Abstract
Bifidobacteria constitute up to 3% of the total microbiota and represent one of the most important health-promoting bacterial groups of the human intestinal microflora. The presence of Bifidobacterium in the human gastrointestinal tract has been directly related to several health-promoting activities; however, to date, no information about the specific mechanisms of interaction with the host is available. In order to provide some insight into the molecular mechanisms involved in the interaction with the host, we investigated whether Bifidobacterium was able to capture human plasminogen on the cell surface. By using flow cytometry, we demonstrated a dose-dependent human plasminogen-binding activity for four strains belonging to three bifidobacterial species: Bifidobacterium lactis, B. bifidum, and B. longum. The binding of human plasminogen to Bifidobacterium was dependent on lysine residues of surface protein receptors. By using a proteomic approach, we identified five putative plasminogen-binding proteins in the cell wall fraction of the model strain B. lactis BI07. The data suggest that plasminogen binding to B. lactis is due to the concerted action of a number of proteins located on the bacterial cell surface, some of which are highly conserved cytoplasmic proteins which have other essential cellular functions. Our findings represent a step forward in understanding the mechanisms involved in the Bifidobacterium-host interaction.
A large population of microorganisms inhabits the human gastrointestinal tract (GIT) and forms a closely integrated unit with the host called the intestinal microbiota. The quantity of living bacteria which compose the human microbiota can range from 1012 to 1014 CFU/g of luminal contents, and the assemblage contains up to 1,000 different species (1, 12, 48). During evolution the association of microbes with tissues of the human GIT resulted in the development of a balanced symbiotic relationship, where the microorganisms profit by the acquisition of nutrients and a stable temperature and in turn provide important health benefits to the host (7). In fact, the intestinal microbiota influences several biochemical, immunological, and physiological features of the human host, resulting in an increase in the host metabolic/digestive capacity and the exclusion of harmful microbes (5, 42, 43).
Up to 3% of the total human intestinal microflora is composed of bacteria belonging to genus Bifidobacterium, which represents one of the most important health-promoting groups of the human microbiota (15, 34, 36, 45). Members of the genus Bifidobacterium are gram-positive, nonmotile, non-spore-forming, anaerobic rods with variable appearance. The presence of Bifidobacterium in the human GIT has been directly related to several health-promoting activities, including maintenance of the normal microflora, immunostimulation and immunomodulation, improvement of lactose utilization, and reduction of serum cholesterol levels (12, 30, 43). Due to their beneficial effects, some Bifidobacterium species have become common components of many dairy and pharmaceutical products. However, our knowledge of the mechanisms involved in the bifidobacterial health-promoting activities is very limited, and in particular, no information about the specific mechanisms of the interaction of bifidobacteria with the host is available.
Several pathogenic bacterial species intervene with the plasminogen (Plg)-plasmin system of the human host (16, 17, 35, 40, 41). In particular, in the human gastrointestinal niche, enteropathogens such as Salmonella enterica, Listeria monocytogenes, Helicobacter pylori, and Escherichia coli, as well as Bacteroides fragilis, an opportunistic pathogen and common member of the normal human gut flora, have been shown to capture human Plg on the cell surface (16, 27, 33, 38, 40). Plg is a single-chain glycoprotein with a molecular mass of 92 kDa and represents the monomeric proenzyme of the serine protease plasmin. Plg comprises an N-terminal, 67- or 77-residue preactivation peptide (∼8 kDa), five consecutive disulfide-bound kringle domains (K1 to K5; 65 kDa), and a serine protease domain (25 kDa). It is abundant in human plasma and extracellular fluids (47), and its active form, plasmin, plays a crucial role in fibrinolysis (6), homeostasis (29), and degradation of the extracellular matrix proteins (44). Because of these characteristics and the broad proteolytic activity of plasmin, the mammalian Plg system offers a high-potential proteolytic system to bacteria for colonization of the human host. In fact, with the recruitment of human Plg on the bacterial cell surface and its subsequent conversion to plasmin, microorganisms acquire a surface-associated proteolytic activity useful for facilitating the migration across physical and molecular barriers and for responding to the nutritional demands during the colonization process (4, 8, 16, 17, 25, 27, 33, 38, 39).
Since pathogens and symbionts of the human GIT colonize the same ecological niche, the hypothesis that they share common molecular mechanisms to initiate and maintain their relationships with the host has been addressed (1, 19, 21-23, 32). According to Ochman and Moran (22), for pathogens and symbionts the biological process for host colonization is largely the same regardless of whether the final outcome of the interaction is harmful, benign, or beneficial to the host. In light of this statement, we investigated whether Bifidobacterium possesses human Plg-binding activity like that reported for some enteropathogens and an opportunistic pathogen of the human gastrointestinal microflora.
The experimental data that we report here provide evidence of a dose-dependent human Plg-binding activity in strains belonging to three bifidobacterial species. Furthermore, in the cell wall fraction of Bifidobacterium lactis BI07 five putative Plg-binding proteins have been identified. Our findings provide insight for understanding the mechanisms involved in the Bifidobacterium-host interaction.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
For this study, four bifidobacterial strains were selected. Bifidobacterium bifidum S16 and Bifidobacterium longum S123 were isolated from human feces, whereas B. lactis DSM10140 (20) and B. lactis BI07 were isolated from dairy products. Bifidobacteria were cultured in MRS medium (Difco) supplemented with 0.05% (wt/vol) l-cysteine at 37°C in anaerobic conditions. Anaerobic conditions were obtained by using Anaerocult A (Merck) in a jar. The bifidobacterial cells were grown for 18 h until they reached the stationary phase.
Flow cytometric analysis of Plg binding in Bifidobacterium.
Bifidobacterial cells (B. lactis BI07, B. lactis DSM10140, B. longum S123, and B. bifidum S16) were grown as described above for 16 to 18 h until they reached the stationary phase. Bacteria were washed in phosphate-buffered saline (PBS), and 5 × 107 bacteria were incubated with different amounts of human Plg (0, 0.5, 1.0, 2.5, 5.0, and 10.0 μg) in 100 μl (final volume) for 30 min at 37°C (5% CO2). Bacteria were washed with PBS containing 0.5% fetal calf serum and incubated with anti-Plg antibodies (Affinity Biologicals, Ontario, Canada) diluted 1:600 in PBS for 20 min at 25°C with constant agitation. Bifidobacterium-bound Plg was detected by incubation with a 1:200 dilution of anti-goat antibodies conjugated with fluorescein isothiocyanate (FITC) (Sigma) in PBS in the dark for 20 min at 25°C with constant agitation. Finally the bacteria were washed with PBS containing 0.5% fetal calf serum and fixed with 3% paraformaldehyde in the dark for 1 h at 4°C. Flow cytometric measurements were obtained by using a FACSCalibur flow cytometer (BD Biosciences). The bacteria were detected using log forward and log side scatter dot plots, and a gating region was set to exclude debris and larger aggregates of bacteria. Ten thousand bacteria were analyzed for fluorescence using log-scale amplification. The geometric mean fluorescence intensity (GMFI) multiplied by the percentage of labeled bacteria was recorded as a measure of binding activity. Data analysis was done with the WinMDI 2.8 software.
For inhibition studies 5 × 107 B. lactis BI07 and B. longum S123 bacteria were incubated with 2.5 μg of Plg in the presence of 1, 10, or 50 μg ɛ-aminocaproic acid (EACA), followed by the protocol described above. In order to study the effect of carboxypeptidase B treatment, 5 × 107 bacteria were incubated with 0, 5, and 25 U carboxypeptidase B (from porcine pancreas; C9584-5MG; Sigma) in PBS buffer for 1 h at 37°C. After washing with PBS buffer, bacteria were incubated with Plg as reported above.
Transmission electron microscopy.
In order to visualize the binding of Plg to B. lactis BI07, we performed preembedding immunogold-labeling experiments using whole bifidobacterial cells.
For immunolocalization of Plg bound to the bacterial surface, 1 × 109 bifidobacterial cells recovered from a stationary-growth-phase culture were incubated with 20 μg of human Plg for 1 h at 25°C, washed several times with PBS, and incubated with 100 μl of goat anti-Plg immunoglobulin G (IgG) antibody (Kordia) diluted 1:500 in PBS-1% bovine serum albumin (BSA) for 1 h at 25°C with constant agitation. After two washes in PBS-1% BSA, bacteria were incubated with a rabbit anti-goat antibody (DakoCytomation) diluted 1:100 in PBS-1% BSA for 45 min at 25°C. After two washes, bacteria were resuspended in 25 μl of anti-rabbit IgG coupled to 10-nm gold particles (Auro Probe; GE Healthcare) diluted 1:5 in PBS-1% BSA and incubated for 30 min at 25°C with constant agitation. Bacteria were then collected and washed two times in PBS-1% BSA, and the sediment was fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 4 h at 4°C.
For transmission electron microscopy processing, the glutaraldehyde-fixed bacteria were washed with 0.15 M cacodylate buffer and postfixed with 1% OsO4 for 1 h at 4°C. Bacteria were then washed, dehydrated in a graded alcohol series, and embedded in araldite (Fluka, Sigma-Aldrich). Ultrathin sections were obtained with a Reichert OMu3 ultramicrotome, counterstained with uranyl acetate and lead citrate, and examined with a Philips 400T transmission electron microscope.
For each sample, four ultrathin sections were examined (two sections per grid), and in each section eight fields were randomly photographed. Gold-labeled cells were counted on photographs.
Fractionation of bifidobacterial cell wall proteins.
The bifidobacterial cell wall proteins were extracted from 50 ml of a bacterial culture in stationary growth phase as reported by Hardie and Williams (13). Bifidobacterial cells were collected by centrifugation for 10 min at 5,000 rpm at 4°C and then washed in 50 mM Tris-HCl (pH 7.6). The bacteria were resuspended in 2 ml of protoplast buffer (50 mM Tris-HCl [pH 7.6], 1 M sucrose, 1.4 mM phenylmethylsulfonyl fluoride, 15 mg/ml lysozyme). The suspension obtained was incubated for 90 min at 37°C. Subsequently, the cell suspension was centrifuged for 3 min at 4,000 rpm at 4°C, and the supernatant, containing the cell wall proteins, was collected and stored at −20°C.
Resolution of Bifidobacterium cell wall proteins by 2DE.
The cell wall fractions were solubilized in IEF solution containing 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and 0.005% (vol/vol) 2-mercaptoethanol for two-dimensional polyacrylamide gel electrophoresis (2DE) analysis. The total protein concentration was calculated by using a PlusOne 2D Quant kit (GE Healthcare).
Isoelectric focusing was carried out using Immobiline DryStrips with a linear pH gradient from 4 and 7 (in 7 cm) on an IPGphor system (GE Healthcare). Forty micrograms of proteins was rehydrated for 12 h in 125 μl of buffer A (8 M urea, 2% [wt/vol] CHAPS, 2% [vol/vol] Ampholine pH 4.0 to 6.5 [GE Healthcare], 10 mM dithiothreitol, 0.8% bromophenol blue) and focused for a total of 10 kV·h. IPG strips were then reduced and alkylated (11) prior to loading onto 12% acrylamide separating gels (length, 8 cm; thickness, 1 mm), and electrophoresis was performed using an SE 250 mini vertical electrophoresis unit (GE Healthcare). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 160 V for 2.5 h. The spots were visualized by staining with Coomassie blue R350 (PhastGel Blue R; GE Healthcare).
Plg overlay assay.
B. lactis BI07 cell wall proteins (40 μg) were subjected to 2DE as reported above and blotted onto a nitrocellulose membrane (Pure nitrocellulose membrane; Bio-Rad) by using a trans-Blot electrophoretic cell (Bio-Rad). After transfer, the membrane was blocked in 10% skim milk, dissolved in 10% PBS, and then incubated with 4 μg/ml human Plg (Sigma-Aldrich) in PBS for 1 h at 25°C. After several washes in PBS, the captured Plg was detected by incubating the membrane with goat anti-Plg IgG antibody (Kordia). The membrane was washed three times in TBS-T (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 0.05% Tween 20) and incubated with peroxidase-conjugated anti-goat IgG (Sigma-Aldrich). After three washes in TBS-T, the membrane was incubated with ECL Plus (GE Healthcare) and the chemiluminescence signal was detected by using a PhosphorImager Storm system (GE Healthcare). To determine the role of the lysine-binding site(s) (LBS) in Plg binding, the experiment was repeated in the presence of EACA. The first and second antibodies alone did not result in nonspecific background binding.
Protein identification using MALDI-TOF MS.
The selected protein spots were excised from the acrylamide gel and subjected to in-gel tryptic digestion and extraction of peptides (37). The extracted peptides were purified with ZipTip (Millipore). Peptide mass fingerprinting maps of tryptic peptides were generated by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with a Voyager-DE Pro Biospectrometry work station (Applied Biosystems). All spectra were obtained in reflectron mode with an accelerating voltage of 20 kV and 40-ns delayed extraction. Internal calibration with peptides arising from trypsin autoproteolysis was performed. Aldente (http://www.expasy.org/tools/aldente) and ProFound (http://129.85.19.192/index.html) database search algorithms were used for identification of the proteins. Proteins with a minimum of four matching peptides were considered positive.
RESULTS
Flow cytometric analysis of Plg binding to Bifidobacterium.
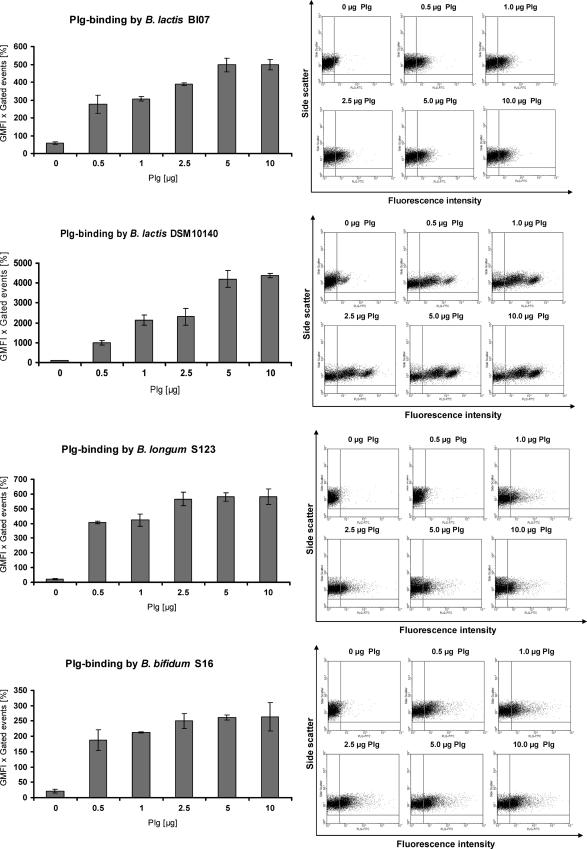
The ability of whole Bifidobacterium cells to bind human Plg was assessed using four bifidobacterial strains: B. lactis BI07, B. lactis DSM10140, B. longum S123, and B. bifidum S16. Bacteria grown to stationary phase were incubated with different amounts of human Plg, and bound Plg was detected using an anti-Plg antibody followed by a secondary FITC-labeled antibody. The increase in fluorescence intensity due to the captured Plg was evaluated by flow cytometric analysis (Fig. 1). The product of the GMFI and the percentage of gated events was significantly enhanced when bifidobacterial cells were incubated with a higher dose of human Plg, indicating that all bifidobacterial strains tested possessed a dose-dependent Plg-binding activity. B. lactis BI07, B. longum S123, and B. bifidum S16 showed comparable Plg-binding activities, while B. lactis DSM10140 was about 10 times more efficient in Plg binding. In control experiments, the anti-Plg antibody and secondary antibody showed no significant binding to the bifidobacterial cells tested.
FIG. 1.
Detection of Plg recruitment by bifidobacterial cells after incubation with 0, 0.5, 1, 2.5, 5, or 10 μg of human Plg by flow cytometry. Plg was detected with polyclonal Plg IgG, followed by an FITC-conjugated second antibody. Countable bifidobacteria were gated using forward and side scatter dot plots, and the shift in fluorescence intensity was evaluated. Binding data are expressed as the GMFI multiplied by the percentage of gated events. The shift in fluorescence is visualized in the dot plots.
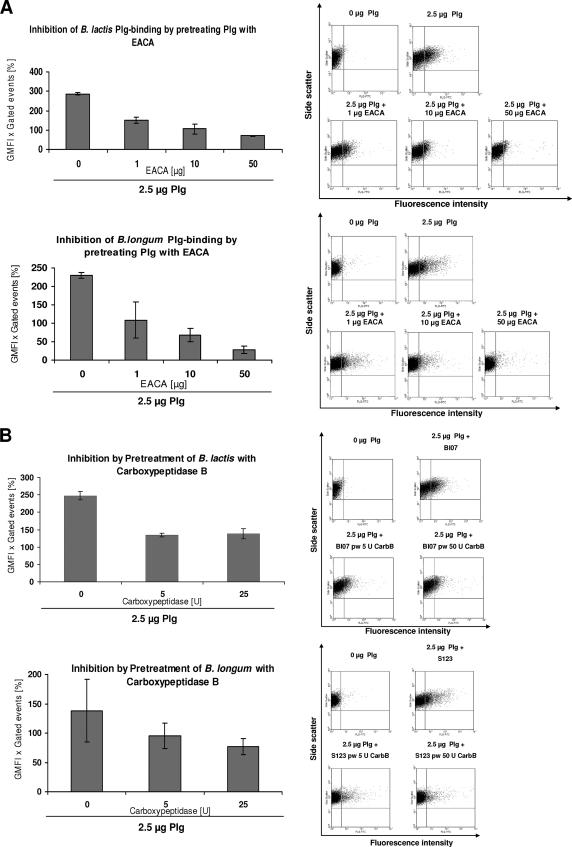
It has been reported that binding of Plg to mammalian species and some bacterial species is mediated by LBS within the kringle domains of Plg which have affinity for the amino acid lysine (40). To investigate the role of LBS for Plg recruitment on the bifidobacterial cell surface, we evaluated the binding of 2.5 μg of human Plg to B. lactis BI07 and B. longum S123 in the presence of 1, 10, and 50 μg of the lysine analog EACA (Fig. 2A). Our results showed that EACA inhibited the Plg binding to the surface of Bifidobacterium in a concentration-dependent manner, with complete inhibition reached when 50 μg of EACA was employed. These data demonstrate that the LBS of the Plg kringle domains are involved in Plg recruitment on the bifidobacterial cell surface.
FIG. 2.
Flow cytometry analysis of inhibition of Plg binding to B. lactis BI07 and B. longum S123 by EACA and after pretreatment with carboxypeptidase B. (A) Bifidobacteria were incubated with 1, 10, and 50 μg EACA and 2.5 μg of Plg. Bacterial surface-bound Plg was detected with FITC-labeled antibody. The results indicate that there was dose-dependent inhibition of Plg binding by EACA. (B) Plg binding to bifidobacteria after treatment of the bacteria with 5 and 25 U carboxypeptidase B. The shift in fluorescence intensity is shown in plots of representative results.
Many of the Plg receptor proteins identified have a carboxy-terminal lysine residue that is involved in binding of Plg (8, 17). To evaluate the impact of the C-terminal lysine(s) in Plg binding to Bifidobacterium, B. lactis BI07 and B. longum S123 cells were treated with increasing concentrations of carboxypeptidase B (0, 5, and 25 U), which is a C-terminal lysine-specific endopeptidase. In flow cytometry analysis, both strains showed a significant reduction in Plg binding after pretreatment with higher doses of carboxypeptidase B. The plateau of Plg binding was reached with 5 U of carboxypeptidase B, a value at which the bifidobacterial Plg-binding activity was reduced to approximately 50% (Fig. 2B). However, treatment with carboxypeptidase B did not completely abolish Plg binding to bifidobacteria. These data suggest that Plg recruitment to the surface of bifidobacteria partially depends on C-terminal lysine residues present in Plg receptors.
Localization of human Plg on the B. lactis BI07 cell surface and detection of putative Plg-binding proteins.
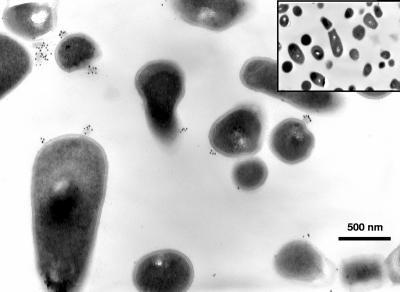
For the visualization of human Plg on the bifidobacterial cell surface, immunoelectron microscopy experiments were carried out with B. lactis BI07 cells. Bacteria grown to stationary phase were incubated with human Plg, washed, and then incubated in preembedding conditions with anti-human Plg antibody, followed by incubation with the secondary antibody labeled with 10-nm gold particles. Ultrathin sections were examined at a magnification of ×13,000 (Fig. 3). Plg aggregates, similar to those reported by Bergmann et al. (4) for Streptococcus pneumoniae, were clearly visible on the B. lactis BI07 cell surface as aggregates of gold particles. In control experiments, bifidobacterial cells incubated with anti-human Plg antibody resulted in no labeling, demonstrating the specificity of the Plg labeling shown in Fig. 3.
FIG. 3.
Visualization of human Plg on the cell surface of B. lactis BI07 by immunoelectron microscopy. Plg was detected on the bacterial cell surface by using anti-Plg antibody and a colloidal gold-coated secondary antibody in preembedding labeling experiments. Plg aggregates associated with the cell surface were visualized as black dots in ultrathin sections of bacterial cells at a magnification of ×13,000. Incubation of B. lactis BI07 cells with anti-Plg antibody resulted in no labeling (inset). In each experimental condition, for eight fields examined per sample, 70% of B. lactis BI07 cells were labeled with gold particles, whereas none of the negative control cells was labeled.
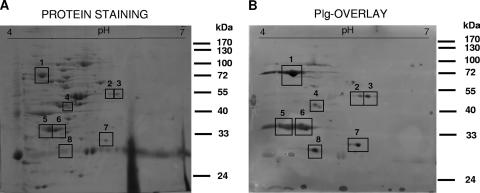
In order to detect possible cell surface protein candidates for the interaction with human Plg, we focused on B. lactis BI07 as a model strain. The cell wall fraction from B. lactis BI07 in stationary phase was purified, and a Plg overlay assay was carried out. The cell wall proteins were resolved in a two-dimensional gel, immobilized on a nitrocellulose membrane, incubated with human Plg, and probed with anti-Plg antibody to identify bound Plg. Binding of Plg to eight B. lactis BI07 cell wall proteins was observed (Fig. 4B), and the coordinates of the major putative Plg-binding proteins detected could be matched to a protein spot on the replica gel stained for proteins (Fig. 4A). Plg binding was completely inhibited in the presence of EACA (data not shown). These data confirm our results obtained by flow cytometry and demonstrate that the interaction between the putative Plg-binding proteins and human Plg is mediated by LBS of Plg.
FIG. 4.
Plg overlay assay carried out with the cell wall fraction of B. lactis BI07 resolved in a two-dimensional gel stained for protein (A) and in a replicate two-dimensional gel used for Plg overlay (B). The squares indicate the major Plg-binding proteins; 4 μg/ml of Plg was applied in the Plg overlay. Plg-binding proteins were detected with anti-Plg antibody and peroxidase-conjugated second antibody. Table1 shows the identities of the putative Plg-binding proteins.
Identification of B. lactis BI07 putative Plg-binding proteins on two-dimensional gels by mass spectrometry.
Spots identified as putative Plg-binding proteins were excised from the gel and subjected to trypsin digestion and MALDI-TOF MS. The peptide fingerprints obtained were scanned with the searching tools Aldente and ProFound, and unambiguous identification was obtained for six of the eight putative Plg-binding proteins previously detected (Table 1). Protein spot 1 (70 kDa, pI 4.5) was identified as DnaK, protein spot 3 (52 kDa, pI 5.5) was identified as glutamine synthetase, protein spot 4 (46 kDa, pI 4.7) was identified as enolase, protein spots 5 and no. 6 (37 kDa, pI 4.6 to 4.7) were identified as bile salt hydrolase, and protein spot 7 (30 kDa, pI 5.4) was identified as phosphoglycerate mutase. With the exception of glutamine synthetase, all the putative Plg-binding proteins identified are predicted to have a C-terminal lysine. The peptides in protein spots 2 and 8 have not been identified yet.
TABLE 1.
Identification of B. lactis BI07 putative Plg-binding proteins
| Spot | Protein | pI | Molecular mass (kDa) | Mass spectrometry peptide coverage (%) |
|---|---|---|---|---|
| 1 | DnaK (gi 23325724) | 4.7 | 67 | 29 |
| 2 | Unidentified | |||
| 3 | Glutamine synthetase (gi 42519488) | 5.3 | 50 | 24 |
| 4 | Enolase (gi 46191081) | 4.7 | 46.6 | 32 |
| 5-6 | Bile salt hydrolase (gi 46486762) | 4.7 | 35 | 31 |
| 7 | Phosphoglycerate mutase (gi 23336625) | 5.3 | 28 | 50 |
| 8 | Unidentified |
DISCUSSION
Using a flow cytometric approach, we demonstrated dose-dependent human Plg-binding activity in four bifidobacterial strains belonging to three different species: B. lactis, B. bifidum, and B. longum. All strains tested showed binding to human Plg, as previously shown by using human bacterial pathogens, such as group G streptococci, E. coli, and Salmonella. The capability of B. lactis BI07 whole cells to bind human Plg was further demonstrated by immunoelectron microscopy. Binding was localized to the cell surface of B. lactis BI07, where Plg aggregates were visible. These hot spots resemble the Plg binding found on the cell surface of S. pneumoniae (4). The binding of human Plg to Bifidobacterium is strongly dependent on lysine residues of surface protein receptors. Flow cytometry experiments demonstrated that there was complete inhibition of Plg recruitment on the bifidobacterial cell surface in the presence of the lysine analog EACA. Although it is generally assumed that the recruitment of Plg on the cell surface is mediated by C-terminal lysines of Plg receptors, carboxypeptidase B treatment of bifidobacterial cells only partially decreased their binding to human Plg. Thus, the existence of an internal Plg-binding motif for at least some Plg receptors displayed on the bifidobacterial cell surface can be assumed. Strengthening our findings, evidence of a non-C-terminal lysine Plg-binding site has been reported for pneumococcal enolase and Prp, a group A streptococcal M protein-related protein (10, 28, 31).
A proteomic approach enabled identification of five putative Plg-binding proteins in the cell wall fraction of the model strain B. lactis BI07: DnaK, glutamine synthetase, enolase, bile salt hydrolase, and phosphoglycerate mutase. This is the first experimental evidence of a potential Plg-binding activity for bile salt hydrolase and glutamine synthetase, whereas DnaK, enolase, and phosphoglycerate mutase have already been identified as human Plg receptors in different microorganisms. In particular, enolase is one of the best-characterized human Plg receptors in prokaryotes and eukaryotes (2-4, 8-10, 16-18, 24-26, 33, 38, 46), while the surface-located DnaK and phosphoglycerate mutase proteins have been demonstrated to bind Plg in the gram-positive bacterium L. monocytogenes (33) and in Candida albicans (8), respectively. Of the putative Plg-binding proteins that have been identified, glutamine synthetase is the only one which lacks C-terminal lysines. Hence, these data suggested the existence of an internal Plg-binding motif in bifidobacterial Plg receptors, which was experimentally proven by flow cytometric analysis. Interestingly, as reported for other gram-positive bacteria (17), at least some of the B. lactis BI07 Plg-binding proteins are highly conserved cytoplasmatic proteins that, when expressed on the bacterial cell wall, acquire a “moonlighting” function different from their well-known activity performed in the cytoplasm (33). None of these enzymes exhibits any detectable export and retention signal, and the mechanisms of secretion and cell anchoring remain to be determined. Taken together, our data demonstrate that Plg binding to B. lactis BI07 cells is due to the concerted action of a number of proteins that often have other essential cellular functions.
In this paper, we demonstrated that the human species B. bifidum and B. longum, as well as B. lactis, which is widely employed as a probiotic in food and pharmaceutical formulas (7, 15), possess significant human Plg-binding activity and show the potential to intervene with the plasmin(ogen) system of the human host. To our knowledge, this is the first experimental evidence of a Plg-binding activity in the health-promoting member of the human intestinal microbiota Bifidobacterium. Recently, Lactobacillus crispatus, a commensal member of the human intestinal microbiota, has been shown to interact with Plg (14), opening the way to compare the Plg/plasmin system in bacterial commensalism and pathogenicity. The ability of Bifidobacterium to intervene with the plasmin(ogen) system of the human host may have a role in facilitating the colonization of the human GIT through degradation of the extracellular matrix and thereby improving the possibility that the bacteria come in contact with the host cells. However, the relevance of recruited bifidobacterial plasmin activity for establishment of colonization of the human GIT has to be studied.
Acknowledgments
We gratefully acknowledge Giacomo Miccoli for the flow cytometry experiments carried out during work on his degree thesis partially supported by a Deutscher Akademischer Austauschdienst Dienst (DAAD) grant.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host/bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann, S., M. Rohde, K. T. Preissner, and S. Hammerschmidt. 2005. The nine residue plasminogen-binding motif of the pneumococcal enolase is the major cofactor of plasmin-mediated degradation of extracellular matrix, dissolution of fibrin and transmigration. Thromb. Haemostasis 94:304-311. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49:411-423. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 5.Borriello, S. P. 1986. Microbial flora of the gastrointestinal tract, p. 1-20. In M. J. Hill (ed.), Microbial metabolisms in the digestive tract. CRC Press, Boca Raton, FL.
- 6.Collen, D., and M. Verstraete. 1975. Molecular biology of human plasminogen. II. Metabolism in physiological and some pathological conditions in man. Thromb. Diath. Haemorrh. 34:403-408. [PubMed] [Google Scholar]
- 7.Collier-Hyams, L. S., and A. S. Neish. 2005. Innate immune relationship between commensal flora and the mammalian intestinal epithelium. Cell. Mol. Life Sci. 62:1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowe, J. D., I. K. Sievwright, G. C. Auld, N. R. Moore, N. A. Gow, and N. A. Booth. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 47:1637-1651. [DOI] [PubMed] [Google Scholar]
- 9.Derbise, A., Y. P. Song, S. Parikh, V. A. Fischetti, and V. Pancholi. 2004. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect. Immun. 72:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehinger, S., W. D. Schubert, S. Bergmann, S. Hammerschmidt, and D. W. Heinz. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343:997-1005. [DOI] [PubMed] [Google Scholar]
- 11.Görg, A., W. Postel, and S. Günther. 1988. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 12.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 13.Hardie, K., and P. Williams. 1998. Introduction: fractionation of bacterial cell envelopes. Methods Microbiol. 27:185-190. [Google Scholar]
- 14.Hurmalainen, V., S. Edelman, J. Antikainen, M. Baumann, K. Lahteenmaki, and T. K. Korhonen. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153:1112-1122. [DOI] [PubMed] [Google Scholar]
- 15.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 16.Lahteenmaki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 13:79-85. [DOI] [PubMed] [Google Scholar]
- 17.Lahteenmaki, K., P. Kuusela, and T. K. Korhonen. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531-552. [DOI] [PubMed] [Google Scholar]
- 18.Lahteenmaki, K., R. Virkola, R. Pouttu, P. Kuusela, M. Kukkonen, and T. K. Korhonen. 1995. Bacterial plasminogen receptors: in vitro evidence for a role in degradation of the mammalian extracellular matrix. Infect. Immun. 63:3659-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 20.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57-64. [Google Scholar]
- 21.Moran, N. A., P. H. Degnan, S. R. Santos, H. E. Dunbar, and H. Ochman. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc. Natl. Acad. Sci. USA 102:16919-16926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 23.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 24.Pancholi, V., P. Fontan, and H. Jin. 2003. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microb. Pathog. 35:293-303. [DOI] [PubMed] [Google Scholar]
- 25.Pancholi, V. 2001. Multifunctional α-enolase: its role in diseases. Cell. Mol. Life Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 27.Parkkinen, J., and T. K. Korhonen. 1989. Binding of plasminogen to Escherichia coli adhesion proteins. FEBS Lett. 250:437-440. [DOI] [PubMed] [Google Scholar]
- 28.Rios-Steiner, J. L., M. Schenone, I. Mochalkin, A. Tulinsky, and F. J. Castellino. 2001. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A streptococcal surface protein. J. Mol. Biol. 308:705-719. [DOI] [PubMed] [Google Scholar]
- 29.Saksela, O., and D. B. Rifkin. 1988. Cell-associated plasminogen activation: regulation and physiological functions. Annu. Rev. Cell Biol. 4:93-126. [DOI] [PubMed] [Google Scholar]
- 30.Salminen, S., E. Isolauri, and E. Salminen. 1996. Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie Leeuwenhoek 70:347-358. [DOI] [PubMed] [Google Scholar]
- 31.Sanderson-Smith, M., M. Dowton, M. Rauson, and M. J. Walker. 2007. The plasminogen-binding group A streptococcal M protein-related protein Prp binds plasminogen via arginine and histidine residues. J. Bacteriol. 189:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasakawa, C., and J. Hacker. 2006. Host-microbe interaction: bacteria. Curr. Opin. Microbiol. 9:1-4. [Google Scholar]
- 33.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 34.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desire, P., Bork, M. Dalley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebbane, F., O. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. USA 103:5526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28:405-440. [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 38.Sijbrandi, R., T. Den Blaauwen, J. R. H. Tame, B. Oudega, J. Luirink, and B. R. Otto. 2005. Characterization of an iron-regulated alpha-enolase of Bacteroides fragilis. Microbes Infect. 7:9-18. [DOI] [PubMed] [Google Scholar]
- 39.Steinert, M., U. Hentschel, and J. Hacker. 2000. Symbiosis and pathogenesis: evolution of the microbe-host interaction. Naturwissenschaften 87:1-11. [DOI] [PubMed] [Google Scholar]
- 40.Sun, H. 2005. The interaction between pathogens and host coagulation system. Physiology 21:281-288. [DOI] [PubMed] [Google Scholar]
- 41.Sun, H., U. Ringdahl, J. W. Homeister, W. P. Fay, N. C. Engleberg, A. Y. Yang, L. S. Rozek, X. Wang, U. Sjobring, and D. Ginsburg. 2004. Plasminogen is a critical factor for group A streptococcal infection. Science 305:1283-1286. [DOI] [PubMed] [Google Scholar]
- 42.Tannock, G. W. 1999. A fresh look at the intestinal microflora, p. 5-14. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific, Norfolk, United Kingdom.
- 43.Tannock, G. W. 1997. Influences of the normal microbiota on the animal host, p. 466-497. In R. I. Mackie, B. A. Withe, and R. E. Isaacson (ed.), Gastrointestinal microbiology. Chapman & Hall, New York, NY.
- 44.Vassalli, J. D., A. P. Sappino, and D. Belin. 1991. The plasminogen activator/plasmin system. J. Clin. Investig. 88:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., R. Schultz, J. Hong, D. L. Cundiff, K. Jiang, and G. A. Soff. 2004. Cell surface-dependent generation of angiostatin 4.5. Cancer Res. 64:162-168. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., D. Seiffert, B. J. Fowler, G. R. Jenkins, T. C. Thinnes, D. J. Loskutoff, R. J. Parmer, and L. A. Miles. 2002. Plasminogen has a broad extrahepatic distribution. Thromb. Haemostasis 87:493-501. [PubMed] [Google Scholar]
- 48.Zoetendal, E. G., E. E. Vaughan, and W. M. De Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]