli MEs characterized in the present work. The scale indicates the number of amino acid residues in each protein. In boxes, the putative conserved domains (CDD) in SfcA and MaeB are indicated above each protein: ME, N-terminal domain; ME, NAD-binding domain and PTA domain. The truncated MaeB proteins Sp1, Sp2, and Sp3 (413, 388, and 468 amino acids, respectively) have the two ME domains, while MaeB-PTA (351 amino acids from the carboxyl-terminal part of MaeB) is composed of only the PTA domain.
Enzymatic-activity assay.
Spectrophotometric assays were utilized to monitor the oxidative decarboxylation of l-malate by SfcA or MaeB using a standard reaction mixture containing 50 mM Tris-HCl, pH 7.5, and 10 or 1 mM MgCl2, 0.5 mM NAD+ or NADP+, and 10 or 40 mM l-malate, respectively, in a final volume of 0.5 ml. The reaction was started by the addition of l-malate, and the absorbance at 340 nm at 30°C was instantly recorded. One unit of the enzyme was defined as the amount of enzyme that catalyzed the production of 1 μmol of NAD(P)H/min. An absorption coefficient of 6.22 mM−1 for NAD(P)H was used in the calculations. The apparent Michaelis-Menten constants of the substrate and cofactors were determined by varying the concentration of one substrate (or cofactor) around its Km value while leaving other components unchanged at saturation concentrations. All substrate concentrations reported refer to the free, uncomplexed reactant concentrations and were calculated taking into account the dissociation constants of the different complexes formed [Mg2+-NAD(P) or Mg2+-malate].
When sigmoidal curves were observed, initial rates were fitted to the Hill equation, while data were further analyzed to calculate the K0.5 value, the substrate concentration at half-maximal velocity, and the Hill coefficient (h), which is a measure of the degree of cooperativity: v = Vmax [malate]h/(K0.5h + [malate]h), where v is velocity.
The rate of the reductive carboxylation of pyruvate by SfcA or MaeB was measured by the reduction in absorbance at 340 nm in a solution containing 50 mM MOPS (morpholinepropanesulfonic acid)-KOH, pH 7.0, 30 mM NaHCO3, 30 mM pyruvate, and 10 or 1 mM MgCl2 and 0.1 mM NADH or NADPH, respectively, in a final volume of 0.5 ml. The linearity of the reaction was monitored to detect any CO2 loss during the assay.
All kinetic parameters were calculated at least in triplicate and adjusted to nonlineal regression using free-concentration values for all substrates (5). All data-fitting procedures were performed with the Sigma Plot 8.0 program.
When different compounds were tested as potential inhibitors or activators of enzyme activity, SfcA (using NAD+ as a cofactor) and MaeB activities were measured at pH 7.5 in the absence or presence of 0.5, 1.0, or 2.0 mM of each effector (OAA, succinate, fumarate, pyruvate, glucose-6P, fructose-6P, aspartate, glycine, glutamate, alanine, or acetyl-P) or 20 or 50 μM of CoA, acetyl-CoA, or palmitoyl-CoA while the malate concentration was maintained at one-fifth of the Km,malate value for each enzyme (0.1 mM for SfcA and 0.6 mM for MaeB). The results are presented as the percentages of activity in the presence of the effectors in relation to the activity measured in the absence of the metabolites. KCl (0.5 or 2 mM) was also tested in order to confirm previous work indicating activation by that cation. The effects of some compounds on the Sp3 activity were measured as described above but in the presence of 1 mM malate (one-fifth of the Km,malate value).
The optimal pH for recombinant MaeB and SfcA reactions was determined using different buffers: 50 mM MES (morpholineethanesulfonic acid) (pH 5.5 to 6.5), 50 mM Tricine-MOPS (pH 7.0 to 7.5), and 50 mM Tris-HCl (pH 7.5 to 8.5), as for the maize recombinant photosynthetic NADP ME (5).
PTA activity was assayed at 30°C by monitoring the thioester bond formation of acetyl-CoA at 233 nm (ɛ233 = 5.55 mM−1 cm−1). The assay mixture contained 50 mM Tris-HCl, pH 7.0, 20 mM KCl, 10 mM lithium acetyl-P, 0.2 mM lithium CoA, and 2 mM dithiothreitol. The UV spectrum of the reaction assay was compared to that of the reaction medium without acetyl-P in order to confirm the presence of a peak at 233 nm due to the formation of acetyl-CoA and the corresponding decrease in the peak at 260 nm due to the disappearance of free CoA. Alternatively, PTA activity was monitored by measuring the Pi-dependent CoA release from acetyl-CoA with Ellman's thiol reagent, 5′5-dithiobis (2-nitrobenzoic acid), as the formation of thiophenolate anion at 412 nm. The assay mixture contained 100 mM Tris-HCl (pH 9.0), 1.5 M KCl, 5 mM MgCl2, 0.1 mM 5′5-dithiobis (2-nitrobenzoic acid), 0.1 mM acetyl-CoA, and 5 mM KH2PO4 (20).
Preparation of antibodies against MaeB.
Polyclonal antibodies against MaeB recombinant ME were obtained by immunization of rabbits with 200 μg of the purified protein in four subcutaneous injections of 50 μg at 15-day intervals. Antibodies were further purified from the crude antiserum (28).
Gel electrophoresis and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 10 or 12% (wt/vol) polyacrylamide gels according to the method of Laemmli (18). Proteins were visualized with Coomassie blue or electroblotted onto a nitrocellulose membrane for immunoblotting. Bound antibodies were detected by incubation with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G according to the manufacturer's instructions (Sigma). Alkaline phosphatase activity was detected colorimetrically or, alternatively, by using a chemiluminescence kit (Immun-Star; Bio-Rad).
Native PAGE was performed according to the method of Davis (4), employing a 6 or 8% (wt/vol) acrylamide separating gel. Electrophoresis was run at 150 V and 10°C. The gels were analyzed by Coomassie staining or Western blotting or assayed for ME activity by incubating them in a solution containing 200 mM Tris-HCl, pH 7.5, 200 mM l-malate, 10 mM Mg2+, 10 mM NADP+ (for MaeB) or NAD+ (for SfcA), 0.1 mg/ml nitroblue tetrazolium, and 5 μg/ml phenazine methosulfate at room temperature.
Gel filtration chromatography.
The molecular masses of recombinant native E. coli MEs were evaluated by gel filtration chromatography on a fast protein liquid chromatography system with a Superdex 200 10/300 GL column (Amersham Biosciences). The column was equilibrated with 25 mM Tris-HCl at pH 7.5 and 10% (vol/vol) glycerol and calibrated using molecular mass standards. The sample and the standards were applied separately in a final volume of 50 μl at a constant flow rate of 1 ml/min.
CD.
Circular-dichroism (CD) spectra of purified MaeB were obtained with a Jasco J-810 spectropolarimeter using 1.0-cm-path-length cell and averaging five repetitive scans between 250 and 200 nm. Typically, 50 μg of the proteins in phosphate buffer (20 mM NaPi, pH 8.0, 5 mM MgCl2) was used for each assay. CD spectra were also obtained at different Mg2+ concentrations.
RESULTS
Purification and characterization of recombinant SfcA and MaeB.
Two genes from the whole E. coli K-12 genome (E. coli genome and proteome database, GenProtEC [http://genprotec.mbl.edu/]; The EcoGene Database of E. coli Sequence and Function [http://ecogene.org/]) possess homology to ME: the sfcA (or maeA) and maeB (or ypfF) genes. Between the predicted two proteins encoded by these genes (SfcA and MaeB), three putative conserved domains can be detected (Fig. 2). Two of these domains are specific for MEs: the N-terminal ME domain (pfam00390) and the ME NAD-binding domain (pfam03949) (Fig. 2) (Conserved Domain Database [CDD] [21]). Interestingly, an extra domain can be found in the carboxyl-terminal part of MaeB (Fig. 2) (COG0280; CDD [21]) with homology to the non-ME-related enzyme PTA, which catalyzes the transfer of an acetyl group to orthophosphate. However, this extra domain can also be found in other MEs from gram-negative bacteria (25, 26, 38).
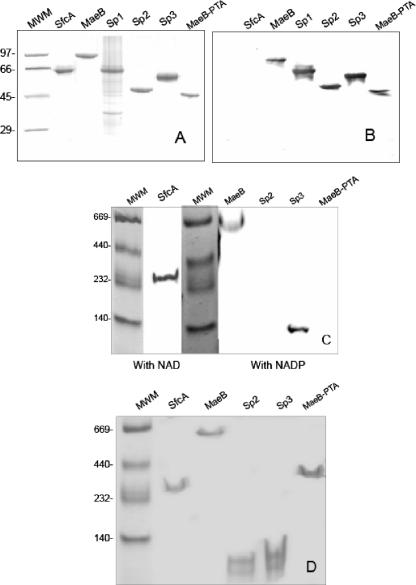
In order to characterize SfcA and MaeB structurally and kinetically, both genes were cloned from E. coli K-12 and fused to an expression vector to obtain the corresponding His-tagged proteins. SfcA and MaeB were overproduced in BL21(DE3) cells to activity levels 500 times higher than those found in nontransformed cells. The typical level of expression for SfcA was 200 mg protein/liter of culture, while that for MaeB was 300 mg protein/liter of culture. The proteins were purified to homogeneity (Fig. 3A), and the monomer molecular masses were 64 kDa for SfcA and 83 kDa for MaeB, both of which correspond to the predicted molecular masses based on their sequences. Polyclonal antibodies obtained and purified against recombinant MaeB did not cross-react with purified recombinant SfcA (Fig. 3B).
FIG. 3.
Characterization of purified recombinant E. coli MEs and deletions. (A) Coomassie-stained SDS-PAGE (5 μg of each protein) of recombinant purified MaeB, SfcA, Sp1 (in this case, a total extract of the insoluble fraction of induced BL21 cells transformed with pET-Sp1), Sp2, Sp3, and MaeB-PTA. (B) Western blot using antibodies against recombinant MaeB. The proteins shown in panel A were transferred for Western analysis. Molecular mass markers (MWM) were run on the left of each gel. (C) Nondenaturing activity gels. Purified SfcA (2 mU) was detected as NAD ME activity, while purified MaeB (2 mU), Sp2 (10 μg), Sp3 (2 mU), and MaeB-PTA (10 μg) were detected as NADP ME activity. Native molecular markers were loaded on the left of each gel and Coomassie blue stained. (D) Coomassie blue-stained native gel (5 μg of each protein) of purified recombinant SfcA, MaeB, Sp2, Sp3, and MaeB-PTA. Native molecular markers were loaded on the left.
Kinetic parameters for the oxidative decarboxylation of malate.
A detailed kinetic characterization was performed for both purified MEs (Table 1). All kinetic parameters for SfcA were determined at pH 7.5, since it was optimal for enzyme activity. With regard to coenzyme specificity, the enzyme clearly preferred NAD+ over NADP+, with a Km value for NAD+ nearly 15 times lower than the K0.5 for NADP+ (Table 1). Interestingly, when SfcA uses NADP+ as a cofactor, the response is nonhyperbolic, with a Hill coefficient of 1.95 (see Fig. S1B in the supplemental material). Although activity with NADP+ was detected, the kcat was almost six times lower than the kcat for NAD+, producing an enzyme with 100-times-higher catalytic efficiency with NAD+ over NADP+ (Table 1). The Km for malate (using NAD+ as a cofactor) was correlated with the Km value previously reported for SfcA (36). For the cofactor Mg2+, a hyperbolic response was obtained (Table 1).
TABLE 1.
Kinetic parameters of recombinant SfcA and MaeB
| Parameter | Valuea
|
|
|---|---|---|
| SfcA | MaeB | |
| Malate oxidative decarboxylation (pH 7.5) | ||
| kcat,NAD (s−1) | 82.7 ± 5.1 | ND |
| Km,NAD (μM) | 68.8 ± 3.2 | ND |
| kcat,NAD/Km,NAD | 1.2 | |
| kcat,NADP (s−1) | 12.4 ± 0.6 | 66.6 ± 2.5 |
| Km,NADP (μM) | 1,000 ± 78b | 41.5 ± 1.5 |
| kcat,NADP/Km,NADP | 0.012 | 1.6 |
| Km,malate (mM) | 0.66 ± 0.04 | 3.41 ± 0.25b |
| Km,Mg (mM) | 2.03 ± 0.1 | 0.19 ± 0.01 |
| Pyruvate reductive carboxylation (pH 7.0) | ||
| kcat (s−1) | 2.93 ± 0.15 | 0.38 ± 0.02 |
| Km,pyruvate (mM) | 2.59 ± 0.1 | 6.21 ± 0.4 |
| kcat,forward/kcat,reverse | 28.2 | 175.2 |
Kinetic values are given as averages ± standard deviations. Each value was averaged over at least two different enzyme preparations. ND, no activity was detected.
Kinetics for these reactions are sigmoidal, and the reported values are K0.5 values, not true Km values. The calculated Hill coefficients were as follows: 1.95 for K0.5,NADP of SfcA and 1.42 for K0.5,malate of MaeB.
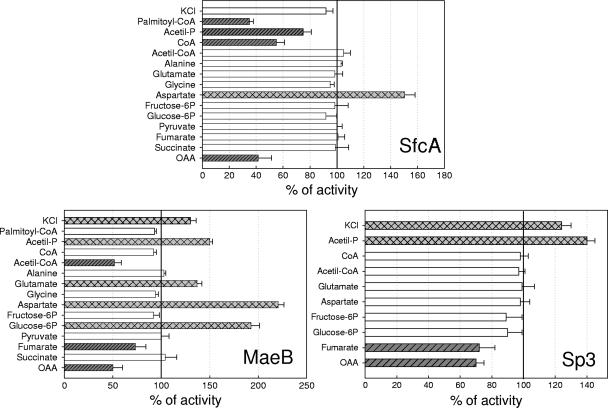
On the other hand, MaeB showed activity only when NADP+ was used as a cofactor (Table 1). Its optimum pH for activity is 7.5, and this value was used for further characterization of the recombinant enzyme. The kcat estimated for MaeB was of the same order of magnitude as the kcat for SfcA using NAD+ as a cofactor but more than five times higher than the value for SfcA using NADP+. The kinetics of MaeB when malate was analyzed as the variable substrate was nonhyperbolic, exhibiting a sigmoidal response with a positive Hill coefficient (Table 1; see Fig. S1B in the supplemental material). The K0.5 for malate was more than five times higher than the Km,malate measured for SfcA. Enzyme activation was detected in the presence of 2 mM KCl, a result not observed with SfcA (no modification of the NAD-dependent activity of SfcA was observed) (Fig. 4).
FIG. 4.
Regulatory properties of recombinant SfcA, MaeB, and Sp3. ME activity was measured for each isoform at pH 7.5 in the presence of 2 mM of each effector indicated on the y axes, with the exception of CoA, acetyl-CoA, and palmitoyl-CoA (50 μM). The malate concentration was kept at approximately one-fifth of the Km value for each isoform (0.1 mM for SfcA, 0.6 mM for MaeB, and 1 mM for Sp3). The results are presented as the percentages of activity in the presence of the effectors in relation to the activity measured in the absence of the metabolites. The assays were done at least in triplicate, and the error bars indicate deviations between the measurements. Significant inhibition is shown by dark-gray bars with hatching. Significant activation is shown by light-gray bars with cross-hatching.
Interestingly, MaeB displayed inhibition at high Mg2+ concentrations (more than 4 mM), something not observed in the case of SfcA. This effect was not due to chelation of the substrates (malate or NADP+), as saturating free concentrations of these substrates were used when measuring activities at high Mg2+ concentrations. Moreover, analysis of the secondary structure of recombinant MaeB by the CD spectrum showed essentially identical spectra when the Mg2+ concentration was increased (not shown), indicating that the decrease in activity of MaeB at high cation concentrations was not due to a modification of the overall structure of the enzyme. Thus, it is possible that a high Mg2+ concentration may block substrate binding in the case of MaeB, which must be taken into account when measuring NADP ME activity in E. coli.
Regulatory properties of MaeB and SfcA.
Several compounds were tested as possible effectors of the enzymatic activity in the direction of the oxidative decarboxylation of malate for MaeB and SfcA. The results obtained (Fig. 4), as a whole, indicate that MaeB, the NADP-specific ME composed of two distinct domains (Fig. 2), is the isoform most highly regulated by the metabolites tested, as either activators or inhibitors. Figure 4 shows the results obtained when the highest concentrations of the metabolites tested in each case were used, although lower concentrations gave the same effects to a lesser extent (not shown).
In the case of SfcA, aspartate was the only metabolite found to activate the enzyme, while CoA, acetyl-P, palmitoyl-CoA, and OAA inhibited enzyme activity (Fig. 4). It is worth mentioning that previous work performed with purified NAD ME from E. coli also showed activation by aspartate and inhibition by CoA (24, 30, 37).
Several compounds were found to increase the activity of MaeB: the amino acids glutamate and aspartate, the sugar-phosphate glucose-6P, and acetyl-P (Fig. 4). On the other hand, fumarate, OAA, and acetyl-CoA inhibited enzyme activity (Fig. 4). Inhibition by OAA and acetyl-CoA was also previously reported for the purified E. coli NADP ME (2, 29).
Reverse reaction catalyzed by MaeB and SfcA.
SfcA and MaeB were tested for their abilities to catalyze the pyruvate reductive carboxylation (reverse reaction; SfcA with NAD+ and MaeB with NADP+). Both enzymes showed maximum activity for this reaction at pH 7.0. The results obtained when comparing forward and reverse reactions (malate oxidative decarboxylation and reductive pyruvate carboxylation, respectively) for both enzymes measured at their optimum pH values indicated that SfcA and MaeB behave differently in terms of reversibility, with SfcA more capable of carboxylating and reducing pyruvate than MaeB (Table 1). When the reverse and the forward reactions for each enzyme were compared, SfcA and MaeB catalyzed the carboxylation reaction at rates nearly 30 times and 200 times lower, respectively, than those for decarboxylation (Table 1). The Km for pyruvate was more than two times lower for SfcA than for MaeB (Table 1).
Native structures of MaeB and SfcA.
The native molecular masses of MaeB and SfcA were calculated by size exclusion chromatography. The values obtained for each enzyme (668.0 ± 9.9 kDa for MaeB and 263.5 ± 3.7 kDa for SfcA) indicate that while SfcA assembles as a tetramer (64 kDa per subunit), MaeB shows a higher native molecular mass, which is in accord with an octamer (83 kDa per subunit).
When both purified proteins were analyzed by native electrophoresis (Fig. 3C and D), the estimated native molecular masses were correlated with the data obtained by size exclusion chromatography. Native activity gels were incubated in the presence of appropriate substrates for detection (SfcA with NAD+ and MaeB with NADP+) (Fig. 3C) or Coomassie stained (Fig. 3D). In the latter case, only those bands previously detected through activity gels appeared. This indicates that no oligomers other than the active ones are formed by each enzyme (Fig. 3C and D).
Characterization of truncated polypeptides from the N terminus of MaeB.
Different truncated polypeptides derived from MaeB were constructed, overexpressed in BL21(DE3) cells, and purified. Polypeptides Sp1, Sp2, and Sp3 were designed in order to contain the two specific domains for ME and to exclude the extra domain in MaeB corresponding to PTA (Fig. 2).
The truncated Sp1 (46 kDa plus 17 kDa encoded by the expression vector) could not be obtained in the soluble fraction and was expressed as inclusion bodies (Fig. 3A). On the other hand, Sp2 and Sp3 could be purified to homogeneity from the soluble fraction of induced cells carrying pET-Sp2 or pET-Sp3. The molecular masses calculated by SDS-PAGE for the purified recombinant proteins after elimination of the His tag used for purification were consistent with the predicted molecular mass calculated from their sequences (53 kDa for Sp3 and 43 kDa for Sp2) (Fig. 3A). When analyzed by Western blotting, Sp1, Sp2, and Sp3 reacted with antibodies against MaeB (Fig. 3B). The purified Sp2 and Sp3 proteins were used for further characterization.
No ME activity could be detected for purified Sp2, even when extremely high protein and/or substrate concentrations were used (Fig. 3C). Conversely, Sp3 showed ME activity at pH 7.5 using NADP+ as a cofactor. The kcat estimated for this truncated version of MaeB was 18 s−1, with a Km,NADP value of 58.8 μM. Based on these data, the catalytic efficiency for Sp3 was 0.30, nearly five times lower than the catalytic efficiency calculated for the complete MaeB (Table 1). The estimated Km,malate (5.2 mM) was very close to the value determined for MaeB (Table 1), although a hyperbolic response was obtained in that case.
The potential metabolic regulation of truncated Sp3 by effectors of MaeB activity was determined (Fig. 4). The results obtained indicated that the activation by glutamate, aspartate, and glucose-6P observed for MaeB was lost in Sp3 (Fig. 4). Activation by KCl was also observed for Sp3, as well as inhibition by fumarate and OAA (Fig. 4). Based on structural similarities, we could infer that these two metabolites are inhibitors due to competition with malate. Interestingly, the inhibition due to acetyl-CoA was lost, while the activation due to acetyl-P was maintained (Fig. 4).
The oligomeric states of Sp3 and Sp2 were analyzed by native electrophoresis and subsequently detected in activity gels and by Coomassie staining in the case of Sp3 or by Coomassie staining alone in the case of Sp2 (Fig. 3C and D). In both cases, the truncated versions of MaeB presented molecular masses in agreement with dimeric states.
Characterization of the carboxyl-terminal domain from MaeB with homology to PTA.
Sequence similarity searches using the Basic Local Alignment Search Tool (BLAST) indicate that the carboxyl-terminal 350-amino-acid sequence from MaeB is 31% identical to the protein EutD from E. coli. This protein is involved in ethanolamine utilization and belongs to the phosphate acetyltransferase family. The results also show that this sequence is 29% identical to PTA from E. coli. (GenProtEC [http://genprotec.mbl.edu/]; EcoGene [http://ecogene.org/]). The similarity between PTA and MaeB spanned the C-terminal amino acid sequences for both proteins, while that for EutD comprised the complete amino acid sequence. Figure 5 shows an alignment among the carboxyl-terminal 351 amino acids from MaeB, the complete sequence from E. coli EutD (338 amino acids), and the carboxyl-terminal 357 amino acids from E. coli PTA (the complete PTA is composed of 713 amino acids). Interestingly, although EutD from E. coli was not characterized, EutD from Salmonella enterica exhibited PTA activity levels similar to those of other PTA enzymes (1). Therefore, it seems probable that the carboxyl-terminal domains of MaeB and PTA correspond to the catalytic domains of PTA enzymes.
FIG. 5.
Comparison of the amino acid sequences from E. coli MaeB, PTA, and EutD. The sequences for E. coli MaeB, PTA, and EutD were aligned using CLUSTAL W (version 1.82). The similarity among these three proteins extended over the approximately 350 carboxyl-terminal amino acids from MaeB and PTA and the complete sequence from EutD. Conserved regions among the three proteins are marked with boxes.
In order to characterize the domain of MaeB with homology to EutD and PTA, the 351 amino acids corresponding to the carboxyl-terminal part of MaeB were cloned in frame into the expression vector pET-32. The His-tagged polypeptide obtained was subsequently purified. This truncated MaeB protein (called MaeB-PTA) (Fig. 2) was overexpressed in BL21(DE3) cells and purified to homogeneity, followed by the removal of the His tag with thrombin (Fig. 3A). The molecular mass of the polypeptide (41 kDa) was in accord with the predicted molecular mass based on the amino acid sequence. MaeB-PTA showed cross-reaction with antibodies against MaeB (Fig. 3B).
Despite the high level of homology between MaeB-PTA and EutD (Fig. 5), the recombinant purified MaeB-PTA did not display PTA activity, although alternative activity measurement protocols were used for the forward and reverse directions. It is worth mentioning that PTA activity measurements were also performed on crude bacterial extracts, using nontransformed E. coli BL21(DE3) cells and BL21(DE3) cells transformed with MaeB or MaeB-PTA. The PTA activity levels were very similar in the three cases. These additional PTA activity measurements were performed because previous reports indicated loss of enzymatic activity during purification (20).
When the oligomeric state of the purified MaeB-PTA was analyzed by size exclusion chromatography, the molecular mass obtained (321.2 ± 2.9 kDa) indicated that the protein was able to assemble as an octamer (a monomer of 41 kDa). This molecular mass correlates with the band detected by Coomassie staining after native electrophoresis (Fig. 3D).
DISCUSSION
The two E. coli recombinant MEs exhibit different kinetic and structural properties and are distinct from previously characterized purified MEs.
The results obtained in the present work indicate that E. coli possesses a NAD(P) ME (SfcA) and a chimerical ME-PTA NADP ME (MaeB). SfcA prefers NAD+ over NADP+, in contrast to MaeB, which is active only with NADP+ (Table 1). The fingerprint regions of NAD(P)-binding sites are characterized by a Gly-rich sequence (GXGXXG), which is the phosphate binding consensus. Two Ala residues (italic) separated by 3 amino acids (GXGXXAXXXA) were suggested as more characteristic of NADP-binding domains, while Gly residues were preferred in NAD-dependent enzymes (32, 40). In agreement with this, the predicted signature motifs for NAD(P)-binding sites identified in SfcA (310-GAGSAGCGI-320) and MaeB (193-GAGAAAIA-202) follow this rule, although the second Ala residue in MaeB is moved two positions in relation to the proposed conserved region for NADP-binding domains, similar to other NADP MEs (5). Multiple-sequence alignment helped us to determine that these signature motifs correspond to regions with Rossman βαβ folds in crystallized eukaryotic MEs (41, 43). Similarly to E. coli, S. meliloti possesses one ME specific for NADP+ and a second ME with a preference for NAD+ over NADP+ (38), although both S. meliloti MEs possess chimerical structures similar to that of MaeB.
The molecular masses for the recombinant SfcA and MaeB determined in the present work (Fig. 3) are in correlation with the values predicted from their sequences. Previous work with purified E. coli NAD and NADP MEs has indicated lower molecular masses (2, 13, 35) and different amino acid compositions (13, 14, 42) than those calculated based on SfcA and MaeB sequences. Thus, it seems that purified E. coli NAD and NADP MEs were proteolized during the long purification procedure, which may explain the differences in structures and kinetic parameters between purified and recombinant proteins. On the other hand, copurification of both MEs and/or copurification with other proteins could account for some differences in metabolic regulation between purified and recombinant MEs.
SfcA and MaeB have distinct metabolic regulation and reversibility.
When the two E. coli MEs were compared, MaeB was the isoform most subject to metabolic regulation (Fig. 4). The enzyme showed inhibition by acetyl-CoA, OAA, and fumarate and activation by amino acids (glutamate and aspartate), sugar phosphates (glucose-6P), and acetyl-P, while SfcA was activated only by aspartate (Fig. 4). Interestingly, while acetyl-P is an inhibitor of SfcA but an activator of MaeB, acetyl-CoA is an inhibitor of MaeB and had no effect on SfcA. Moreover, CoA had no effect on MaeB but is an inhibitor of SfcA (Fig. 4). Therefore, the results indicate that when the acetyl-CoA concentration increases and thus the CoA concentration decreases, SfcA is active, while the increase in acetyl-P is accompanied by an increase in MaeB activity. Altogether, the results suggest that the regulation of MaeB and SfcA activities seems to be correlated with the activation of the reverse or forward PTA reaction that is responsible for the interconversion of acetyl-CoA and acetyl-P (Fig. 1B).
The activation of MaeB by amino acids and sugar-phosphate indicates that the enzyme is most active in the presence of high-energy bonds (acetyl-P is a high-energy phosphate compound) and/or high concentrations of carbon-containing molecules. The positive regulation by amino acids and sugar phosphates is in accord with its participation in gluconeogenesis. Previous work has shown that both MaeB and SfcA are induced in E. coli during acetate metabolism (27). Deletion mutants in the two possible pathways to provide PEP for gluconeogenesis (PckA or ME-PpsA) (Fig. 1B) have shown that both pathways can alternatively serve to deliver carbon from TCA to gluconeogenesis (27). Further deletions in either one or both MEs have shown that the activity of either SfcA or MaeB can sufficiently catalyze the malate-to-pyruvate conversion (15, 27). Nevertheless, although the disruption in MEs has no phenotype in balanced growth during glucose-to-acetate transition, due to the presence of PckA, the sfcaA maeB double mutant showed a severe lag phase, which could not be rescued by PckA overexpression, suggesting that MEs play roles other than carbon flux to PEP (15) (Fig. 1B).
In the present work, allosteric regulation of both E. coli MEs by acetate derivatives was found. The most intriguing case is acetyl-P, which reciprocally regulates both isoforms (Fig. 4). Acetyl-P has been suggested to be a global signal in E. coli and related bacteria that conveys information to various two-component regulatory systems (9, 22, 39). Its levels go up and down in response to the C source and the nutritive state of the cell, reaching very high concentrations (even more than 1 mM) under certain conditions (23). Although it is clear that under gluconeogenic conditions each ME alone can fulfill the pyruvate requirements (15, 27) (Fig. 1B), the regulatory differences between MaeB and SfcA shown in the present work indicate a preference for one ME isoform over the other under certain physiological conditions (e.g., in the presence of high concentrations of acetyl-P, MaeB activity is clearly favored over SfcA). It remains to be proven if acetyl-P, along with the differential regulation of SfcA and MaeB activities, also mediates a signal for selective induction/repression of E. coli MEs.
With regard to the type of interaction of acetyl-CoA and acetyl-P with MaeB, we thought that, as these compounds are substrates or products of the PTA reaction, they probably interact with the carboxyl-terminal domain of MaeB (Fig. 2). Nevertheless, the truncated version of MaeB retaining ME activity (Sp3) is not inhibited by acetyl-CoA but is still activated by acetyl-P (Fig. 4). Thus, the interaction of acetyl-P with MaeB is not due to interaction with the domain homologous to PTA (Fig. 2).
Finally, MaeB and SfcA presented differences in reversibility, with SfcA more able to catalyze the carboxylation of pyruvate than MaeB (Table 1). Therefore, SfcA is more useful than MaeB as a tool in the production of succinic acid through metabolic-engineering techniques (12, 36). Higher reverse-reaction rates employing genetically engineered SfcA would create an even better tool for this purpose. With this aim, it will also be interesting to compare SfcA with the NADP ME from the archaebacterium Thermococcus kodakarensis, which showed no preference for either the direct or forward direction and presented a very high Km for malate (10).
The PTA domain in MaeB is not active but seems to be involved in allosteric regulation.
In the present work, we found that, although MaeB seems to contain the catalytic domain of PTA, the domain is not able to catalyze the PTA reaction (interconversion of acetyl-CoA and acetyl-P) at all. Deletion of this PTA domain in MaeB does not abolish ME activity, as Sp3 was able to catalyze the oxidative decarboxylation of malate at rates comparable to those with MaeB. In the chimerical MEs from S. meliloti, it was also verified that the PTA domain is not essential for the ME reaction (25, 26). On the other hand, Sp2 does not retain ME activity, probably because some essential residue(s) is lost in this truncated protein.
We wondered about the significance of the PTA domain in MaeB, taking into account that its occurrence in different MEs from several gram-negative bacteria suggests its functional relevance in these organisms. In the present work, we found that this domain is involved in the regulation of MaeB by several metabolites: inhibition by acetyl-CoA and activation by glutamate, aspartate, and glucose-6P. The results obtained suggest that these effectors interact allosterically with the PTA domain of MaeB (Fig. 2), as Sp3 maintains ME activity with only five-times-lower catalytic efficiency and no regulation by these compounds. The control of MaeB may also be due to oligomerization, as the C-terminal PTA domain in MaeB seems to drive the formation of MaeB octamers (Fig. 3). Thus, the association of the two domains in MaeB may facilitate the metabolic control of this intriguing enzyme.
Finally, the high sequence similarity between the C-terminal domains of MaeB and PTA, which is also very similar to EutD (Fig. 5), indicated to us that the PTA-like domain in MaeB might be functional. However, we found that it was not. Further experiments are needed to determine which critical amino acid residue(s) is responsible for the interconversion between acetyl-CoA and acetyl-P and to verify if it is possible to restore the PTA activity in MaeB.
Supplementary Material
Acknowledgments
This work was funded by grants from CONICET and Agencia Nacional de Promoción Científica y Tecnológica. M.F.D. and C.S.A. are members of the Researcher Career of CONICET, and F.P.B. is a fellow of the same institution.
Footnotes
Published ahead of print on 8 June 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, D. A., and R. A. Cook. 1981. Role of metal cofactors in enzyme regulation. Differences in the regulatory properties of the Escherichia coli nicotinamide adenine dinucleotide phosphate specific malic enzyme depending on whether Mg2+ or Mn2+ serves as divalent cation. Biochemistry 20:2503-2512. [DOI] [PubMed] [Google Scholar]
- 3.Chang, G.-G., and L. Tong. 2003. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 42:12721-12733. [DOI] [PubMed] [Google Scholar]
- 4.Davis, B. J. 1964. Disc electrophoresis. II. Method and application to human serum proteins. Ann. N. Y. Acad. Sci. 121:404-427. [DOI] [PubMed] [Google Scholar]
- 5.Detarsio, E., M. C. Gerrard Wheeler, V. A. Campos Bermúdez, C. S. Andreo, and M. F. Drincovich. 2003. Maize C4 NADP-malic enzyme: expression in E. coli and characterization of site-directed mutants at the putative nucleotide binding sites. J. Biol. Chem. 278:13757-13764. [DOI] [PubMed] [Google Scholar]
- 6.Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K.-I. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 149:2331-2343. [DOI] [PubMed] [Google Scholar]
- 7.Drincovich, M. F., P. Casati, and C. S. Andreo. 2001. NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll, B. T., and T. M. Finan. 1997. Properties of NAD and NADP-dependent malic enzymes of Rhizobium (Sinohirzobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 143:489-498. [DOI] [PubMed] [Google Scholar]
- 9.Fredericks, C. E., S. Shibata, S. Aizawa, S. A. Reimann, and A. J. Wolfe. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734-747. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, W., Y. S. Ismail, T. Fukui, H. Atomi, and T. Imanaka. 2005. Characterization of an archaeal malic enzyme from the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Archaea 1:293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gourdon, P., M.-F. Baucher, N. D. Lindley, and A. Guyonvarch. 2000. Cloning the malic enzyme from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66:2981-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, S. H., and S. Y. Lee. 2001. Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol. Bioeng. 74:89-95. [DOI] [PubMed] [Google Scholar]
- 13.Iwakura, M., J. Hattori, Y. Arita, M. Tokushige, and H. Katsuki. 1979. Studies on regulatory functions of malic enzymes. VI. Purification and molecular properties of NADP-linked malic enzyme from Escherichia coli W. J. Biochem. 85:1355-1365. [PubMed] [Google Scholar]
- 14.Iwakura, M., M. Tokushige, and H. Katsuki. 1979. Studies on regulatory functions of malic enzymes: structural and functional characteristics of sulfhydryl groups in NADP-linked malic enzyme from Escherichia coli W. J. Biochem. 86:1239-1249. [DOI] [PubMed] [Google Scholar]
- 15.Kao, K. C., L. M. Tran, and J. C. Liao. 2005. A global regulatory role of gluconeogenic genes in Escherichia coli revealed by transcriptome network analysis. J. Biol. Chem. 280:36079-36087. [DOI] [PubMed] [Google Scholar]
- 16.Kawai, S., H. Suzuki, K. Yanamoto, M. Inui, H. Yukawa, and H. Kumagai. 1996. Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing its gene. Appl. Environ. Microbiol. 62:2692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, K., S. Doi, S. Negoro, I. Urabe, and H. Okada. 1989. Structure and properties of malic enzyme from Bacillus stearothermophilus. J. Biol. Chem. 264:3200-3205. [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lerondel, G., T. Doan, N. Zamboni, U. Sauer, and S. Aymerich. 2006. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J. Bacteriol. 188:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundie, L. L., and J. G. Ferry. 1989. Activation of acetate by Methanosarcina thermofila. Purification and characterization of phosphotransacetylase. J. Biol. Chem. 264:18392-18396. [PubMed] [Google Scholar]
- 21.Marchler-Bauer, A., J. B. Anderson, P. F. Cherokuki, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 24.Milne, J. A., and R. A. Cook. 1979. Role of metal cofactors in enzyme regulation. Differences in the regulatory properties of the Escherichia coli nicotinamide adenine dinucleotide specific malic enzyme depending on whether Mg2+ or Mn2+ serves as divalent cation. Biochemistry 18:3604-3610. [DOI] [PubMed] [Google Scholar]
- 25.Mitsch, M. J., R. T. Voegele, A. Cowie, M. Osteras, and T. M. Finan. 1998. Chimeric structure of the NAD(P)- and NADP-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti. J. Biol. Chem. 273:9330-9336. [DOI] [PubMed] [Google Scholar]
- 26.Mitsch, M. J., A. Cowie, and T. M. Finan. 2007. Malic enzyme cofactor and domain requirements for symbiotic N2 fixation by Sinorhizobium meliloti. J. Bacteriol. 189:160-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh, M.-K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 28.Saigo, M., F. Bologna, V. G. Maurino, E. Detarsio, C. S. Andreo, and M. F. Drincovich. 2004. Maize recombinant non-C4 NADP-malic enzyme: a novel dimeric enzyme with high specific activity. Plant Mol. Biol. 55:97-107. [DOI] [PubMed] [Google Scholar]
- 29.Sanwal, B. D., and R. Smando. 1969. Malic enzyme of Escherichia coli. Diversity of the effectors controlling enzyme activity. J. Biol. Chem. 244:1817-1823. [PubMed] [Google Scholar]
- 30.Sanwal, B. D. 1970. Regulatory characteristics of the diphosphopyridine nucleotide-specific malic enzyme of Escherichia coli. J. Biol. Chem. 245:1212-1216. [PubMed] [Google Scholar]
- 31.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 32.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 33.Sedmak, J. J., and S. E. Grossberg. 1977. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem. 79:544-552. [DOI] [PubMed] [Google Scholar]
- 34.Sender, P. D., M. G. Martín, S. Peirú, and C. Magni. 2004. Characterization of an oxaloacetate decarboxylase that belongs to the malic enzyme family. FEBS Lett. 570:217-222. [DOI] [PubMed] [Google Scholar]
- 35.Spina, J., H. J. Bright, and J. Rosenbloom. 1970. Purification and properties of l-malic enzyme from Escherichia coli. Biochemistry 9:3794-3801. [Google Scholar]
- 36.Stols, L., and M. I. Donnelly. 1997. Production of succinic acid through overexpression of NAD-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeo, K., T. Murai, J. Nagai, and H. Katsuki. 1967. Allosteric activation of DPN-linked malic enzyme from Escherichia coli by aspartate. Biochem. Biophys. Res. Commun. 29:717-722. [DOI] [PubMed] [Google Scholar]
- 38.Voegele, R. T., M. J. Mitsch, and T. M. Finan. 1999. Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1432:275-285. [DOI] [PubMed] [Google Scholar]
- 39.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 40.Wierenga, R. K., M. C. de Maever, and W. G. J. Hol. 1985. Interaction of pyrophosphate moieties with alpha-helixes in dinucleotide-binding proteins. Biochemistry 24:1346-1357. [Google Scholar]
- 41.Xu, Y., G. Bhargava, H. Wu, G. Loeber, and L. Tong. 1999. Crystal structure of human mitochondrial NAD(P)-dependent malic enzyme: a new class of oxidative decarboxylases. Structure 7:877-889. [PubMed] [Google Scholar]
- 42.Yamaguchi, M., M. Tokushige, and H. Katsuki. 1973. Studies on regulatory functions of malic enzymes. Purification and molecular properties of nicotinamide adenine dinucleotide-linked malic enzyme from Escherichia coli. J. Biochem. 73:169-180. [PubMed] [Google Scholar]
- 43.Yang, Z., H. Zhang, H.-H. Hung, C.-C. Kuo, L.-C. Tsai, H. S. Yuan, W.-Y. Chou, G.-G. Chang, and L. Tong. 2002. Structural studies of the pigeon liver cytosolic NADP-dependent malic enzyme. Protein Sci. 11:332-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.