Abstract
Both Rhodobacter capsulatus PII homologs GlnB and GlnK were found to be necessary for the proper regulation of nitrogenase activity and modification in response to an ammonium shock. As previously reported for several other bacteria, ammonium addition triggered the AmtB-dependent association of GlnK with the R. capsulatus membrane. Native polyacrylamide gel electrophoresis analysis indicates that the modification/demodification of one PII homolog is aberrant in the absence of the other. In a glnK mutant, more GlnB was found to be membrane associated under these conditions. In a glnB mutant, GlnK fails to be significantly sequestered by AmtB, even though it appears to be fully deuridylylated. Additionally, the ammonium-induced enhanced sequestration by AmtB of the unmodifiable GlnK variant GlnK-Y51F follows the wild-type GlnK pattern with a high level in the cytoplasm without the addition of ammonium and an increased level in the membrane fraction after ammonium treatment. These results suggest that factors other than PII modification are driving its association with AmtB in the membrane in R. capsulatus.
In Rhodobacter capsulatus, a non-sulfur purple phototrophic bacterium, molybdenum-containing nitrogenase is one of two enzymatic complexes capable of reducing dinitrogen to ammonium. Mo-nitrogenase is constituted of two proteins: a dinitrogenase (MoFe protein) containing the active site for N2 fixation and a dinitrogenase reductase (Fe protein) responsible for providing electrons to the MoFe protein. The reduction of 1 mol of N2 requires the transfer of 8 mol electrons and the hydrolysis of 20 to 30 mol of MgATP. This highly energy-demanding process is tightly regulated by fixed nitrogen (ammonium) availability. (In an aqueous solution, at neutral pHs, both ammonia [NH3] and ammonium [NH4+] are present. For simplification, the term “ammonium” is used throughout for both ammonia and ammonium.) Regulation of nitrogenase in R. capsulatus occurs on three levels (11, 41). At the transcriptional level, the NtrB/NtrC two-component system controls nifA transcription. In turn, NifA induces the expression of the other nif genes, including the Mo-nitrogenase structural genes (34, 39). Apparently, NtrY, through cross talk, can substitute for NtrB in this process (12). At the posttranslational level, NifA activity is regulated, being active only in the absence of fixed nitrogen. As well, at a third level, Mo-nitrogenase activity is regulated, being switched off within 5 min after an ammonium shock by either a dinitrogenase reductase ADP-ribosylation-dependent or -independent mechanism (17, 27, 28, 40, 45, 59). Two proteins, DraT and DraG, are implicated in the regulation of Fe protein through covalent modification: DraT mediates ADP-ribosylation, whereas DraG removes the ADP-ribosyl residue (37, 48).
GlnB and GlnK, the two PII homologs of R. capsulatus, play central roles in the transcriptional and posttranslational regulation of Mo-nitrogenase (11, 41). GlnB plays a predominant role in regulation at the transcriptional level, and GlnK cannot fully substitute for GlnB (11). In the presence of ammonium, GlnB prevents expression from Ntr promoters by binding to NtrB (44) and regulating its activity by inhibiting its activation of NtrC (41). Both GlnB and GlnK may be involved in R. capsulatus in the posttranslational control of NifA activity. Previously it has been shown that the presence of GlnK is sufficient to mediate the ammonium-induced inhibition of NifA activity; it was not possible to evaluate the potential role of GlnB (11). However, it is likely that both GlnB and GlnK are capable of binding to NifA and inhibiting its activity since both proteins have been shown, by yeast two-hybrid studies, to interact directly with the transcriptional activator, NifA (44).
In Escherichia coli, GlnB and GlnK are key regulatory proteins for various pathways of fixed-nitrogen assimilation. The uridylylation state of these trimeric proteins reflects the intracellular concentration of glutamine (26), with GlnB and GlnK becoming uridylylated on tyrosine 51 in the T-loop when the concentration of fixed nitrogen is low (3, 51). GlnK of E. coli, as in R. capsulatus, is encoded on the same operon as AmtB, a homotrimeric ammonium channel protein of the Mep/Amount family (4, 52, 53). AmtB is presumed to act as a channel for NH3 in a mechanism, whereby NH4+ is first bound and deprotonated to allow passage of NH3. In addition to AmtB, R. capsulatus has the capacity to synthesize a second Amt-like protein, AmtY (62). In contrast to amtB, the amtY gene is not associated with a PII-encoding gene. Ammonium shock initiates the sequestration of GlnK and, to a lesser extent, GlnB by AmtB (9, 13, 25). Deuridylylation of GlnK is thought to be necessary for its capture by AmtB. The GlnK-AmtB interaction regulates ammonium transport by AmtB (9) and prevents titration of GlnB (6).
AmtB (62) and GlnB and GlnK (11) of R. capsulatus have previously been implicated in the posttranslational regulation of Mo-nitrogenase. Indeed, in either an amtB or glnB glnK mutant, nitrogenase activity is not switched off by ammonium addition, and dinitrogenase reductase is not ADP-ribosylated (11, 62). Also, an interaction between GlnB/GlnK and DraT has been demonstrated by analysis with the yeast two-hybrid system (44). PII proteins and members of the Mep/Amount family play important roles in nitrogenase activity regulation in other bacteria such as Azospirillum brasilense and Rhodospirillum rubrum (30, 63, 64). In these organisms, addition of ammonium triggers a membrane sequestration of DraG dependent on AmtB and PII homologs (23, 56). Moreover, in A. brasilense, pull-down assays have shown binding between deuridylylated GlnB and DraT and between uridylylated/deuridylylated GlnZ and DraG (24).
Here we report the effect of amtB, amtY, glnB, glnK, and glnK51 (encoding GlnK-Y51F) mutations on membrane sequestration of PII homologs in R. capsulatus. Our results suggest that in this organism the modification states of GlnB and GlnK are of limited importance for their interaction with AmtB. We have also observed that GlnB and GlnK may be involved in regulating the uridylylation/deuridylyation state of each other.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Appropriate antibiotics were added when necessary. E. coli strains were cultivated in LB medium. R. capsulatus strains were maintained in YPS medium (57). For nitrogenase-derepressing conditions for β-galactosidase assays, the RCV medium (57) contained 20 mM serine as a nitrogen source. For all other experiments, cultures were grown anaerobically to early stationary phase at 30°C under light in RCV medium without fixed nitrogen (60). Conjugal transfer of plasmid from E. coli S17.1 to R. capsulatus was achieved by filter mating (39). Standard methods for DNA isolation and manipulation were performed as described by Sambrook et al. (49).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17.1 | RP4-2 Tc::Mu Km::Tn7 | 50 |
| BL21(DE3) | Host for overexpression of GlnBHis6 and GlnKHis6 | Novagen |
| R. capsulatus | ||
| B10S | Wild-type Smr | 32 |
| BSRUB13 | ΔglnK | This work |
| PHU332 | glnB::Kmr | P. Hübner, Basel, Switzerland |
| RCAY22 | amtY::Gmr | 62 |
| RCAY63 | amtB::Kmr | 62 |
| SG21 | glnK::Gmr | This work |
| TR3 | glnB51 orf3528::Gmr | This work |
| Plasmids | ||
| pAP2 | pJB3TC20 derivative carrying glnK51 (GlnK-Y51F) and 300 bp of upstream sequence | This work |
| pAY98 | pMECA derivative carrying glnKamtB; Apr | A. Yakunin |
| pBSRUB13 | pNIRUB145 derivative carrying lacZc expressed from a Km cassette; Gmr | This work |
| pBSRUB19 | pPHU231 derivative carrying amtB and most of glnK | This work |
| pBSRUB21-I | pBSRUB19 derivative carrying amtB expressed from a Km cassette | This work |
| pET-22b(+) | T7 expression vector; Apr | Novagen |
| pJB3TC20 | Broad-host-range vector; mob Apr Tcr | 5 |
| pK18 | Cloning vector; Kmr | 46 |
| pML5 | Broad-host-range lacZ-fusion vector; mob Tcr | 35 |
| pNIRUB60-I | pUC18 derivative carrying aphII (Kmr) of transposon Tn5 | This work |
| pNIRUB142 | pML5 derivative carrying lacZc expressed from a Km cassette | This work |
| pNIRUB143 | pSUP202 derivative carrying most of amtB | This work |
| pNIRUB144 | pNIRUB143 derivative carrying glnK::GmramtB | This work |
| pNIRUB145 | pNIRUB144 derivative with 228-bp deletion in glnK | This work |
| pPHU231 | Broad-host-range vector; mob Tcr | 47 |
| pSG5IIa | pUC18 derivative carrying glnK::Gmr; Apr | This work |
| pSG18 | pK18 derivative carrying most of amtB | This work |
| pSGNJ1 | pSVB10 derivative carrying glnK-amtB; Apr | This work |
| pSUP202 | Mobilizable cloning vector; mob Tcr Apr Cmr | 50 |
| pSV5 | pET-22b(+) derivative carrying glnBHis6 | This work |
| pSV6 | pET-22b(+) derivative carrying glnKHis6 | This work |
| pSVB10 | Cloning vector; Apr | 2 |
| pTR9 | pK18 derivative carrying 5′ part of glnB and orf3528 | This work |
| pTR10 | pUC18 derivative carrying 3′ part of glnB | This work |
| pTR11 | pTR9 derivative carrying Gmr from pWKR440 inserted into orf3528 | This work |
| pTR12 | pTR10 derivative; mob Tcr | This work |
| pTR14-I | pTR12 derivative carrying glnB51 (GlnB-Y51F) and orf3528::Gmr | This work |
| pUC18 | Cloning vector; Apr | 55 |
| pWKR56-I | Mobilizable cloning vector; mob Tcr | 32 |
| pWKR440 | pACYC derivative carrying Gmr Tcr | 11 |
Construction of plasmids.
pAP2 contains glnK with a Y51F substitution and 300 bp of upstream sequence. Mutagenesis was performed by overlap extension PCR using pAY98 as a template. The forward primer was 5′-CGCCTGCAGTGATGCGTAGC-3′, and the reverse primer was 5′-CCCTGGATCCCCTGTGGTTCTT-3′ (PstI and BamHI sites underlined). Mutagenic primers were 5′-ATTCACCGCGAATTCGGCGCC-3′ and 5′-CGCGGCGCCGAATTCGCGG-3′ (EcoRI site underlined, mutagenic bases in boldface). PCR product was cloned into pJB3TC20, resulting in pAP2.
pSV5 and pSV6 contain, respectively, glnBHis6 and glnKHis6. PCR products were obtained by using chromosomal DNA as a template with primer pairs 5′-CATATGAAGAAGGTCGAGGCGATC-3′ and 5′-CTCGAGCGCGTCCTCGCCGGTCTC-3′ for glnB (NdeI and XhoI sites underlined) and 5′-CATATGGTGAAACTCATCATTGCAGCG-3′ and 5′-GCGGCCGCCAGCGCTTCGTCGCCAGCTTC-3′ for glnK (NdeI and NotI sites underlined). Fragments were blunt end cloned into the SmaI site of pK18. A 0.35-kb NdeI-XhoI fragment carrying glnB and a 0.35-kb NdeI-NotI fragment carrying glnK were subsequently cloned into pET-22b(+) (Novagen, Darmstadt, Germany), resulting in pSV5 and pSV6, respectively.
pBSRUB21-I contains amtB under the control of the constitutive promoter of aphII (Kmr gene). A 2-kb PCR product encompassing glnK and amtB was obtained by using chromosomal DNA as a template with primer pair 5′-TCGGCTGGAATCGGTTTTTGACTG-3′ and 5′-TGATTTCGAGGCGCTGATGTGGAT-3′. The PCR product was blunt-end cloned into the SmaI site of pSVB10, resulting in pSGJN1. Subsequently, a 1.9-kb XhoI-EcoRI fragment from pSGJN1 carrying amtB and a part of glnK was cloned into pPHU231, resulting in pBSRUB19. A 1.9-kb BamHI fragment bearing the aphII gene from pNIRUB60-I was cloned into pBSRUB19, resulting in pBSRUB21-I.
Construction of R. capsulatus ΔglnK mutant BSRUB13.
A 1.2-kb SalI-XhoI fragment from pSG18 carrying most of amtB was cloned into pSUP202, resulting in pNIRUB143. A 4.7-kb SalI fragment from pSG5IIa carrying the rest of amtB and glnK was cloned into pNIRUB143, resulting in pNIRUB144. Plasmid pNIRUB144 was cut with XhoI-BglII (both sites are within glnK). Sticky ends were filled in by treatment with Klenow fragment, and blunt ends were ligated, leading to the deletion of 228 bp in the glnK coding region, resulting in pNIRUB145. A 12.6-kb BamHI fragment carrying E. coli lacZ constitutively expressed from the Km cassette from pNIRUB142 was cloned into pNIRUB145, resulting in pBSRUB13. Plasmid pBSRUB13 was introduced by conjugation into R. capsulatus strain SG21, in which the glnK gene is interrupted by a gentamicin resistance cassette (glnK::Gm). Selection for Kmr resulted in a strain carrying pBSRUB13 integrated in the chromosome via a single-crossover event. One Kmr exconjugant was grown in the absence of antibiotics prior to plating on nonselective X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing plates. Double-crossover recombination events were identified by loss of a LacZ+ phenotype, and subsequently Lac− strains were checked for sensitivity towards kanamycin and gentamicin. Deletion in glnK in BSRUB13 was verified by Southern hybridization.
Construction of R. capsulatus mutant TR3 (GlnB-Y51F).
Mutant strain TR3 contains a glnB51 mutant allele (encoding GlnB-Y51F) in the chromosome instead of the glnB wild-type gene and a Gm cassette in orf3528, located upstream of glnB. A 0.6-kb DNA fragment encompassing the 3′ end of glnB and a portion of glnA, located downstream of glnB, was PCR amplified using chromosomal DNA as the template and primer pair 5′-TATCGCGGGGCCGAATTCGTCGTCGA-3′ and 5′-TGATTACGAATTAAGCTTCCGGCCCG-3′ (EcoRI and HindIII sites underlined). The amplification product was cut with EcoRI and HindIII and cloned into pUC18, resulting in pTR10. An 8.5-kb HindIII fragment carrying a Tcr gene and the mob site from pWKR56-I was cloned into pTR10, resulting in pTR12. In parallel, a 1.5-kb DNA fragment encompassing the 5′ end of glnB and orf3528, located upstream of glnB, was PCR amplified using chromosomal DNA as a template and primer pair 5′-GGTACCGAGCTCGAATTCGCCCTGAT-3′ and 5′-GAAGTCGACGACGAATTCGGCCCCGC-3′ (EcoRI sites underlined). The amplification product was cut with EcoRI and cloned into pK18, resulting in pTR9. A 45-bp XhoI fragment from the orf3528 coding region was exchanged against the 2.6-kb XhoI Gm cassette from pWKR440, resulting in pTR11. Next, the 4.1-kb EcoRI fragment from pTR11 was cloned into pTR12, resulting in pTR14-I, thereby combining the two parts of the glnB gene. The glnB allele in pTR14-I differs from the wild-type gene in two nucleotides (generating an EcoRI site) and thus codes for a GlnB-Y51F mutant protein. After conjugational transfer of pTR14-I into R. capsulatus, selection for Gmr and loss of the vector-encoded Tcr identified double-crossover events leading to orf3528::Gm mutants. Depending on the individual crossover sites, some of the orf3528::Gm mutant strains additionally carried the GlnB-Y51F substitution (caused by the introduction of the EcoRI site within glnB). These GlnB-Y51F substitution strains were identified by Southern blot analysis using genomic DNA cut with EcoRI and a glnB-specific probe. One strain carrying glnB51 (GlnB-Y51F substitution) was selected for further studies and designated TR3.
GlnBHis6 and GlnKHis6 overexpression, purification, and antibody production.
E. coli strain BL21(DE3) carrying either plasmid pSV5 (glnBHis6) or pSV6 (glnKHis6), respectively, was grown at 30°C in 1 liter of selective LB medium until an optical density at 580 nm (OD580) of 0.5 was reached. Synthesis of the recombinant proteins was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. After further incubation at 30°C for 3 h, cells were harvested, washed with 50 ml buffer A (50 mM NaH2PO4, 300 mM NaCl [pH 8]) containing 20 mM imidazole, and resuspended in 20 ml buffer A (with 20 mM imidazole). Cells were disrupted in a French press cell (at 2,000 lb/in2). The lysate was centrifuged at 22,548 × g for 10 min at 4°C to remove cell debris. The recombinant proteins from the supernatant were purified using Ni-nitrilotriacetic acid columns (QIAGEN, Hilden, Germany) preequilibrated with buffer A containing 50 mM imidazole. Recombinant proteins were eluted from the column using buffer A containing 500 mM imidazole. GlnBHis6and GlnKHis6 were sent to Eurogentec (Köln, Germany) to immunize rabbits. Specificity of the antibodies was verified with the appropriate R. capsulatus mutants.
In vivo nitrogenase activity, dinitrogenase reductase ADP-ribosylation, and β-galactosidase assay.
In vivo nitrogenase activity was measured by the acetylene reduction method as described previously (62). For the dinitrogenase reductase ADP-ribosylation results shown in Fig. 1G, 50 μl of culture samples was removed at the indicated time, treated, loaded on low-cross-linker gels, immunoblotted, and detected by chemiluminescence, all as described previously (62). β-Galactosidase activity was measured as previously described (22, 42).
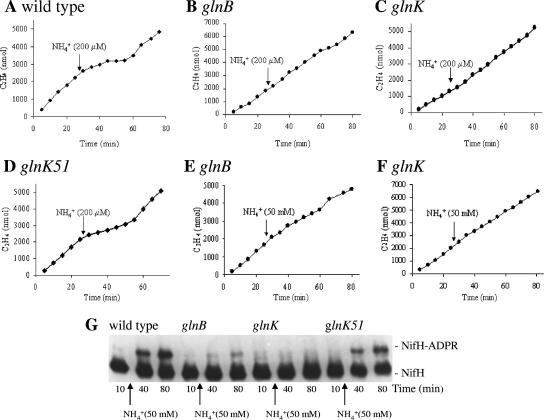
FIG. 1.
Nitrogenase switch-off and ADP-ribosylation of NifH in glnB and glnK mutants. R. capsulatus cells were grown under N2-fixing conditions. Gas samples were withdrawn at the indicated times and assayed for in vivo nitrogenase activity by the acetylene reduction method. Acetylene was added at time 0. For panels A to D, NH4Cl was added to 200 μM at 25 min. (A) Wild-type (B10S). (B) glnB (PHU332). (C) glnK (BSRUB13). (D) glnK51 (GlnK-Y51F; BSRUB13/pAP2). For panels E and F, NH4Cl was added to 50 mM at 25 min. (E) glnB (PHU332). (F) glnK (BSRUB13). The ADP ribosylation state of NifH was monitored for the wild type (B10S) and glnB (PHU332), glnK (BSRUB13), and glnK51 (GlnK-Y51F; BSRUB13/pAP2) mutants (G). Where indicated, NH4Cl was added to 50 mM at 25 min.
Ammonium treatment and cellular fractionation.
Two separate cultures of the same strain were grown photoheterotrophically in the absence of oxygen to early stationary phase in 320 ml of RCV medium. NH4Cl was added to 1 mM to one of the duplicate cultures 15 min prior to harvest. The cells were harvested (10,000 × g, 30 min, 4°C), resuspended in 10 ml of SP buffer (23), and frozen. After thawing, the cells were sonicated and cellular debris was removed by centrifugation (10,000 × g, 30 min, 4°C). The supernatant was processed according to Coutts et al. (9), with the exceptions that 10 ml of clarified lysate was ultracentrifuged using a Beckman L8-60 M with a Beckman 70.1 Ti rotor and the 1 ml at the top of the ultracentrifuge tube after the first ultracentrifugation was considered as the cytoplasmic fraction. The pellet containing the membrane fraction obtained after two washes was resuspended in 1 ml of SP buffer. Fractionation experiments were repeated at least twice for each strain.
Western blot analysis.
For all polyacrylamide gel electrophoresis (PAGE), 5 μg of total protein was loaded per well. Protein concentration was measured by the Bradford reaction (7). Sodium dodecyl sulfate (SDS)-PAGE was conducted with 15% polyacrylamide gels and native PAGE with 10% polyacrylamide gels. After transfer to polyvinylidene difluoride (PVDF) membranes (Roche, Mississauga, Ontario, Canada), immunoblots were probed with either anti-GlnB or anti-GlnK antibody. Signals were detected with the ECL enhanced chemiluminescence system of Amersham (Oakville, Ontario, Canada).
RNA extraction and qPCR.
Total RNA was extracted with TRIzol (Invitrogen, Burlington, Ontario, Canada). cDNA was synthesized with the QuantiTect reverse transcripition kit (QIAGEN, Mississauga, Ontario, Canada). The QuantiTect Sybr green PCR kit (QIAGEN, Mississauga, Ontario, Canada) and Rotor-Gene 6000 cycler were used to amplify and quantify PCR products from glnB and glnK by quantitative PCR (qPCR). Expression of PII homologs was normalized with rpoZ expression, a gene encoding the ω-subunit of RNA polymerase. Relative levels of expression of glnB and glnK were calculated by the 2−ΔΔCT method (36). The following primers were used for gene amplification: glnB, 5′-AGCCGTTCAAGCTCGATGAAGTGA-3′ and 5′-AACCATCTCGATCTTCACCTTGGG-3′; glnK, 5′-TGATGGTGACGGAAATCAAGGGCT-3′ and 5′-CAAGCTTCACCTTCGGCACGAAAT-3′; and rpoZ, 5′-TGACAATGACAAGAACCCGGTGGT-3′ and 5′-TCTGGGTCTGGTTGCTTTCGATCA-3′. Experiments were done in triplicate.
RESULTS
Effects of glnB and glnK mutations on the posttranslational regulation of Mo-nitrogenase activity and Fe protein ADP-ribosylation.
A previous study showed the abolition of Mo-nitrogenase posttranslational regulation and of ADP-ribosylation of dinitrogenase reductase (NifH) in a glnB glnK strain (11). However, AmtB is required for these processes (62), and the glnB glnK mutant used in the previous study was probably polar on amtB expression, compromising the interpretation of these results. To further pursue the characterization of the roles of PII homologs in R. capsulatus, the effects of individual mutations in either glnB or glnK were examined. The glnB mutant contained a glnB gene interrupted by an antibiotic cassette (Table 1). To generate a glnK mutant that was not polar on the downstream amtB, a markerless in-frame deletion mutant of glnK was constructed (Materials and Methods). Southern and Western blot analyses demonstrated the inactivation of glnK (not shown). As expected, this mutation was not polar on amtB, as methylammonium transport requires the presence of AmtB, which cannot be replaced by AmtY (62), and the glnK strain showed high levels of [14C]methylammonium transport (not shown).
R. capsulatus possesses two different systems implicated in the regulation of nitrogenase activity: one that is linked to ADP-ribosylation of the Fe protein and one that is independent of this covalent modification (59). Both responses can be provoked by the addition of ammonium to the medium, and the relative importance of the two responses varies with growth conditions (59). However, AmtB plays a key role since AmtB− strains are deficient in both responses (62). The potential ammonium-induced switch-off of nitrogenase activity and ADP-ribosylation of the Fe protein were examined in these glnB and glnK mutant strains (Fig. 1). There was no nitrogenase switch off in the glnB strain when subjected to the addition of 200 μM NH4+ (Fig. 1B), and this strain was even insensitive to 50 mM NH4+ (Fig. 1E). When ADP-ribosylation of the Fe protein in response to a 50 mM NH4+ addition was examined, only a moderate response after 80 min was noted for the glnB strain, whereas the wild-type strain showed appreciable Fe protein modification after 40 min and even more substantial modification after 80 min (Fig. 1G). Under these conditions, apparent modification was less than 100%; hence, both responses, ADP-ribosylation-dependent and -independent, appeared to be operative. (Only one subunit per dimer is modified; therefore, 100% modification should result in two Fe protein bands of equal intensity.) Therefore, GlnB appears necessary for both nitrogenase switch-off and Fe protein modification in R. capsulatus. Surprisingly, like GlnB, GlnK is also necessary for the NH4+-induced switch-off of Mo-nitrogenase activity and ADP-ribosylation of Fe-protein. Thus, in the glnK strain, the addition of 200 μM ammonium does not suppress acetylene reduction by the Mo-nitrogenase (Fig. 1C). Even a 50 mM ammonium shock has no effect on Mo-nitrogenase activity or on the ADP-ribosylation state of NifH in a glnK mutant (Fig. 1F and G).
To explore the potential need for GlnK modification in the regulation of these processes, a mutant strain carrying glnK51 (encoding GlnK-Y51F) was examined. This analysis is potentially complicated by the fact that the glnK51 allele was expressed from a plasmid and therefore might have different expression levels which could affect protein-protein interactions important to its functioning. To address this issue, we performed qPCR analysis of the relative expression levels of GlnK-Y51F (from the plasmid pAP2) and wild-type GlnK (from the chromosome) under the nitrogen-limited growth conditions used here. Expression levels of the two alleles were nearly the same since glnK51 (GlnK-Y51F) is expressed 1.2 (±0.3)-fold higher than glnK; thus, the results obtained with the GlnK and GlnK-Y51F strains can be directly compared. In the strain carrying the GlnK-Y51F allele, BSRUB13/pAP2, which lacks the wild-type GlnK, the mutant GlnK cannot be modified by uridylylation due to the replacement of the amino acid at the site of modification, tyrosine 51, with phenylalanine. However, the regulation of nitrogenase activity (switch-off) (Fig. 1D) and nitrogenase modification (Fig. 1G) are nearly normal in this strain. At lower doses of ammonium, normal demodification was shown to occur in this strain (not shown). This strongly argues that the modification status of GlnK is not critical to its function in regulating these processes.
AmtB-dependent membrane sequestration of GlnB and GlnK.
The interaction of PII homologs with the membrane, dependent on proteins of the Amt family, has previously been demonstrated in both gram-positive and gram-negative bacteria (9, 10, 25). For example, in R. rubrum, a non-sulfur purple bacterium, GlnJ is sequestered by the membrane in an AmtB-dependent manner (AmtB1) 30 min after the addition of 10 mM ammonium (56). Here, cell fractionation experiments were carried out to study the possible sequestration of the two R. capsulatus PII homologs, GlnB and GlnK, by the two Amt family members of R. capsulatus, AmtB and AmtY. Western blot analysis was performed on the cytoplasmic and membrane fractions obtained from parallel cultures—one harvested with no treatment and the other harvested 15 min after the addition of ammonium to 1 mM (Fig. 2). Antisera to GlnB and GlnK showed no cross-reactivity to the heterologous PII proteins when extracts of deletion strains were examined (data not shown). Before the addition of NH4+, the membrane fraction was devoid of GlnB, whereas with the culture to which ammonium had been added, there was relatively more GlnB in the membrane fraction (Fig. 2A). Cell fractionation analysis also showed greatly increased membrane association of GlnK in the ammonium-treated culture. Comparison of the relative intensities of the membrane fraction with the cytoplasmic fraction suggests that there is a greater fraction of the GlnK pool associated with the membrane in the ammonium-treated cells than is the case with GlnB (Fig. 2B). Previously, the two E coli PII paralogues, GlnB and GlnK, have been shown to form heterotrimers (14, 54). As well, yeast two-hybrid studies have demonstrated an interaction between R. capsulatus GlnB and GlnK (44). Therefore, possible heterotrimer formation is of potential concern in interpreting the results presented here. However, qPCR analysis shows that in the wild-type strain there is 100 (±30)-fold greater expression of GlnK than GlnB under the nitrogen-limiting conditions used here. Therefore, potential GlnB-2GlnK heterotrimers would represent an insignificant fraction of the total GlnK population and can be ignored in the analysis of the GlnK results. However, as the resolution of native gel electrophoresis of the R. capsulatus PII paralogs (see below) is insufficient to differentiate between potential heterotrimers and different modified forms of GlnB, we cannot rule out the possibility of heterotrimers influencing the results observed with anti-GlnB antisera (see Fig. 2A, for example). Indeed, GlnB-GlnK heterotrimers may be responsible for some of the membrane-associated GlnB observed here.
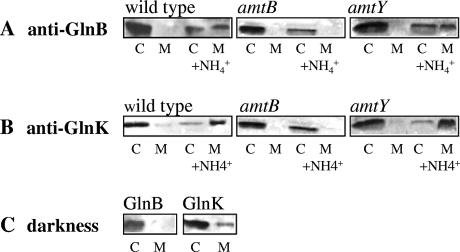
FIG. 2.
Role of AmtB and AmtY in the membrane sequestration of GlnB and GlnK. Two parallel cultures of the R. capsulatus wild type (B10S) and amtB (RCAY63) and amtY (RCAY22) mutants were grown under N2-fixing conditions. NH4Cl (1 mM) was added to one of the parallel cultures 15 min prior to harvest. Cytoplasmic (C) and membrane (M) fractions were subjected to SDS-PAGE (5 μg of total protein was loaded per well) followed by Western blotting with anti-GlnB (A) or anti-GlnK antibody (B). Wild-type (B10S) cells grown under N2-fixing conditions was collected 30 min after being exposed to darkness. Cytoplasmic and membrane fractions were subjected to SDS-PAGE (5 μg of total protein was loaded per well) and Western blotting with anti-GlnB or anti-GlnK antibody (C).
In an R. capsulatus amtB mutant, there is no capture of either of the two PII homologs by the membrane, indicating that membrane sequestration is necessarily dependent upon AmtB (Fig. 2A and B). This suggests that AmtY, an R. capsulatus homolog of AmtB, is incapable of participating in the ammonium-induced membrane sequestration of the PII proteins. Indeed, whether GlnB (Fig. 2A) or GlnK is examined (Fig. 2B), an amtY strain presents the same phenotype as the wild-type strain. This is not surprising since a mutation in amtY does not affect methylammonium uptake, a hallmark of bona fide AmtB proteins, and AmtY, as shown previously, is not implicated in the posttranslational regulation of the Mo-nitrogenase (62).
In addition to ammonium shock, darkness triggers Mo-nitrogenase switch-off and ADP-ribosylation of dinitrogenase reductase in R. capsulatus (61). Knowing that darkness still induces this phenomenon in an amtB mutant (62), we checked if this stimulus has an impact on the membrane sequestration of the R. capsulatus PII homologs. As expected, after 30 min of darkness, the relative distribution of GlnB and GlnK appears unchanged: i.e., there seems to be no darkness-induced sequestration (Fig. 2C).
GlnB is necessary for the membrane sequestration of GlnK.
In an A. brasilense glnB mutant, GlnK is always uridylylated and does not interact with the membrane even upon addition of ammonium (23). We show here that in R. capsulatus, GlnB is also necessary for the membrane sequestration of GlnK (Fig. 3B). As noted before, there is relatively more GlnK in the membrane fraction, and less in the cytoplasmic fraction, in the wild-type strain receiving an ammonium shock compared to no treatment. However, in a glnB strain, there appears to be no change of the localization of GlnK upon ammonium shock (Fig. 3B). On the other hand, contrary to what has been reported for the A. brasilense glnB mutant (23), the R. capsulatus glnB mutant, after 15 min of exposure to 1 mM of ammonium, displays an almost complete deuridylylation of GlnK, as revealed by native PAGE (Fig. 3E). In fact, GlnB appears to be necessary for the efficient uridylylation of GlnK since intermediary forms of the protein (GlnK-1UMP, GlnK-2UMP, and GlnK-3UMP) are observed in absence of NH4+. In contrast, in the wild type, only GlnK-3UMP, the fully uridylylated form, is present in the absence of ammonium. Since the ammonium-treated culture possesses a large fraction of completely deuridylylated GlnK, but the membrane fraction is almost devoid of GlnK, it would appear that the absence of UMP is not sufficient for membrane localization.
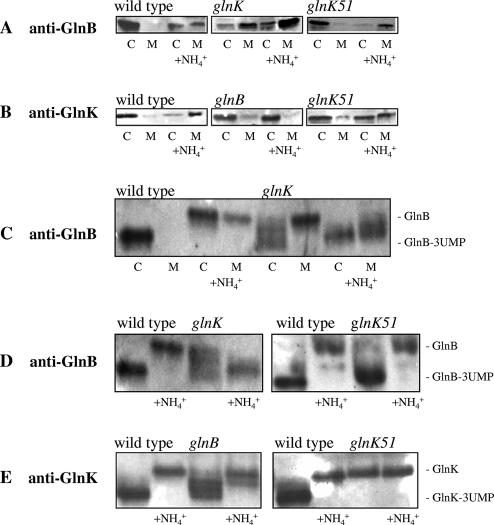
FIG. 3.
Interaction of PII homologs with the membrane in glnK and glnB mutants. Two parallel cultures of the R. capsulatus wild type (B10S) and glnB (PHU332), glnK (BSRUB13), and glnK51 (GlnK-Y51F; BSRUB13/pAP2) mutants were grown under N2-fixing conditions. NH4Cl (1 mM) was added to one of the parallel cultures 15 min prior to harvest. Cytoplasmic (C) and membrane (M) fractions were subjected to SDS-PAGE (5 μg of total protein loaded per well) and Western immunoblotting with anti-GlnB (A) or anti-GlnK (B) antibody. Cytoplasmic (C) and membrane (M) fractions of the wild type (B10S) and glnK mutant (BSRUB13) were subjected to native PAGE (5 μg of total protein loaded per well) followed by Western immunoblotting with anti-GlnB antibody (C). Whole-cell extracts from cultures of the wild type (B10S) and glnB (PHU332), glnK (BSRUB13), and glnK51 (GlnK-Y51F; BSRUB13/pAP2) mutants exposed or not for 15 min to 1 mM NH4Cl were analyzed by native PAGE (5 μg of total protein loaded per well) and Western blotting (5 μg of total protein was loaded per well) with anti-GlnB (D) or anti-GlnK (E) antibody.
GlnK prevents efficient AmtB-dependent membrane sequestration of GlnB.
One of the roles of GlnK of E. coli appears to be to prevent the titration of GlnB by AmtB. In an E. coli glnK mutant, Ntr-promoted genes are not downregulated by the addition of ammonium, whereas in a glnK amtB double mutant, the inhibition efficiency is comparable to that of the wild type (6). Since in an E. coli glnK background, GlnB is the only PII homolog present to control NtrB activity, the absence of inhibition of Ntr-promoted gene expression is explained by the fact that GlnB is not present in the cytoplasm to assume its normal regulatory function with respect to fixed nitrogen (43).
In R. capsulatus, one function of GlnB is to control the synthesis of the nitrogen fixation regulatory protein NifA. GlnK does not participate in the regulation of NifA synthesis but appears to be capable of the posttranslational modulation of NifA activity. Therefore, GlnB− strains, which constitutively express NifA, can still regulate nitrogenase synthesis in response to fixed nitrogen (11). Two lines of evidence establish that GlnK of R. capsulatus also acts to prevent significant sequestration of GlnB by AmtB. First, cell fractionation experiments were performed to explore the impact of a glnK mutation on the relationship between AmtB and GlnB. An examination of the relative intensities of GlnB in the membrane fraction compared to the cytoplasmic fraction of the culture without ammonium treatment shows that in a glnK strain there is a relatively greater amount of the GlnB pool found in the membrane fraction compared to the wild type (Fig. 3A). The relative intensity of the membrane-associated GlnB to the cytoplasmic pool of GlnB was even greater in the culture that had received an ammonium shock (Fig. 3A).
Second, the constitutive expression of amtB (driven by the aphII promoter) in a glnK strain grown in RCV medium containing 20 mM NH4+ allows the synthesis of an active Mo-nitrogenase (Table 2). At this ammonium concentration, there is no Mo-nitrogenase expression in the wild type or in a glnK mutant since AmtB is expressed only at a very low level and GlnB, free in the cytoplasm, can suppress the NtrB-mediated phosphorylation of NtrC (41). NtrC-P induces the expression of nifA coding for the activator of Mo-nitrogenase structural genes (15, 21). Thus, in agreement with the results of the cell fractionation study, it would seem that in a glnK mutant PaphII::amtB strain growing in an ammonium-rich medium, AmtB probably sequesters GlnB, preventing it from interacting with NtrB and NifA.
TABLE 2.
Activity of Mo-nitrogenase in the presence of a constitutively expressed AmtB
| Strain | Relevant genotype | Nitrogenase activitya
|
|
|---|---|---|---|
| +NH4+b | −NH4+c | ||
| B10S | Wild type | 3.0 ± 1.6 | 619 ± 80.5 |
| PHU332 | glnB | 0.7 ± 0.1 | 725 ± 13.0 |
| BSRUB13 | glnK | 1.9 ± 0.3 | 599 ± 20.7 |
| B10S(pBSRUB21-I) | PaphII::amtB | 0.8 ± 0.1 | 657 ± 55.5 |
| PHU332(pBSRUB21-I) | glnB PaphII::amtB | 1.2 ± 0.1 | 572 ± 20.1 |
| BSRUB13(pBSRUB21-I) | glnK PaphII::amtB | 70 ± 4.6 | 718 ± 13.0 |
Activities are given in nmol of ethylene produced/h/mg protein. Values are means of at least three independent experiments ± standard deviations.
Strains were cultivated in RCV medium plus 20 mM NH4+.
Strains were cultivated in RCV medium plus 9.5 mM serine.
The uridylylation/deuridylylation status of the PII homologs and its impact on PII homolog localization.
In E. coli, deuridylylation of GlnK is thought to trigger its binding to AmtB (25). Purification of an E. coli GlnK-AmtB complex (GlnK-AmtBEc) reveals that only completely deuridylylated GlnK is copurified with AmtBEc (13). A previous study has also shown that GlnK-Y51FEc, a GlnK variant that cannot be uridylylated, is sequestered by AmtBEc even before ammonium shock (25). In accordance with this, ammonium conduction through AmtB in E. coli is blocked in a strain carrying GlnK-Y51F even under nitrogen-deficient conditions (25). Thus, it has been suggested that membrane sequestration of GlnK is governed by its uridylylation status (25).
The importance of the uridylylation state of GlnK and GlnB of R. capsulatus for their AmtB-dependent membrane sequestration was examined further. A number of observations suggest that factors other than uridylylation status drive PII membrane association in R. capsulatus. For one thing, as noted above, in a glnB strain, native PAGE analysis shows that GlnK, although apparently almost entirely in the fully deuridylylated form after an ammonium shock (Fig. 3E), is not appreciably membrane associated (Fig. 3B). (The primary purpose of Fig. 3C, D, and E is to show the modification status of the PII proteins. The apparently greater amount of the uridylylated form may be due to the antibody giving a more intense signal with this form than with the deuridylylated form. When the experiment show in Fig. 3E was performed by splitting the same culture in two and treating one-half with ammonium, the uridylylated form seen in the untreated sample was again more intense than the deuridylylated form found in the ammonium-treated sample.) We next examined the effect of introducing a Y51F mutant allele of GlnK by using plasmid pAP2 carrying glnK51 (coding for a GlnK-Y51F variant) to complement the glnK strain BSRUB13. As noted before, qPCR showed that this allele was expressed at approximately the same level as the wild-type allele. As to be expected, replacement of tyrosine 51 by phenylalanine prevents the uridylylation of GlnK-Y51F in the absence of ammonium and its migration on native PAGE gels was the same whether or not the culture had been treated with ammonium (Fig. 3E). A variety of the physiological properties of the R. capsulatus GlnK-Y51F-containing strain were examined. The PII variant strain was found to present many of the same characteristics as the wild-type strain. As already noted, the pattern of Mo-nitrogenase activity switch-off in response to the addition of 200 μM ammonium in the GlnK-Y51F strain was nearly identical to that of the wild-type GlnK strain and nearly normal dinitrogenase reductase ADP ribosylation was restored to a glnK strain (Fig. 1). The pattern of the modification status of GlnB with and without the addition of ammonium was found to be the same in the GlnK-Y51F-containing strain as in the wild type (Fig. 3D); thus, in this regard GlnK-Y51F restores normal GlnK function in a glnK background. In addition, methylammonium uptake activity was within the range found for wild-type strain B10S (not shown). This was surprising since, based on previous studies with E. coli, it might be expected that GlnK-Y51F, unable to be modified, would bind to AmtB under all conditions, rendering it inactive as an ammonium (methylammonium) channel.
The localization of GlnK-Y51F in cultures without, and with, the addition of ammonium was examined. Indeed, although traces of the PII homolog variant were present in the membrane of cultures to which ammonium had not been added, the relative intensity of GlnK-Y51F in the cytoplasmic fraction was high and appears to be almost of the same relative intensity with respect to the membrane fraction as the wild type (Fig. 3B). In the culture to which ammonium had been added, there was appreciable localization of the GlnK-Y51F pool in the membrane fraction. The relative intensity of the membrane fraction compared to the cytoplasmic does not seem to be as great as for the wild-type GlnK, suggesting that the mutant protein may have a lower affinity for the membrane than the wild-type protein. Nevertheless, the ammonium-induced change in the membrane localization of GlnK-Y51F strongly argues that deuridylylation of GlnK is not the main signal that provokes the AmtB-dependent membrane sequestration of GlnK in R. capsulatus.
Binding of GlnB to the membrane after an ammonium shock may be of little physiological relevance since in the presence of GlnK most of the PII binding sites of AmtB would be occupied by GlnK. Nevertheless, it was of mechanistic interest to further examine the influence of the modification status of GlnB on its membrane sequestration in a glnK strain. Interestingly, in this mutant background the modification status of GlnB appears to be deregulated. Without the addition of ammonium, the range of GlnB isoforms goes from deuridylylated to fully uridylylated, whereas in the culture to which ammonium was added, apparent full uridylylation is the only modification pattern observed (Fig. 3D). Since in a glnK background GlnB interacts strongly with the membrane, we checked the uridylylation states of the different fractions, cytoplasmic and membrane, by native PAGE. (Native PAGE has previously been used to examine the uridylylation status of membrane-associated GlnK in E. coli [9] and Klebsiella pneumoniae [33].) Without ammonium addition, all of the GlnB isoforms, except the completely deuridylylated one, are present in the cytoplasmic fraction (Fig. 3C). However, in the culure with an ammonium shock, partially uridylylated GlnB is found in the membrane fraction. Since these isoforms are located in the cytoplasm without the stimulus, this suggests that a signal other than uridylylation state must induce its membrane sequestration.
To further pursue this, it would be interesting to examine the localization of an unmodifiable GlnB in a glnK background. However, this is not possible with the strains presently on hand. In a strain carrying glnB51 (wild type for glnK) in which wild-type GlnB has been replaced with a GlnB-Y51F substitution, expression of a plasmid-encoded glnK-lacZ fusion is only at 3% of the wild-type level with serine as the fixed nitrogen source. Also, in this strain, levels of nifH-lacZ and nifA-lacZ expression are lower and GlnK is not detectable by Western blotting (unpublished data). These results support the notion that fully deuridylylated GlnB shuts down expression from NtrC-activated promoters by binding to NtrB, thus favoring the dephosphorylation and deactivation of NtrC-P. Therefore, since glnB and, especially, glnK-amtB are transcribed from Ntr promoters, it is futile to attempt fractionation experiments with the strain encoding GlnB-Y51F.
DISCUSSION
PII proteins serve a variety of functions in the regulation of cellular nitrogen metabolism. In many bacteria, there are at least two PII homologs present, usually GlnB and GlnK. Within the same species, they appear in some cases to be functionally redundant: in others, they have been shown to be functionally distinct. For example, in the nitrogen-fixing enteric bacterium K. pneumoniae, GlnK can replace GlnB in most of its role in regulating nitrogen metabolism (18, 20) but GlnB cannot replace GlnK in modulating the activity of NifL (1). As well, the same PII homolog, GlnB, for example, may vary in its mode of action from species to species. For example, under nitrogen-limited conditions GlnB is required for NifA activity in the purple non-sulfur bacterium R. rubrum (65), whereas it is dispensable under the same conditions in the purple non-sulfur bacterium R. capsulatus (11).
The gene for the PII homolog GlnK is usually found in an operon with the gene encoding an AmtB protein of the Amount/MEP family. Crystal structures have recently shown that trimeric E. coli GlnK forms a complex with trimeric AmtB largely through interactions mediated by the GlnK T-loop (8, 16). Different bacteria have been found to have different complements of PII and AmtB homologs, and in some cases evidence is accumulating that they are involved in the posttranslational regulation by ADP-ribosylation of Mo-nitrogenase following an ammonium shock. In the nitrogen-fixing betaproteobacterium Azoarcus sp. strain BH72, there are three PII homologs, GlnB, GlnK, and GlnY, and a single AmtB. Regulation of nitrogenase is complex, with nitrogenase modification in response to ammonium requiring both GlnB and GlnK, as well as AmtB, whereas nitrogenase activity switch-off appears to require only GlnK and AmtB (38). In A. brasilense, two PII homologs are present (GlnB and GlnZ), as well as a single AmtB. GlnB is required for ammonium-induced Fe protein ADP-ribosylation (30), and GlnZ is needed for Fe protein demodification (31). Regulation may be through the formation of specific protein-protein complexes between GlnB and DraT and GlnZ and DraG (24). In R. rubrum, which contains two AmtB homologs (AmtB1 and AmtB2) and three PII homologs (GlnB, GlnK, and GlnJ), it has been shown that proper regulation of nitrogenase modification requires only AmtB1 and either GlnB or GlnJ (56, 64). In other words, the functioning of these two R. rubrum PII homologs appears to be redundant in this case. Thus, in general, there is participation by PII homologs and AmtB in regulating nitrogenase switch-off and the modification of the Fe protein, but specific details on the number and types of PII homologs involved appear to be different, depending upon the organism. The mechanistic particulars behind these differences are as yet unclear.
Previously, it has been shown that, in R. capsulatus, AmtB is required for nitrogenase switch-off and covalent modification (62) and that a strain doubly mutated in glnB and glnK was also defective in this regard (11). Here we have shown that both GlnB and GlnK are necessary for the proper regulation of both nitrogenase activity switch-off and Fe protein modification. How they function together is unclear at present. Heterotrimer formation might be a factor, but this is somewhat unlikely as we have shown here that glnK expression is 100-fold higher than that of glnB. One reasonable hypothesis is that the absence of one protein might perturb metabolite pools, which in turn could affect the behavior of the other PII homolog. As well, the modification state of one or both of the PII homologs may be important in the regulation of nitrogenase activity and ADP-ribosylation and our results strongly suggest that one PII homolog might participate in the proper regulation of the modification state of the other.
Here we have shown that the PII homologs of R. capsulatus, GlnB and GlnK, are sequestered by the membrane in an AmtB-dependent way upon addition of ammonium, with significant binding of GlnB in response to an ammonium shock only in the absence of GlnK. Not surprisingly, this stimulus also provokes deuridylylation of GlnB and GlnK in the wild-type background. In E. coli, it has been shown that it is the completely deuridylyated GlnK that binds to the membrane in an AmtB-dependent manner and it has been thought that it is the PII modification state that governs its localization (25). In accordance with this, the E. coli PII variant GlnK-Y51F is always membrane associated independent of the nitrogen status of the cell (25). Here, contrary to what has been observed in E. coli, cell fractionation analysis of an R. capsulatus glnK51 mutant shows that GlnK-Y51F localization is nonetheless dependent upon the cellular nitrogen status. As well, in a glnB strain GlnK is deuridylylated in response to an ammonium shock but is not significantly sequestered by the AmtB in the membrane. These results strongly suggest that in R. capsulatus it is something other than the deuridylylation of GlnK that triggers its capture by AmtB in the membrane.
Therefore, how the level of fixed nitrogen influences PII membrane sequestration in R. capsulatus is uncertain. It is known that in some cases PII protein activity may also be controlled by noncovalent binding of ATP and 2-oxoglutarate (29, 58), and in fact binding of GlnK with some protein effectors, for example, K. pneumoniae GlnK with NifL (18) and A. brasilense GlnZ with DraG (24), appears to be indifferent to modification state. A recent in vitro study has shown that the presence of ATP is necessary for the constitution and dissociation of a GlnK-AmtB complex in E. coli, whereas 2-oxoglutarate is essential for its dissociation (13). Indeed, this explains why GlnK-AmtB membrane-bound complexes can be isolated and studied, as we have done here with electrophoretic analysis. Essentially, since dissociation requires the presence of both ATP and 2-oxoglutarate, and these are drastically diluted during processing, the complexes remain “locked in.” The notion that membrane localization may be in part driven by interaction with metabolites is supported by a recent crystallographic study of an AmtB-GlnK complex where it was suggested that there are two sites on GlnK where 2-oxoglutarate could bind and affect the ability of GlnK to interact with AmtB (16). As well, GlnK in Bacillus subtilis is differentially membrane associated, depending upon the cellular nitrogen status, and its localization appears to be independent of modification and possibly influenced by ATP levels (19). It is possible that metabolite concentrations, and changes in their levels are the major driving forces for PII localization in R. capsulatus. Further investigation is required to verify this hypothesis. As well, evidence is mounting for key roles for AmtB and PII proteins in the control of the activities of the enzymes involved in NifH modification, DraG and DraT, with AmtB and a PII protein being required for the ammonium-induced membrane sequestration of DraG in both A. brasilense (23) and R. rubrum (56). Additional work is necessary to further decipher the exact roles of these players in the regulation of nitrogenase in R. capsulatus.
Acknowledgments
This research was supported by a grant from the Natural Sciences and Engineering Council (P.C.H.).
We thank the following persons for plasmid and strain construction: Silke Groß (pSG plasmids), Nazila Isakovic (pNIRUB plasmids), Amélie Pelletier (pAP2), Tanja Rehmann (pTR plasmids), Britta Schubert (pBSRUB13; R. capsulatus BSRUB13), and Sven Vermöhlen (pSV5 and pSV6). We thank Rita Simo-Guemfing for the result concerning the glnB mutant presented in Fig. 3B.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Arcondėguy, T., W. C. Van Heeswijk, and M. Merrick. 1999. Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniae NifL-dependent nitrogen control. FEMS Microbiol. Lett. 180:263-270. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, W., and A. Pühler. 1988. A family of high-copy-number plasmid vectors with single end-label sites for rapid nucleotide sequencing. Gene 70:171-179. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson, M. R., and A. J. Ninfa. 1999. Characterization of the GlnK protein of Escherichia coli. Mol. Microbiol. 32:301-313. [DOI] [PubMed] [Google Scholar]
- 4.Blakey, D., A. Leetch, G.H. Thomas, G. Coutts, K. Findlay, and M. Merrick. 2002. Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem. J. 364:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blatny, J. M., T. Brautaset, H.C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blauwkamp, T. A., and A. J. Ninfa. 2003. Antagonism of PII signalling by the AmtB protein of Escherichia coli. Mol. Microbiol. 48:1017-1028. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Conroy, M. J., A. Durand, D. Lupo, X.-D. Li, P. A. Bullough. F. K. Winkler, and M. Merrick. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. USA 104:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutts, G., G. Thomas, D. Blakey, and M. Merrick. 2002. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Detsch, C., and J. Stülke. 2003. Ammonium utilization in Bacillus subtilis: transport and regulatory functions of NrgA and NrgB. Microbiology 149:3289-3297. [DOI] [PubMed] [Google Scholar]
- 11.Drepper, T., S. Groβ, A.F. Yakunin, P.C. Hallenbeck, B. Masepohl, and W. Klipp. 2003. Role of GlnB and GlnK in ammonium control of both nitrogenase systems in the phototrophic bacterium Rhodobacter capsulatus. Microbiology 149:2203-2212. [DOI] [PubMed] [Google Scholar]
- 12.Drepper, T., J. Wiethaus, D. Giaourakis, S. Groβ, B. Schubert, M. Vogt, Y. Wiencek, A.G. McEwan, and B. Masepohl. 2006. Cross-talk towards the response regulator NtrC controlling nitrogen metabolism in Rhodobacter capsulatus. FEMS Microbiol. Lett. 258:250-256. [DOI] [PubMed] [Google Scholar]
- 13.Durand, A., and M. Merrick. 2006. In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J. Biol. Chem. 281:29558-29567. [DOI] [PubMed] [Google Scholar]
- 14.Forchhammer, K., A. Hedler, H. Strobel, and V. Weiss. 1999. Heterotrimerization of PII-like signalling proteins: implications for P-II-mediated signal transduction systems. Mol. Microbiol. 33:338-349. [DOI] [PubMed] [Google Scholar]
- 15.Foster-Hartnett, D., and R. G. Kranz. 1992. Analysis of the promoters and upstream sequences of nifA1 and nifA2 in Rhodobacter capsulatus: activation requires ntrC but not rpoN. Mol. Microbiol. 6:1049-1060. [DOI] [PubMed] [Google Scholar]
- 16.Gruswitz, F., J. O'Connell III, and R. M. Stroud. 2007. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc. Natl. Acad. Sci. USA 104:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallenbeck, P. C., C. M. Meyer, and P. M. Vignais. 1982. Nitrogenase from the photosynthetic bacterium Rhodopseudomonas capsulata: purification and molecular properties. J. Bacteriol. 149:708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, L., E. Soupene, A. Ninfa, and S. Kustu. 1998. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol. 180:6661-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich, A., K. Woyda, K. Brauburger, G. Meiss, C. Detsch, J. Stülke, and K. Forchhammer. 2006. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J. Biol. Chem. 281:34909-34917. [DOI] [PubMed] [Google Scholar]
- 20.Holtel, A., and M. Merrick. 1989. The Klebsiella pneumoniae PII protein (glnB gene product) is not absolutely required for nitrogen regulation and is not involved in NifL-mediated nif gene regulation. Mol. Gen. Genet. 217:474-480. [DOI] [PubMed] [Google Scholar]
- 21.Hübner, P., B. Masepohl, W. Klipp, and T. A. Bickle. 1993. nif gene expression studies in Rhodobacter capsulatus: ntrC-independent repression by high ammonium concentration. Mol. Microbiol. 10:123-132. [DOI] [PubMed] [Google Scholar]
- 22.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huergo, L. F., E. M. Souza, M. S. Araujo, F. O. Pedrosa, L. S. Chubatsu, M. B. Steffens, and M. Merrick. 2006. ADP-ribosylation of dinitrogenase reductase in Azospirillum brasilense is regulated by AmtB-dependent membrane sequestration of DraG. Mol. Microbiol. 59:326-337. [DOI] [PubMed] [Google Scholar]
- 24.Huergo, L. F., L. S. Chubatsu, E. M. Souza, F. O. Pedrosa, M. B. Steffens, and M. Merrick. 2006. Interactions between PII proteins and the nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Lett. 580:5232-5236. [DOI] [PubMed] [Google Scholar]
- 25.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 279:8530-8538. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of Ntr gene transcription in Escherichia coli. Biochemistry 37:12795-12801. [DOI] [PubMed] [Google Scholar]
- 27.Jouanneau, Y., C. M. Meyer, and P. M. Vignais. 1983. Regulation of nitrogenase activity through iron protein interconversion into an active and inactive form in Rhodopseudomonas capsulata. Biochim. Biophys. Acta 749:318-328. [Google Scholar]
- 28.Jouanneau, Y., C. Roby, C. M. Meyer, and P. M. Vignais. 1989. ADP-ribosylation of dinitrogenase reductase in Rhodobacter capsulatus. Biochemistry 267:780-787. [Google Scholar]
- 29.Kamberov, E. S., M. R. Atkinson, and A. J. Ninfa. 1995. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 270:17797-17807. [DOI] [PubMed] [Google Scholar]
- 30.Klassen, G., E. M. Souza, M. G. Yates, L. U. Rigo, R. M. Costa, J. Inaba, and F. O. Pedrosa. 2005. Nitrogenase switch-off by ammonium ions in Azospirillum brasilense requires the GlnB nitrogen signal-transducing protein. Appl. Environ. Microbiol. 71:5637-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klassen, G., E. M. de Souza, M. G. Yates, L. U. Rigo, J. Inaba, and F. de Oliveira Pedrosa. 2001. Control of nitrogenase reactivation by the GlnZ protein in Azospirillum brasilense. J. Bacteriol. 183:6710-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klipp, W., B. Masepohl, and A. Pühler. 1988. Identification and mapping of nitrogen fixation genes of Rhodobacter capsulatus: duplication of a nifA-nifB region. J. Bacteriol. 170:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klopprogge, K., R. Grabbe, M. Hoppert, and R. A. Schmitz. 2002. Membrane association of Klebsiella pneumoniae NifL is affected by molecular oxygen and combined nitrogen. Arch. Microbiol. 177:223-234. [DOI] [PubMed] [Google Scholar]
- 34.Kranz, R. G., and D. Foster-Hartnett. 1990. Transcriptional regulatory cascade of nitrogen-fixation genes in anoxygenic photosynthetic bacteria: oxygen- and nitrogen-responsive factors. Mol. Microbiol. 4:1793-1800. [DOI] [PubMed] [Google Scholar]
- 35.Labes, M., A. Pühler, and R. Simon. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for Gram-negative bacteria. Gene 89:37-46. [DOI] [PubMed] [Google Scholar]
- 36.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 37.Lowery, R. G., and P. W. Ludden. 1988. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem. 15:16714-16719. [PubMed] [Google Scholar]
- 38.Martin, D. E., and B. Reinhold-Hurek. 2002. Distinct roles of PII-like signal transmitter proteins and amtB in regulation of nif gene expression, nitrogenase activity, and posttranslational modification of NifH in Azoarcus sp. strain BH72. J. Bacteriol. 184:2251-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masepohl, B., W. Klipp, and A. Pühler. 1988. Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus. Mol. Gen. Genet. 212:27-37. [DOI] [PubMed] [Google Scholar]
- 40.Masepohl, B., R. Krey, and W. Klipp. 1993. The draTG gene region of Rhodobacter capsulatus is required for posttranslational regulation of both the molybdenum and the alternative nitrogenase. J. Gen. Microbiol. 139:2667-2675. [DOI] [PubMed] [Google Scholar]
- 41.Masepohl, B., T. Drepper, A. Paschen, S. Groβ, A. Pawlowski, K. Raabe, K.-U. Riedel, and W. Klipp. 2002. Regulation of nitrogen fixation in the phototrophic purple bacterium Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 4:243-248. [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 44.Pawlowski, A., K.-U. Riedel, W. Klipp, P. Dreiskemper, S. Groβ, H. Bierhof, T. Drepper, and B. Masepohl. 2003. Yeast two-hybrid studies on interaction of proteins involved in regulation of nitrogen fixation in the phototrophic bacterium Rhodobacter capsulatus. J. Bacteriol. 185:5240-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierrard, J., P. W. Ludden, and G. P. Roberts. 1993. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J. Bacteriol. 175:1358-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 47.Reyes, F., M. D. Roldan, W. Klipp, F. Castillo, and C. Moreno-Vivian. 1996. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductases. Mol. Microbiol. 19:1307-1318. [DOI] [PubMed] [Google Scholar]
- 48.Saari, L. L., E. W. Triplett, and P. W. Ludden. 1984. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem. 25:15502-15508. [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Simon, R., U. Priefer, and A. Pühler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 51.Son, H. S., and S. G. Rhee. 1987. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII protein and nucleotide sequence of its structural gene. J. Biol. Chem. 262:8690-8695. [PubMed] [Google Scholar]
- 52.Thomas, G., G. Coutts, and M. Merrick. 2000. The glnKamtB operon. A conserved gene pair in prokaryotes. Trends Genet. 16:11-14. [DOI] [PubMed] [Google Scholar]
- 53.Van Heeswijk, W. C., S. Hoving, D. Molenaar, B. Stegeman, D. Kahn, and H. V. Westerhoff. 1996. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 21:133-146. [DOI] [PubMed] [Google Scholar]
- 54.Van Heeswijk, W. C., D. Wen, P. Clancy, R. Jaggi, D. L. Ollis, H. V. Westerhoff, and S. G. Vasudevan. 2000. The Escherichia coli signal transducers PII (GlnB) and GlnK form heterotrimers in vivo: fine tuning the nitrogen signal cascade. Proc. Natl. Acad. Sci. USA 97:3942-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 56.Wang, H., C. C. Franke, S. Nordlund, and A. Noren. 2005. Reversible membrane association of dinitrogenase reductase activating glycohydrolase in the regulation of nitrogenase activity in Rhodospirillum rubrum: dependence on GlnJ and AmtB1. FEMS Microbiol. Lett. 253:273-279. [DOI] [PubMed] [Google Scholar]
- 57.Weaver, P. F., J. D. Wall, and H. Gest. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207-216. [DOI] [PubMed] [Google Scholar]
- 58.Xu, Y., E. Cheach, P. D. Carr, W. C. Van Heeswijk, H. V. Westerhoff, S. G. Vasudevan, and D. L. Ollis. 1998. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol. 282:149-165. [DOI] [PubMed] [Google Scholar]
- 59.Yakunin, A. F., and P. C. Hallenbeck. 1998. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase responses to NH4+ addition. J. Bacteriol. 180:6392-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yakunin, A. F., T. V. Laurinavichene, A. A. Tsygankov, and P. C. Hallenbeck. 1999. The presence of ADP-ribosylated Fe protein of nitrogenase in Rhodobacter capsulatus is correlated with cellular nitrogen status. J. Bacteriol. 181:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yakunin, A. F., and P. C. Hallenbeck. 2000. Regulation of nitrogenase activity in Rhodobacter capsulatus under dark microoxic conditions. Arch. Microbiol. 173:366-372. [DOI] [PubMed] [Google Scholar]
- 62.Yakunin, A. F., and P. C. Hallenbeck. 2002. AmtB is necessary for NH4+-induced nitrogenase switch-off and ADP-ribosylation in Rhodobacter capsulatus. J. Bacteriol. 184:4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2001. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: roles in sensing ammonium and energy status. J. Bacteriol. 183:6159-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., D. M. Wolfe, E. L. Pohlmann, M. C. Conrad, and G. P. Roberts. 2006. Effect of AmtB homologues on the post-translational regulation of nitrogenase activity in response to ammonium and energy signals in Rhodospirillum rubrum. Microbiology 152:2075-2089. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2000. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J. Bacteriol. 182:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]