Abstract
We present a comprehensive analysis of carbohydrate uptake systems of the soil bacterium Mycobacterium smegmatis and the human pathogen Mycobacterium tuberculosis. Our results show that M. smegmatis has 28 putative carbohydrate transporters. The majority of sugar transport systems (19/28) in M. smegmatis belong to the ATP-binding cassette (ABC) transporter family. In contrast to previous reports, we identified genes encoding all components of the phosphotransferase system (PTS), including permeases for fructose, glucose, and dihydroxyacetone, in M. smegmatis. It is anticipated that the PTS of M. smegmatis plays an important role in the global control of carbon metabolism similar to those of other bacteria. M. smegmatis further possesses one putative glycerol facilitator of the major intrinsic protein family, four sugar permeases of the major facilitator superfamily, one of which was assigned as a glucose transporter, and one galactose permease of the sodium solute superfamily. Our predictions were validated by gene expression, growth, and sugar transport analyses. Strikingly, we detected only five sugar permeases in the slow-growing species M. tuberculosis, two of which occur in M. smegmatis. Genes for a PTS are missing in M. tuberculosis. Our analysis thus brings the diversity of carbohydrate uptake systems of fast- and a slow-growing mycobacteria to light, which reflects the lifestyles of M. smegmatis and M. tuberculosis in their natural habitats, the soil and the human body, respectively.
The growth and nutritional requirements of mycobacteria have been intensely studied since the discovery of Mycobacterium tuberculosis (32). This resulted in an overwhelming body of literature on the physiology of mycobacterial metabolism in the years before the dawn of molecular biology (20, 53, 54). Carbon metabolism of mycobacteria has attracted renewed interest since the discovery that M. tuberculosis relies on the glyoxylate cycle for survival in mice (36, 41). This observation indicates that M. tuberculosis uses lipids as the main carbon source during infection. On the other side, genes that encode a putative disaccharide transporter were essential for M. tuberculosis during the first week of infection, indicating that M. tuberculosis may switch its main carbon source from carbohydrates to lipids with the onset of the adaptive immune response (61). However, the nutrients and the corresponding uptake proteins are unknown for M. tuberculosis inside the human host. Surprisingly, this is also true for M. tuberculosis growing in vitro and for Mycobacterium smegmatis, which is often used as a fast-growing, nonpathogenic model organism to learn more about basic mycobacterial physiology. There is no doubt that the uptake pathways have been adapted to the habitats of M. tuberculosis and M. smegmatis, the human body and soil, respectively. Thus, much can be learned about the lifestyles of both organisms by a comparison of the complements of specific nutrient uptake proteins. Previously, 38 ATP-binding cassette (ABC) transport proteins have been identified in M. tuberculosis by bioinformatic analysis, 4 of which were assigned a role in carbohydrate import (9).
The goal of this study was to compile a complete list of potential carbohydrate uptake systems of M. smegmatis and M. tuberculosis based on in silico analyses of their genomes. While M. tuberculosis has only 5 recognizable carbohydrate import systems, M. smegmatis has 28 of such transporters at its disposal. In particular, we show that the genome of M. smegmatis encodes a phosphotransferase system (PTS) that plays a fundamental role in the global control of sugar metabolism in both gram-negative and gram-positive bacteria (12). Furthermore, we show by reverse transcription (RT)-PCR and uptake experiments that the PTS genes and other selected systems are functionally expressed in M. smegmatis.
MATERIALS AND METHODS
Chemicals and enzymes.
Chemicals were purchased from Merck, Roth, or Sigma at the highest purity available. Enzymes for DNA restriction and modification were obtained from New England Biolabs, MBI Fermentas, and Boehringer. Oligonucleotides were obtained from MWG-Biotech AG.
Bacterial strains and growth conditions.
M. smegmatis mc2155 was grown in liquid cultures using Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol and 0.05% Tween 80 or minimal Hartmans-de Bont (HB) medium (65) at 37°C. M. smegmatis mc2155 was grown on plates using Middlebrook 7H10 medium supplemented with 0.5% glycerol.
Computer analyses and screening strategies.
Protein sequences of known carbohydrate uptake systems were used to screen the genome sequence of M. smegmatis mc2155 at the BLAST server of the National Center for Biotechnology Information (NCBI) at the National Institutes of Health, Bethesda, MD (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), using TBLASTN. A data file containing the preliminary genome sequence of M. smegmatis mc2155 containing 7,278,076 nucleotides was obtained from The Institute for Genomic Research (TIGR) (www.tigr.org). This file was loaded into the ARTEMIS software available at the Wellcome Trust Sanger Institute (www.sanger.ac.uk) to annotate gene and protein sequences. The open reading frames (ORFs) and their adjacent genes were checked by visual inspection to detect the most likely start codon that is preceded by a ribosome binding site. Sizes of ORFs were further established by a consideration of codon bias analysis as implemented in Artemis and by multiple sequence alignments with well-characterized homologs. For the latter, the M. smegmatis ORFs were subjected to general PBLAST data bank searches at the NCBI website (www.ncbi.nlm.nih.gov) to detect closely related sequences. Finally, the identified genes were cross-checked with the primary annotation protein list (http://cmr.tigr.org). To find the most representative homologs, we used single genome protein BLAST, which is available for well-characterized bacterial species of diverse phylogenetic origins: Escherichia coli, Bacillus subtilis, M. tuberculosis (all accessible at http://genolist.pasteur.fr), and Streptomyces coelicolor (http://avermitilis.ls.kitasato-u.ac.jp). Prediction of the possible substrate(s) was based on the following criteria: (i) protein identity of the M. smegmatis protein was more than 35% to the homologous protein of E. coli or B. subtilis, more than 50% to Streptomyces, or more than 60% to M. tuberculosis; (ii) more than one gene of the operon was conserved; and, most importantly, (iii) a solid biochemical analysis of the homologous protein was available. Sequence alignments were conducted with CLUSTALW by applying predefined algorithms available from the European Bioinformatics Institute at The European Molecular Biology Laboratory (www.ebi.ac.uk/clustalw).
Growth of M. smegmatis on sugars.
A 4-ml culture of M. smegmatis mc2155 was grown in HB minimal medium supplemented with 0.05% Tween 80 and 1% sugars as the single carbon source (65). Cells were passed through a filter with a pore size of 5 μm to remove cell clumps and were then inoculated into 50 ml HB medium. The 50-ml cultures were grown until an optical density at 600 nm (OD600) of 1 was reached. The bacteria were harvested at 4,000 rpm at 4°C for 10 min and washed twice with minimal medium without a carbon source. The pellet was resuspended in 5 ml HB medium and diluted to an OD600 of 0.02 in 50 ml HB medium containing 1% carbon source. Growth rates were determined in three independent cultures by OD600 measurements every 3 h.
Reverse transcriptase PCR.
Cells of M. smegmatis mc2155 were grown in HB minimal medium with 50 mM glycerol. In other cultures, 50 mM of the carbon source of interest was added to examine gene-specific induction. The cultures were harvested at mid-exponential phase and subjected to total RNA preparation using a procedure that we described previously (68). Quantitative RT-PCR experiments were conducted as described previously (73). RT-PCRs were performed with gene-specific oligonucleotides, which were AACTGTGCTTTCTCAACCG and ATGGCGTCGAGTTGGTGC for ptsI, TCACCGTCGGATCTGCCGTCG and ACCAGTTCGGCAACCTTGGC for ptsH, AGGCATCAACGTGGCCAAGG and ACCGCGTGATCGCATCGAGCG for fruK, ACCGAGTTCCTGTTCCTCG and CGAGCGTCGTGACCATCG for ptsI, GGGCATCCTCACGTCAGG and CAGCAGGTCGATCAGACC for ptsG, TCAGACCGTGACCATCACG and TGGACCAGCACTCCCAC for crr, and GCAAGGTGCTTCCGTTCAGC and CGAGACCGATGATCACCG for glpF1. The assay mixture contained 100 ng of RNA and 5 pmol of each primer in a 20-μl volume. Samples of 4 μl of each reaction mixture were taken at appropriate PCR cycles (cycles 21 to 36 depending on the appearance of a signal in the linear range), and amplification products were separated and visualized on a 1% agarose gel. RT-PCR experiments without prior RT were performed to ensure the exclusion of DNA contamination. The quality of the RNA preparations was checked by the presence of equal amounts of 16S rRNA, which is constitutively expressed. Data were verified in two independent experiments.
Transport assays.
Sugar uptake measurements were carried out as previously described (67). To reduce aggregation and clumping, all M. smegmatis cells were filtered through a 5-μm-pore-size filter (Sartorius) and regrown for 2 days at 37°C before inoculating 100-ml cultures (69). Cells were grown in the presence of 0.2 or 0.4% of the respective carbon source compared to glycerol as the standard carbon source and harvested by centrifugation (1,250 × g at 4°C for 10 min) when they had reached the mid-exponential phase at an OD600 of between 0.5 and 0.7. The cells were washed once in 2 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.5)-0.05 mM MgCl2 and resuspended in the same buffer. Radiolabeled [14C]fructose, [14C]glucose, and [14C]glycerol were added to the cell suspension to obtain final sugar concentrations of 20 μM, 20 μM, and 100 μM, respectively, and a radioactivity of 100,000 cpm per ml cells. The mixtures were incubated at 37°C for glycerol uptake and 25°C for glucose and fructose uptake assays. Cell suspensions (between 0.2 and 1 ml) were filtered through a 0.45-μm-pore-size filter (Sartorius) and washed with 0.1 M LiCl, and the radioactivity was determined by using a liquid scintillation counter (Beckman). All experiments were reproduced by at least one biological replicate.
RESULTS
Genome analysis reveals 28 putative carbohydrate uptake systems in M. smegmatis.
The sequence of the almost finished genome of M. smegmatis mc2155 was searched for homologs of well-characterized bacterial sugar transport systems to identify possible carbohydrate uptake systems. Table 1 shows a list of 28 putative carbohydrate permeases: 19 of the ABC family, 3 of the PTS family, 1 of the major intrinsic protein family (MIP), 4 of the major facilitator superfamily (MFS), and one of the sodium solute superfamily (SSS) (6, 47-49, 56).
TABLE 1.
Sugar transport systems of M. smegmatis
| Family and predicted substrate(s) | Gene designation(s) | Locus tag(s)a | Representative homolog(s)b or description | Reference(s) and/or source(s)c |
|---|---|---|---|---|
| ABC | ||||
| β-Glucosides, chitobiose, disaccharides | nagB1 bglA bglR sugK bglEFGK | 0501-0508 | nagB (SCO5236), bgl (SCO0670), deoR, rbsK E. coli; abcEFG genes are distantly related to many ABC permease genes; msiK (SCO4240) | 6, S, C |
| α-Galactosides, melibiose | agaRZSXPAEFGK mspB | 0509-0517 | SCO5848-SCO5851, SCO0538-SCO0541, msiK (SCO4240) | 6, 11, S |
| Unknown | abcEFGK | 0553-0556 | Distant similarity to many ABC permease genes | S, C |
| Ribose, xylose | rbsA1C1B1 gatY sugK rbsR1 pfkB | 1372-1378 | SCO6009-SCO6011, gatY (SCO5852), pfkB (SCO3197); distantly related to ribose and xylose ABC transporters | 3, 6, S, C, T, B |
| Xylose | xylF2G2H2 | 1704-1706 | E. coli xylFGH | 17, 70, C |
| Arabinose | pho araR araGFKEBDA | 1707-1715 | E. coli ytf operon, B. subtilis araABD | 60, B, C |
| Ribose, ribonucleosides | gap pgk tpiA secG urf rscA deoC rbsC2A2 rbsR2 sugK sugD rbsB2 ppc pgl opcA zwf tal tkt | 3084-3103 | rbsH (SCO2747), rbsA (SCO2746), E. coli rbsB | 3, 6, S, C |
| Unknown | abcKGFE fabG sugK | 3108-3113 | Distant similarity to many ABC permease genes | 6, S, C |
| Unknown | abcR sugK abcEFGK1K2 sugK rpiB | 3264-3272 | Distant similarity to many ABC permease genes, SCO0580, rpiB E. coli | 6, 66, S, C |
| Ribose | rbsB3R3C3A3 | 3598-3602 | SCO2747, B. subtilis ribose operon | 75 |
| Sorbitol | yphREKFB | 3998-4002 | yph operon E. coli | 6, C |
| Ribose | rbsA4C4B4R4 | 4170-4174 | E. coli ribose operon rbsCBR | 3, C |
| Unknown | uspGFE | 4466-4468 | M. tuberculosis usp operon | 9, T |
| Unknown | abcRFGE | 4655-4658 | Distant similar to many ABC permease genes | S, C |
| Unknown | sugKGFE | 5058-5061 | sug operon M. tuberculosis | 9, T |
| β-Xyloside | bglG bxlRAEFG | 5142-5147 | S. coelicolorbxlEFG2 | 6 |
| Sugar alcohol | smoKGFER | 5571-5574 | S. coelicolor smo operon | 6 |
| Xylose | xylG1F1E1A1R1 | 6018-6022 | E. coli xyl operon | 70 |
| Ribose | rbsR5P5K5G5F5E5 | 6798-6805 | SCO0723, SAV5702, SCO2747 | 6, S, C |
| PTS | ||||
| Fructose | ptsH fruAKR ptsI | 0084-0088 | S. coelicolor ptsH fruAKR ptsI | 6, 45, S |
| Glucose, trehalose N-acetylglucosamine | ptsG crr nagB2A ptsR | 2116-2120 | S. coelicolor nagE2 crr nagAB | 5, 44 |
| Dihydroxyacetone | ptsT dhaLKFR | 2121-2125 | S. coelicolor gyl operon, E. coli dhaKLM | 6, 25, S, C |
| MIP | ||||
| Glycerol | glpK2RFK1D | 6756-6760 | S. coelicolor gyl operon | 6, S |
| SSS | ||||
| Galactose | galPRTK | 3689-3692 | S. coelicolor gal operon | 6 |
| MFS | ||||
| Unknown | sugP1 | 2966 | Distant similarity to sugar permeases of the MFS | S, C, B, T |
| Unknown | sugP2 | 4078 | ||
| Glucose | glcP | 4182 | S. coelicolor glcP | 73 |
| Unknown | sugP3 | 5559 |
Only the numbers of the locus tags (msmeg_XXXX) of the M. smegmatis mc2155 genome are shown. Numbers are according to the revised annotation (www.tigr.org).
The column contains information on representatives homologs for which experimental information is available.
The following genome servers were used for BLASTP analysis: http://genolist.pasteur.fr/SubtiList (B) for B. subtilis, http://genolist.pasteur.fr/Colibri (C) for E. coli, www.avermitilis.ls.kitasato-u.ac.jp/blast_local/index.html (S) for S. coelicolor, and http://genolist.pasteur.fr/Tuberculist (T) for M. tuberculosis.
ABC systems.
Operons for carbohydrate-specific transporters of the ABC family always contain a gene encoding a sugar-specific periplasmic binding protein (6, 9). We have conducted TBLASTN analyses of the M. smegmatis genome with known binding proteins of ABC systems, such as the maltose- and ribose-specific binding proteins MalE and RbsB, respectively, of E. coli and the cellobiose binding protein of Streptomyces reticuli (19, 20, 62). We found 18 genes for ABC-type sugar binding proteins in the genome of M. smegmatis mc2155. All were adjacent to ABC permease genes (Fig. 1 and Table 1). For all sugar transport systems, a substrate was predicted when the following criteria were met (6): (i) a solid biochemical analysis of the homologous proteins was available, (ii) the identity of the M. smegmatis protein to the homologous proteins of E. coli or B. subtilis was greater than 35%, and that to Streptomyces was greater than 35% and 50%, respectively, and (iii) more than one gene of the operon was conserved, and (iv) the encoded proteins had the same carbohydrate specificity. In this way, we predict that M. smegmatis can transport via ABC permeases, β-glucosides such as chitobiose, α-galactosides (melibiose), β-xylosides (xylobiose), xylose, arabinose, and sugar alcohols. In addition, we propose that M. smegmatis has several ABC systems for ribose or ribose-like substrates such as ribonucleosides, ribitol, or xylitol (Table 1).
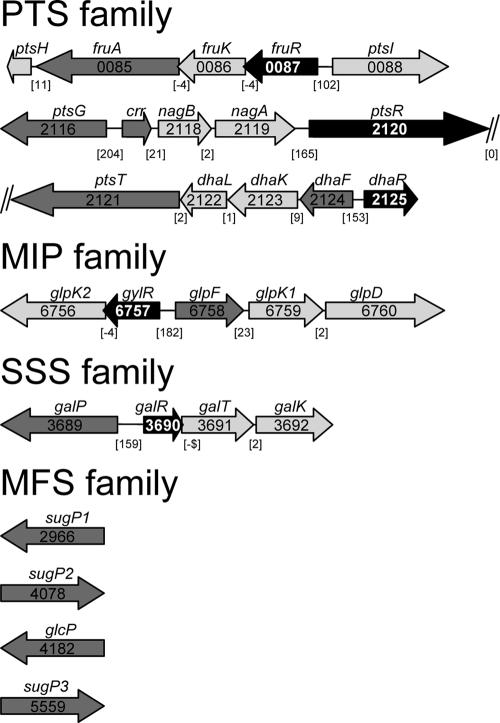
FIG. 1.
Genetic organization of M. smegmatis carbohydrate transporters of the ABC family. The arrows indicate the lengths and transcriptional orientations of annotated genes and predicted ORFs. Genes encoding transport systems are depicted in dark gray, carbohydrate metabolic genes are colored in light gray, and regulatory genes are highlighted in black, while other genes are white. Genes are shown by their number, with the prefix “msmeg_.” The gene names are assigned according to the annotations given by TIGR (http://www.tigr.org) and by us. Numbers in brackets refer to the intergenic distance between two genes. General gene designations are as follows: urf, unknown reading frame; sugD, sugar dehydrogenase; sugK, sugar kinase; sugP, sugar permease; abcE, unspecified substrate binding protein of an ABC permease; abcF and abcG, unspecified membrane proteins of an ABC permease.
(i) β-Glucosides (msmeg_0501 to msmeg_0508).
The first sugar transport cluster in the genome contains genes for a glucosamine isomerase (nagB1), a β-glucosidase (bglA), a sugar kinase (sugK), and a complete ABC permease, the latter with distant similarities to other ABC importers. Due to the presence of the metabolic genes, whose products BglA and NagB exhibit 31% and 32% protein identity to homologs of B. subtilis and E. coli, we suggest that the permease catalyzes the uptake of β-glucosides such as cellobiose (38, 72). It should be noted as well that the closest homologs were the S. coelicolor proteins SCO5236 (NagB homolog with 50% identity) and SCO6670 (BglA homolog with 54% identity).
(ii) α-Galactosides (msmeg_0509 to msmeg_0517).
A gene cluster directly downstream of the putative β-glucoside cluster may be responsible for the uptake of α-galactosides. It comprises genes encoding an α-galactosidase (msmeg_0514), a tagatose-bisphosphate aldolase (agaZ), and a putative isomerase (agaS) besides the genes for the ABC permease, agaEFG. The operon is likely controlled by AgaR, a regulator of the DeoR family (11, 51). Downstream of the aga region is the gene mspB, which encodes a porin (69), which may indicate that this porin is required for the entry of the substrate transported by AgaEFG. The proteins share, as many other proteins from M. smegmatis do, the highest identity to proteins from S. coelicolor (39 to 71%) (Table 1).
(iii) Ribose and ribose-like carbohydrates (msmeg_1372 to msmeg_1378, msmeg_3090 to msmeg_3095, msmeg_3598 to msmeg_3602, msmeg_4170 to msmeg_4174, and msmeg_6798 to msmeg_6804).
Several of the analyzed ABC systems of M. smegmatis revealed similarities to the ribose ABC permeases of E. coli and B. subtilis (3, 75). The best candidates for a ribose-specific ABC permease seem to be msmeg_3090, msmeg_3091, and msmeg_3095. The latter one is the substrate binding protein that shares the highest identity (30%) to the ribose-specific periplasmic binding protein RbsB. Interestingly, this putative ribose permease is embedded within a cluster of genes involved in central carbon metabolism, such as glycolysis. The other four ABC systems with similarities to ribose permeases may also transport ribose or a ribose derivative (Table 1).
(iv) Xylose and β-xylosides (msmeg_1704 to msmeg_1706, msmeg_5142 to msmeg_5147, and msmeg_6018 to msmeg_6022).
Xylose is usually taken up by an ABC permease and further metabolized by isomerization (xylose isomerase [XylA]) and phosphorylation (xylulokinase [XylB]) to enter the pentose-phosphate shunt. The region msmeg_6018 to msmeg_6022 was designated the xylGFEAR1 regulon since it encodes proteins sharing identities of 29% to 38% with the corresponding proteins of E. coli (7, 20). The substrate binding protein XylF1 has the highest score, with 38% identity to the E. coli homolog (70). The regulon further contains the metabolic xylA1 gene. The missing XylB may be encoded by msmeg_3257, having 50% and 32% identity to XylB of Corynebacterium glutamicum and E. coli, respectively (31, 33). A second potential operon for a xylose ABC transporter is encoded by msmeg_1704 to msmeg_1706 (xylFGH2). XylF2, XylG2, and XylH2 share 38% to 48% identity to the corresponding E. coli proteins.
A predicted β-xyloside permease is encoded by msmeg_5142 to msmeg_5147. We have designated the genes bxlRAEFG according to their close relationship to the bxlEFG2 operon of S. coelicolor, which encodes the transporter for xylobiose (6, 27). The amino acid identities are in the range of 45% to 79%.
(v) Arabinose (msmeg_1708 to msmeg_1715).
We detected an araBDA operon that is most similar to the arabinose metabolic genes of B. subtilis (40). AraB (l-ribulokinase), AraD (l-ribulose-5-phosphate-4-epimerase), and AraA (l-arabinose isomerase) show 42 to 48% identity to the corresponding proteins of B. subtilis. The adjacent ABC permease (msmeg_1709 to msmeg_1712) could be the uptake system for arabinose. Its gene products, including the juxtaposed regulator, exhibit residues that are up to 43% identical to an unknown gene cluster of E. coli that is designated ytfRT yjfF. This suggests that the E. coli locus encodes a sugar-specific permease. Transport of arabinose has so far been described only for the E. coli proton symporter AraE of the MFS (34, 47).
(vi) Sugar alcohols (msmeg_3998 to msmeg_4002 and msmeg_5571 to msmeg_5575).
Sugar alcohols like mannitol, glucitol (sorbitol), and xylitol are frequently consumed by bacteria (52). We found two loci that could encode a sugar alcohol-specific transport system. The operon comprising msmeg_3998 to msmeg_4002 is homologous to the yph operon of E. coli. Although the permease proteins do not allow substrate prediction, they are associated with a sugar alcohol dehydrogenase (msmeg_4002), which is 44% identical to YphC and more distantly related to GutB of B. subtilis (sorbitol dehydrogenase) and GatD (galactitol-1P dehydrogenase) of E. coli (43). The second region, msmeg_5571 to msmeg_5575, is homologous to the smo operon of S. coelicolor, encoding a possible permease for sugar alcohols. Here, the two substrate-specific binding SmoE proteins share 52% identity (6).
(vii) Less-well-defined ABC-type sugar transporters.
We further found six gene loci encoding ABC transport systems for carbohydrates. The deduced substrate binding proteins were homologous to known sugar substrate binding proteins in the range of about 30% protein identity. Due to this and due to the absence of adjacent metabolic genes, prediction of substrates was not possible. Among those are the only two permeases that are common to M. smegmatis and M. tuberculosis, SugABC and UspABC (see below).
(viii) Lipid anchors of sugar binding proteins.
In gram-negative bacteria, the substrate binding proteins of ABC transporters are soluble proteins that appear to move freely through the periplasmic space. By contrast, it has been shown for gram-positive bacteria that the substrate binding proteins are covalently anchored to the outside of the cell membrane by fatty acids (63). Lipidation occurs via esterification of a conserved cysteine at the N terminus of the processed protein. Strikingly, all sugar binding proteins of M. smegmatis (Table 2) and M. tuberculosis (9) have a predicted lipoprotein signal peptide.
TABLE 2.
N-terminal sequences of the periplasmic binding proteins of ABC sugar transporters in M. smegmatisa
| Protein | Locus tag | N-terminal sequence (no. of residues) |
|---|---|---|
| * | ||
| BglE | msmeg_0505 | MTRTRLFRFGSAVASTLTVAALALSACAPGPSGDSGSSPAPTGEVSKDI (49) |
| AgaE | msmeg_0515 | MIRRWLCLAVVTAVACLLTACGGGSSSSGPVEIAVWHGYQDTE (43) |
| AbcE | msmeg_0553 | VTSPAFTRRRALQLLGLAGGAAMLAPALAACGSSGGNSALAADAPVSGRFEGV (53) |
| RbsB1 | msmeg_1374 | VNRKRLMLAAGVVALALPMAACTSSKPQADESSETPAAAGEAPA (44) |
| XylF1 | msmeg_1704 | MRKLTWLAALLAALAMAMTLSGCGRSAEGGGGGDGDAKGTVGIAM (45) |
| AraE | msmeg_1712 | VRKMFAAAIGVVAVAAAVTACGSGKAPGSEGGSAPDGALTLGF (43) |
| RbsB2 | msmeg_3095 | VSFAKALSGIALGAAMALSFTGCSVPGDDAAQNAPVVDGALKIGF (45) |
| AbcE | msmeg_3111 | MKIPQLRRRRRRPAITAITAMSVAAGLVLSGCAGTGGPANDEATSGVGDVPTDT (54) |
| AbcE | msmeg_3266 | MMSRESQPGLHRQLSRRNMLAAMGLAGAAAVSLPVLSACGVGGRTNAPNGASEVTGGFDWR (61) |
| RbsB3 | msmeg_3599 | MRLGTTAFAIASATALGLGLTACGAGDPAANSDTTRIGVTVYDMS (45) |
| YphE | msmeg_3999 | MPRSLRRRAVRFATLMLVAALAVSGCSRIGENGRIAVVYLNAEGFYAG (48) |
| RbsB4 | msmeg_4172 | MKIRNILPILVCTTCAVAMTACSSSVNNNADPSDTAAPATNVEV (44) |
| AbcE | msmeg_4658 | MRLSRLVAAAGVGVLMLGASACSSTGGKPDSSGGGDMGAGTADT (44) |
| SugE | msmeg_5061 | VRARRLCAAAVAAMAAASMVSACGSQTGGIVINYYTPANEEATFK (45) |
| BxlE | msmeg_5145 | VDLKSVDANVVESKADFLPSTSRRAFLAAALSVPLLGALAACGSSGPSRSGGGGGGAPGAASYW (64) |
| SmoE | msmeg_5574 | MKRLRRLAACIAAAGLTATAGCAGAGTLGATDQTVTIAMVSNSQ (44) |
| XylE2 | msmeg_6020 | VKRTSTLLVTAVVGLGLTLTACGSDSGSNAGSAEGGSGGKIGVI (40) |
| RbsB5 | msmeg_6804 | MFRKVTRNTRTVGAALMAGSLVLGMTACGGSGSDGVKVGLITKTDSNPYF (50) |
The sequences were aligned according to their signal peptide cleavage site as predicted by SignalP (4). The N-terminal cysteine of the mature protein, which is the catalytic residue of the fatty acid anchor, is marked with an asterisk.
PTS permeases. (i) Glucose, trehalose, GlcNAc, and dihydroxyacetone (msmeg_2116 to msmeg_2125).
Two loci encoding components of a PTS were discovered in the genome of M. smegmatis. This is interesting, since the existence of PTS genes is in contrast to previous reports in which no evidence for PTS proteins in M. smegmatis was found in biochemical experiments (14, 15, 54, 59). Genes for a PTS of the glucose-sucrose subfamily (Fig. 2) were detected in a cluster comprising msmeg_2116 to msmeg_2120 (52). The two divergently transcribed genes msmeg_2116 (ptsG) and msmeg_2117 (crr) encode the IIBC and IIA permeases. The deduced protein sequence of IIBC (496 amino acids [aa]) shares 49% identity with the N-acetylglucosamine (GlcNAc)-specific IIBGlcNAc and IICGlcNAc of S. coelicolor A3(2) and 42 to 43% identity to PTS permeases of E. coli and B. subtilis that transport glucose, GlcNAc, trehalose, or β-glucosides, respectively (52). The IIA protein of M. smegmatis is 50% identical to the S. coelicolor homolog IIACrr (30). Two metabolic genes for the metabolism of GlcNAc are situated downstream of the crr gene. They encode a putative glucosamine-6-phosphate deaminase, NagB2, and a GlcNAc deacetylase, NagA, that share 43% and 33% identity to the characterized E. coli homologs and 59% and 47% to the ones from S. coelicolor, respectively (1). The gene locus further contains a putative regulatory gene, ptsR, whose product shares low similarities to uncharacterized regulators of S. coelicolor and M. tuberculosis. Thus, PtsR might be a possible regulator of the PTS genes.
FIG. 2.
Genetic organization of M. smegmatis carbohydrate transporters of the PTS, MIP, SSS, and MFS protein families. For an explanation of the data, see the legend to Fig. 1.
Downstream of ptsR is a gene for a multiphosphoryl PTS phosphotransferase. The gene, which we have designated ptsT, encodes a protein of 808 aa that comprises all three general phosphotransferases of a PTS: IIA (aa 1 to 150), HPr (aa 151 to 251), and EI (aa 251 to 808). The protein resembles the multiphosphoryl phosphotransferase MTP from Rhodobacter capsulatus (76), which drives fructose uptake by phosphorylation of the respective FruA PTS permease, in length and domain structure. Hence, PtsT may be involved in the phosphorylation of the adjacent PTS permease PtsG. In addition, PtsT is a homolog of the multiphosphoryl phosphotransferase DhaM of E. coli. DhaM undergoes phosphoenolpyruvate-dependent autophosphorylation and then phosphorylates ADP through its enzyme IIA domain. This ATP molecule is used by the dihydroxyacetone kinase DhaKL for substrate phosphorylation (21, 25). Strikingly, homologs of dhaKL are, as in E. coli, juxtaposed with ptsT (dhaM). An adjacent permease gene, which we have termed dhaF, does not exist in E. coli. We predict that PtsT serves as the shuttle for the transfer of phosphate to DhaKL kinase, which in turn phosphorylates dihydroxyacetone molecules that are imported through the MIP family facilitator DhaF.
(ii) Fructose (msmeg_0084 to msmeg_0088).
The second PTS locus comprises genes for a fructose-specific PTS composed of EI (ptsI), HPr (ptsH), and IIABCFru (fruA) (Fig. 2B). The locus further consists of a gene coding for a regulator of the DeoR family (51) and a fruK gene coding for a protein similar to fructose-1-phosphate kinases (45). The gene order fruR-fruK-fruA is the same as what we previously found in S. coelicolor, while the fru operons of other bacteria are under the control of different regulators (52). The similarities of proteins encoded by genes in these operons are highest between M. smegmatis and S. coelicolor, with identities of 51% for the DeoR-type regulator, 43% for FruK, and 51% for FruA (IIABCFru).
MIP permeases.
Sugar permeases of the MIP family were screened by BLASTP searches with the glycerol facilitator protein sequences of E. coli and S. coelicolor. We found a protein, msmeg_6758 (GlpF), with identity scores of 42% and 37% to the respective pendants from E. coli and S. coelicolor. msmeg_6758 is situated in an operon with genes for two glycerol kinases (glpA1 and glpA2) and a glycerol-3-phosphate dehydrogenase (glpD) (Fig. 2). A putative regulatory gene (glpR) that is homologous to the glycerol operon regulator gene gylR of S. coelicolor is found upstream (26). Hence, the genes msmeg_6756 to msmeg_6760 are the best candidates to encode proteins for glycerol uptake and metabolism. As described above for PTS, we identified DhaF (msmeg_2124) as being a second MIP family member that may serve as a facilitator for dihydroxyacetone in conjunction with PTS proteins.
MFS permeases.
Well-characterized permeases of the MFS family are the xylose symporter XylE of E. coli and the glucose-specific symporter GlcP from S. coelicolor and the cyanobacterium Synechocystis (34, 73, 77). We found four homologs (msmeg_2966, msmeg_4098, msmeg_4182, and msmeg_5559), of which msmeg_4182 exhibited 53% identity to glucose symporters (Fig. 2). msmeg_4182 is surrounded by genes that are not related to sugar metabolism, and there is no putative regulatory gene in the vicinity. Hence, glcP might encode a glucose permease that is expressed constitutively in a monocistronic operon. The other three candidates showed very distant similarities to sugar transporters, thus making a substrate prediction impossible.
Growth of M. smegmatis on carbohydrates as single carbon sources.
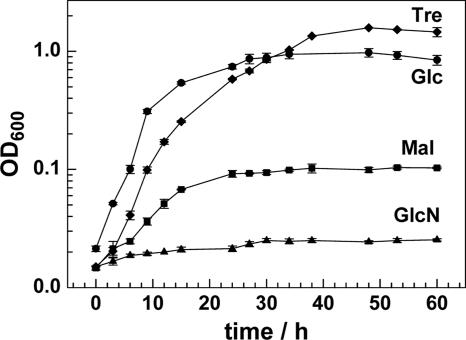
According to our genome analysis, M. smegmatis possesses uptake systems for a large number of sugars. This is consistent with very early findings that M. smegmatis can grow on many monosaccharides, organic acids, and sugar alcohols (20). In those studies, oxygen consumption of starved bacteria with low endogenous respiration was measured after the addition of single carbon sources to bacteria suspended either in phosphate buffer or in phosphate saline (57). However, in those experiments, M. smegmatis may have lacked other elements that may be required to consume the substrate of interest. Therefore, we conducted growth experiments in HB minimal medium, which contains all essential nutrients and trace elements (65), and the carbohydrate of interest as the sole carbon source. M. smegmatis readily grew in HB minimal medium with glucose, glycerol, xylose, fructose, ribose, arabinose, or trehalose (Fig. 3 and Table 3). Potential uptake systems were assigned in this study for all of these mono- and disaccharides (see Fig. 5A). These results demonstrated that functional uptake systems exist for these sugars and that these sugars are metabolized by M. smegmatis. The generation times ranged from 132 to 174 min, which is similar to growth rates of M. smegmatis in Middlebrook 7H9 medium that contains glycerol as the main carbon source (69). The monosaccharides GlcNAc, glucosamine, and galactose; the disaccharides maltose, sucrose, and lactose; and the trisaccharide raffinose were not utilized. This may indicate the lack of uptake and/or lack of metabolic enzymes. Particularly interesting was the growth of M. smegmatis on maltose as a sole carbon source. M. smegmatis stopped growing at a very early stage at an OD600 of 0.1 (Fig. 3). The poor growth of M. smegmatis on maltose was not altered when the amount of maltose in the medium was increased from 1% to 10%, indicating that a putative, minor contaminating carbon source did not did not cause the initial residual growth of M. smegmatis. Similar results were obtained for galactose, lactose, and sucrose (not shown). The reason for this phenomenon is unknown.
FIG. 3.
Growth of M. smegmatis in minimal medium with different carbon sources. The growth of M. smegmatis mc2155 at 37°C in HB medium containing 1% glucose (circles), 1% trehalose (diamonds), 1% maltose (squares), and 1% GlcNAc (triangles) was measured by determining the OD600 of the cultures. The values are the means of three independent experiments. For some data points, the standard deviations were smaller than the symbol size, and therefore, the error bars are invisible.
TABLE 3.
Growth of M. smegmatis on various sugars as a sole carbon sourcea
| Sugar | Generation time (min) | Growthb |
|---|---|---|
| LB | 156 ± 24 | ND |
| Glycerol | 174 ± 6 | + |
| d-Xylose | 192 ± 18 | + |
| d-Ribose | 497 ± 36 | −/+ |
| l-Arabinose | 154 ± 10 | + |
| d-Fructose | 174 ± 18 | + |
| d-Glucose | 132 ± 12 | + |
| Trehalose | 150 ± 12 | ND |
Cultures of M. smegmatis mc2155 were grown on HB minimal medium containing glucose. Cell were washed twice in HB medium, resuspended, and diluted to an OD600 of 0.02 in HB medium containing 1% of the respective sugars. The generation times were calculated as described in Materials and Methods. Growth in full medium (LB, lysogeny broth) was used as a control. ND, not determined.
Data reported previously by Izumori et al. (29).
FIG. 5.
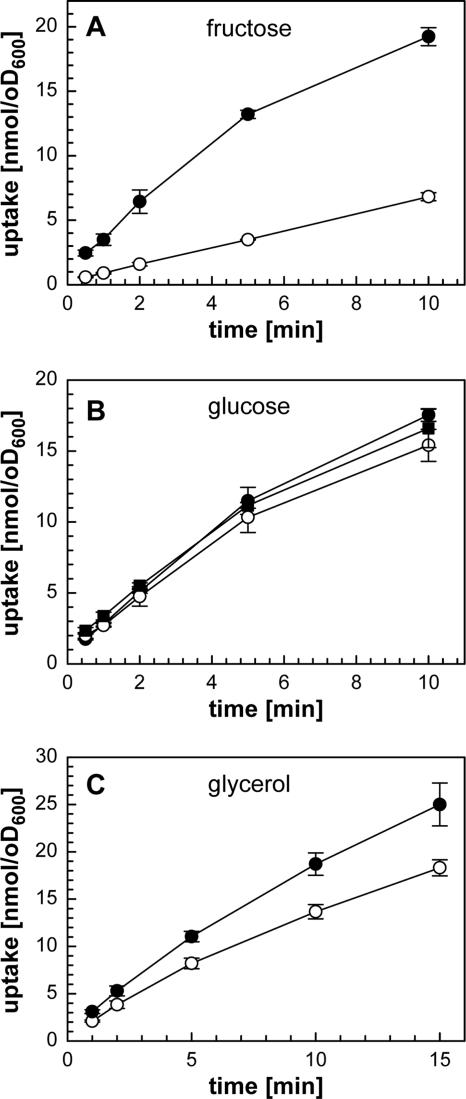
Kinetics and inducibility of fructose, glucose, and glycerol uptake by M. smegmatis. (A) M. smegmatis mc2155 was grown in Middlebrook 7H9 medium in the presence of 2% glycerol (open circles) or 2% fructose (closed circles). Accumulation of [14C]fructose was measured at 25°C at a final fructose concentration of 20 μM. (B) M. smegmatis mc2155 was grown in Middlebrook 7H9 medium in the presence of 2% glycerol (open circles), 2% glucose (closed circles), and 2% glycerol plus 2% glucose (closed squares). Accumulation of [14C]glucose was measured at 25°C at a final glucose concentration of 20 μM. (C) M. smegmatis mc2155 was grown in HB medium containing 0.4% glucose (open circles) and 0.4% glycerol (closed circles) to an OD600 of 0.9. Accumulation of [14C]glycerol was measured at 37°C at a final concentration of 100 μM glycerol. For A and B, the standard deviations are indicated by error bars and represent the means of three independent experiments. For C, samples were taken in duplicates from two biological replicates.
Analysis of the expression of the fructose, glucose, and glycerol import systems.
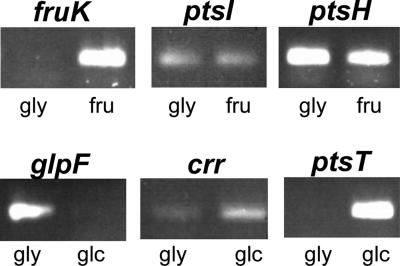
In order to validate the predicted assignment of inner membrane transport systems to a specific sugar, we examined the transcription of selected genes by semiquantitative RT-PCR. First, the transcription of the genes necessary for fructose uptake and utilization were analyzed. The gene msmeg_0086, predicted to encode a fructose-1-phosphate kinase (FruK), showed enhanced mRNA levels in cells grown in the presence of fructose (Fig. 4). Transcription of the adjacent genes ptsH and ptsI, encoding general components of the PTS, was not enhanced by fructose. Transport experiments with [14C]fructose were done to examine whether fructose induces the expression of its uptake system. Figure 5A clearly shows that fructose uptake is enhanced in M. smegmatis when grown in the presence of 2% fructose. This demonstrates that fructose uptake is inducible in M. smegmatis. These results confirm the prediction that msmeg_0085 and msmeg_0086 represent a fruAK operon in which fruA encodes a fructose-specific enzyme IIA permease, as shown previously for S. coelicolor (45). Expression of this operon is most likely controlled by the adjacent DeoR-like regulator that we have thus designated FruR (msmeg_0087).
FIG. 4.
Expression analysis of selected genes of M. smegmatis. The figure shows 1% agarose gels with PCR products from semiquantitative RT-PCR experiments. For each gene, samples were taken periodically along the PCR. The depicted bands show products from the same cycle for each gene, when the amplification was in the linear range. RT-PCR experiments without a reverse transcriptase reaction delivered no signal (negative control). Data were obtained from two biological replicates. gly, glycerol; fru, fructose; glc, glucose.
Two systems were predicted for the uptake of glucose: the symporter GlcP and a glucose-specific PTS. The transcription of crr and ptsT was induced in cells grown in the presence of glucose (Fig. 4), indicating that their assignments as glucose-specific PTS enzyme IIA and tripartite PTS phosphotransferase are correct. Uptake of [14C]glucose by M. smegmatis was not altered when cells were grown in the absence or presence of 2% glucose (Fig. 5B). One possibility to explain this result is that glcP is expressed constitutively. However, the glcP mRNA was not detected in RT-PCR experiments for unknown reasons, in contrast to mRNA of control genes such as sigA (not shown). An alternative explanation is that the rate-limiting step for glucose uptake is diffusion across the outer membrane of M. smegmatis. Indeed, porin mutants of M. smegmatis show a significantly impaired glucose uptake (67, 69). To date, it is unknown whether porin-mediated diffusion of glucose is the rate-limiting step in wild-type M. smegmatis. The fact that uninduced glucose uptake is as fast as the induced uptake of fructose (Fig. 5) argues in favor of a constitutively expressed inner membrane transporter for glucose. Further experiments with mutants lacking either GlcP or PtsG are required to examine whether glucose transport is regulated in M. smegmatis.
The number of transcripts of the predicted glycerol facilitator gene glpF (msmeg_6758) was increased in cells grown in the presence of 0.4% glycerol compared to that in cells grown in the presence of 0.4% glucose (Fig. 4). Furthermore, the uptake of [14C]glycerol was slightly increased in M. smegmatis cells grown in the presence of 0.4% glycerol compared to that in cells grown in the presence of 0.4% glucose (Fig. 5C). These results confirmed the annotation of the msmeg_6758 gene as glpF.
Carbohydrate transporters of M. tuberculosis H37Rv.
We evaluated the complement of the putative carbohydrate uptake systems in the slow-growing pathogenic strain M. tuberculosis H37Rv (13). Our analysis resulted in the identification of four ABC-type systems and one permease of the MFS class (Fig. 6B). Some of these ABC transporters were described previously in an in silico analysis of the M. tuberculosis genome in a more global context (9, 15). It is striking that M. tuberculosis is poorly equipped with carbohydrate transport systems in comparison to M. smegmatis mc2155 (Fig. 6). Two of the operons, the lpgY sugABC and the uspABC operons, are highly conserved between the two species. The proteins of the ABCSug and of the ABCUsp systems share between 62% and 80% similar amino acids, compared to only 25 to 30% similar aminoacids for the UgpABCE and Rv2038c-Rv2041c systems. The similarities of all four ABC systems to known transporters outside the genus Mycobacterium is so low (<25%) that substrates of these transporters cannot be predicted.
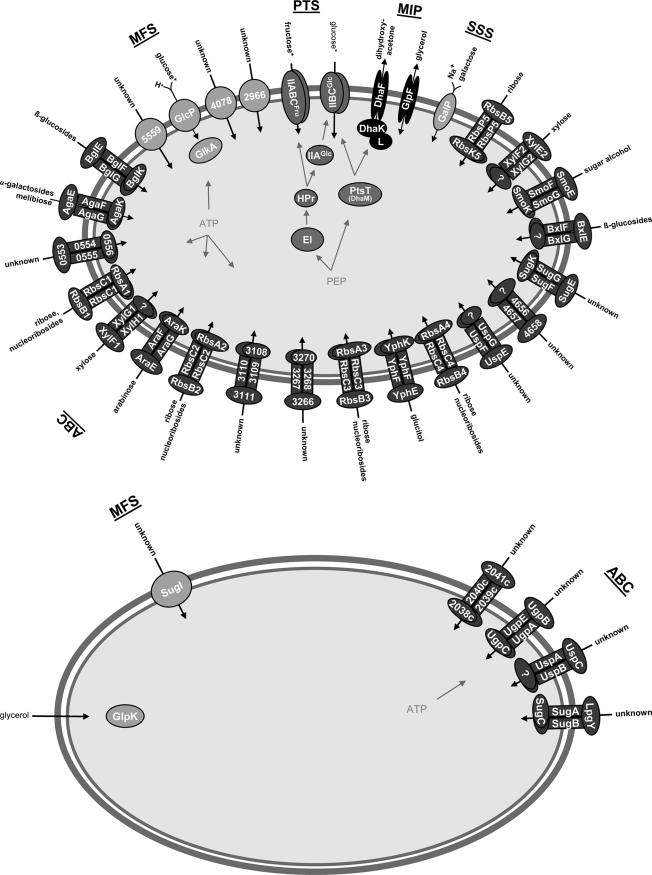
FIG. 6.
Sugar transport systems of M. smegmatis and M. tuberculosis. Shown are the permeases of the ABC, PTS, MIP, MFS, and SSS families. The derived putative substrates are inferred from in silico analyses in combination with experimental data.
The SugI porter of the MFS class shows distant sequence similarity to the glucose permease GlcP (28%) of S. coelicolor and to the galactose (GalP) (24%) and arabinose (AraE) (24%) transporters of E. coli. Thus, the system is likely to transport a monosaccharide.
Glycerol is used as the standard carbon source to grow M. tuberculosis. We did not detect any putative uptake system for this carbohydrate. Since M. tuberculosis grows with a generation time of 24 h, and it has been shown that glycerol can directly diffuse through lipid membranes both in vitro (50) and in vivo (22), it is conceivable that the rate of glycerol intake by passive diffusion may be sufficient for growth. Incoming glycerol will then be converted by glycerol kinase (GlpK) into glycerol-3-phosphate to enter the route of central carbon metabolism (Fig. 6B). M. tuberculosis has one putative glycerol kinase that shows a high similarity to the two glycerol kinases of M. smegmatis (77% protein identity for msmeg_6759 and 57% for msmeg_6756) and to the two glycerol kinases from S. coelicolor SCO0509 (75%) and SCO1660 (59%).
DISCUSSION
Identification of carbohydrate transporters in M. smegmatis.
It has been widely documented that M. smegmatis can grow on many carbon sources such as polyols, pentoses, and hexoses (20, 23, 28). In this study, we identified multiple inner membrane transport systems for all three of these classes of carbohydrates by bioinformatic analysis (Table 1). This provides the molecular basis for the adaptability of M. smegmatis to different environments in the soil and water. Often, our integrated bioinformatic approach enabled us to propose a specific substrate for particular uptake proteins (Table 1). Since the specificity of transport proteins can be altered by the modification of a few residues, the suggested substrates rather represent a hypothesis for experiments such as transport measurements with gene deletion mutants or analysis of the induction of gene expression. Analysis of the induction of both the transport activity and transcription of genes confirmed the substrate predictions for a fructose- and glucose-specific PTS as well as for the predicted glycerol operon. Since glucose uptake was constitutive, at least one more system for glucose transport must occur in M. smegmatis. This was predicted to be GlcP. Indeed, cloning and heterologous expression of glcP in E. coli revealed that it is a glucose-specific permease (data not shown).
Global control of carbon metabolism in M. smegmatis and M. tuberculosis.
The discovery of homologs of all components of a PTS in M. smegmatis contradicts a previous report that did not find biochemical evidence for the existence of a PTS (59) and many repeating statements (14, 15, 54). Components of the PTS play a key role in the global control of sugar metabolism to achieve the hierarchical utilization of carbon sources in bacteria (12), where two different mechanisms have evolved. In E. coli and other closely related gram-negative bacteria, the enzyme IIAGlc is dephosphorylated mainly under repressing conditions and mediates inducer exclusion. Under nonrepressing conditions, phosphorylated IIAGlc stimulates cyclic AMP (cAMP) synthesis and thereby triggers the activation of catabolite-repressed genes by a global regulator, the cAMP-dependent catabolite activator protein. In low-G+C-content gram-positive bacteria, HPr is a central switch of carbon catabolite repression. Under repressing conditions, HPr is phosphorylated mainly at serine 46 by a unique HPr kinase/phosphatase mediating inducer exclusion and carbon catabolite repression/activation (12, 37, 55). Under nonrepressing conditions, HPr is phosphorylated at histidine 15 and activates PTS-dependent sugar transport, glycerol kinase, and substrate-specific regulators. The apparent absence of a protein in M. smegmatis similar to the HPr kinase/phosphatase argues against the mechanism found in low-G+C-content gram-positive bacteria. On the other hand, M. smegmatis apparently does not produce proteins with significant sequence similarities to the cAMP receptor protein (CRP) (catabolite activator protein) of E. coli, which is crucial for carbon catabolite repression in gram-negative bacteria.
By contrast, the coordination of the few operons involved in the uptake and degradation of carbohydrates by M. tuberculosis may not require a global control mechanism, as suggested by the lack of PTS homologs. Alternatively, a completely different mechanism for the global control of carbon metabolism may have evolved in M. tuberculosis to adapt to its specific environment inside the phagosome of human macrophages. Indeed, M. tuberculosis has eight orthologs of CRP-like transcriptional regulators (35), one of which, Rv3676, was experimentally described (2). Furthermore, the large number and the different subcellular localization of the 15 putative nucleotide cyclases in M. tuberculosis imply that this organism may have the ability to sense and respond to many intracellular and extracellular signals through a second messenger system based on cyclic nucleotide monophosphates (35). This is in strong contrast to E. coli and other gram-negative bacteria, which have only one CRP and one adenylate cyclase. CRP homologs have been identified in streptomycetes (18), where the regulator plays a role in germination, and in corynebacteria, where CRPs have been associated with global carbon regulation (39). Although the potential mechanisms of global control of carbon metabolism in both M. smegmatis and M. tuberculosis are not evident from the bioinformatic analysis of their genomes, these findings provide hypotheses for further experiments.
Utilization of galactose by M. smegmatis and M. tuberculosis.
M. smegmatis did not grow on d-galactose as a sole carbon source. This result is in agreement with previous reports (28, 29). Surprisingly, our analysis identified the genes msmeg_3689 to msmeg_3692 as being an operon encoding, among others, a putative galactose transport protein of the SSS with more than 50% amino acid identity to the putative galactose transporter GalP of S. coelicolor (6). Furthermore, it was shown that d-galactose is taken up by M. smegmatis and that this transport activity is inducible by d-galactose (28). However, the putative gal operon of M. smegmatis contains only two of three essential genes (galKTE) of the Leloir pathway, which is used by E. coli and many other bacteria to grow on galactose (24). The first reaction in this pathway is catalyzed by galactokinase, which phosphorylates free galactose to galactose-1-phosphate. In the next steps, galactose-1-phosphate uridylyltransferase transfers the UDP residue from UDP-glucose to galactose-1-phosphate, and UDP-galactose-4-epimerase catalyzes the reversible conversions of UDP-galactose and UDP-glucose. The galE gene encoding the epimerase is missing. These results appear to be counterintuitive: why should M. smegmatis take up a sugar, modify it, and not use it as a carbon or energy source? One explanation may be provided by the fact that d-galactose is a major constituent of the cell wall of mycobacteria. The arabinogalactan polymer is composed of d-arabinose and d-galactose, both in their furanose ring form, and is an essential part of the mycobacterial cell wall by serving as an anchor for the covalent attachment of the peptidoglycan and the mycolic acids (10). Ethambutol is a potent tuberculosis drug and acts by inhibiting the synthesis of arabinogalactan (71). Based on these observations, we suggest the following sequence of reactions: d-galactose is taken up by M. smegmatis via GalP (msmeg_3689), phosphorylated in the cytoplasm by the galactokinase GalK (msmeg_3692), and uridinylated by the galactose-1-phosphate uridyltransferase GalT (msmeg_3691) to yield UDP-galactose. The ring contraction of UDP-galactopyranose to UDP-galactofuranose is catalyzed by the essential enzyme UDP-galactopyranose mutase Glf (46). UDP-galactofuranose is then likely to be transported across the cytoplasm membrane via intermediate binding to decaprenyl phosphate as in other bacteria to be available for the synthesis of arabinogalactan (16). It should be noted that no homologs of the putative galactose transporter GalP of M. smegmatis (Table 1) were found in M. tuberculosis. This suggests that M. tuberculosis has no access to d-galactose in its natural environment, the phagosome of macrophages, and instead synthesizes d-galactose from other sugars. Indeed, in addition to galK and galT, the genome of M. tuberculosis contains three galE genes (Rv3634c, Rv0501, and Rv0536), which are probably used to convert glucose to galactose for biosynthesis purposes. However, it is unclear how M. smegmatis synthesizes galactose in the absence of this sugar and GalE. M. smegmatis may either contain an undetected enzyme with glucose-galactose epimerase activity or use an alternative biosynthetic route.
Utilization of disaccharides by M. smegmatis and M. tuberculosis.
M. smegmatis did not grow on lactose, maltose, and sucrose as a sole carbon source. Franke and Schillinger previously obtained the same result for lactose and maltose but observed respiration of M. smegmatis in the presence of sucrose (23). According to our bioinformatic analysis, M. smegmatis has at least three inner membrane transport systems with significant similarities to other bacterial disaccharide transporters (Table 1). However, the substrate specificities of the transporters encoded within the loci msmeg_0501 to msmeg_0508 and msmeg_0509 to msmeg_0518 are not known. Growth of bacteria on disaccharides as sole carbon sources requires enzymes that cleave the disaccharide and release the monosaccharides for further metabolization. The absence of proteins similar to known bacterial β-d-galactosidases (LacZ of E. coli, BgaB of Bacillus circulans, MbgA of Bacillus megaterium, and LacA of S. coelicolor) provides a molecular explanation for the inability of M. smegmatis to utilize lactose as a sole carbon source. By contrast, M. smegmatis has six homologs (msmeg_3184, msmeg_3576, msmeg_4916, msmeg_4917, msmeg_4696, and msmeg_6515) of MalL of B. subtilis, which hydrolyzes maltose, longer maltodextrines up to maltohexaose, isomaltose, and sucrose (64), and of the cytoplasmic trehalase TreC of E. coli, which cleaves trehalose-6-phosphate (58). It is conceivable that these enzymes are used in trehalose metabolism, considering the unusual importance of trehalose in mycobacteria (42, 74) and the observation that trehalose was the only disaccharide that was used by M. smegmatis as a sole carbon source. However, it cannot be excluded that some of the enzymes with similarities to TreC and MalF have roles in pathways distinct from trehalose metabolism.
The SugABC sugar transport system was shown to be essential for the virulence of M. tuberculosis in mice (61). Previously, it was suggested that this permease may transport maltose or maltodextrins (8, 9). However, the similarities of both ABCSug and the corresponding substrate binding protein LpgY to the maltose transporters and periplasmic maltose binding proteins MalE of E. coli and S. coelicolor are very low (<25%). Thus, it is questionable whether maltose is the substrate of ABCSug. These doubts are supported by the fact that neither M. smegmatis, which has a highly similar ABCSug system, nor M. tuberculosis (20) grows on maltose as a sole carbon source. It has to be noted that similar uncertainties exist for the substrate specificities of the four other carbohydrate uptake systems of M. tuberculosis, including the ABCUsp transporter, which was proposed to transport sn-glycerol-3-phosphate based on low protein similarities (9, 15).
The analysis of the carbohydrate uptake proteins in the genomes of M. smegmatis and M. tuberculosis provides the molecular basis for the very early phenotypic observations that saprophytic mycobacteria have a much broader spectrum of substrates, which they can use as sole carbon and energy sources (20). It is striking that the genome of M. tuberculosis has only five recognizable permeases for carbohydrate uptake. This suggests that the phagosome does not provide an environment rich in diverse sugars. Hence, an experimental analysis of the substrate specificity of the inner membrane carbohydrate transporters of M. tuberculosis is likely to reveal the carbon sources available in the phagosome of human macrophages.
Acknowledgments
This work was supported by grant AI06432 from the National Institutes of Health to M.N. and by grants SFB473 and Graduiertenkolleg GK805 of the Deutsche Forschungsgemeinschaft.
Sequencing of M. smegmatis mc2155 was accomplished by TIGR with support from National Institute of Allergy and Infectious Diseases (NIAID). We thank Natalie Wood, Flavia Pimentel-Schmitt, Hildegard Stork, and Ying Wang for technical assistance.
Footnotes
Published ahead of print on 8 June 2007.
REFERENCES
- 1.Alvarez-Anorve, L. I., M. L. Calcagno, and J. Plumbridge. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 187:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, G., L. A. McCue, and K. A. McDonough. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, A. W., S. D. Buckel, J. M. Groarke, J. N. Hope, D. H. Kingsley, and M. A. Hermodson. 1986. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J. Biol. Chem. 261:7652-7658. [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bertram, R., M. Schlicht, K. Mahr, H. Nothaft, M. H. Saier, Jr., and F. Titgemeyer. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Borich, S. M., A. Murray, and E. Gormley. 2000. Genomic arrangement of a putative operon involved in maltose transport in the Mycobacterium tuberculosis complex and Mycobacterium leprae. Microbios 102:7-15. [PubMed] [Google Scholar]
- 9.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24:449-467. [DOI] [PubMed] [Google Scholar]
- 10.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 11.Brinkkötter, A., H. Kloss, C. Alpert, and J. W. Lengeler. 2000. Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli. Mol. Microbiol. 37:125-135. [DOI] [PubMed] [Google Scholar]
- 12.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 13.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 14.Connell, N. D., and H. Nikaido. 1994. Membrane permeability and transport in Mycobacterium tuberculosis, p. 333-349. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, DC.
- 15.Content, J., M. Braibant, N. Connell, and J. A. Ainsa. 2005. Transport processes, p.379-404. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs (ed.), Tuberculosis and the tubercle bacillus, ASM Press, Washington, DC.
- 16.Crick, D. C., S. Mahapatra, and P. J. Brennan. 2001. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11:107R-118R. [DOI] [PubMed] [Google Scholar]
- 17.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 18.Derouaux, A., S. Halici, H. Nothaft, T. Neutelings, G. Moutzourelis, J. Dusart, F. Titgemeyer, and S. Rigali. 2004. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. J. Bacteriol. 186:1893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duplay, P., H. Bedouelle, A. Fowler, I. Zabin, W. Saurin, and M. Hofnung. 1984. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J. Biol. Chem. 259:10606-10613. [PubMed] [Google Scholar]
- 20.Edson, N. L. 1951. The intermediary metabolism of the mycobacteria. Bacteriol. Rev. 15:147-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erni, B., C. Siebold, S. Christen, A. Srinivas, A. Oberholzer, and U. Baumann. 2006. Small substrate, big surprise: fold, function and phylogeny of dihydroxyacetone kinases. Cell. Mol. Life Sci. 63:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eze, M. O., and R. N. McElhaney. 1981. The effect of alterations in the fluidity and phase state of the membrane lipids on the passive permeation and facilitated diffusion of glycerol in Escherichia coli. J. Gen. Microbiol. 124:299-307. [DOI] [PubMed] [Google Scholar]
- 23.Franke, W., and A. Schillinger. 1944. Zum Stoffwechsel der saeurefesten Bakterien I. Orientierende aerobe Reihenversuche. Biochem. Zeitung. 319:313-334. [Google Scholar]
- 24.Frey, P. A. 1996. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10:461-470. [PubMed] [Google Scholar]
- 25.Gutknecht, R., R. Beutler, L. F. Garcia-Alles, U. Baumann, and B. Erni. 2001. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindle, Z., and C. P. Smith. 1994. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 12:737-745. [DOI] [PubMed] [Google Scholar]
- 27.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 28.Izumori, K., Y. Ueda, and K. Yamanaka. 1978. Pentose metabolism in Mycobacterium smegmatis: comparison of l-arabinose isomerases induced by l-arabinose and d-galactose. J. Bacteriol. 133:413-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumori, K., K. Yamanaka, and D. Elbein. 1976. Pentose metabolism in Mycobacterium smegmatis: specificity of induction of pentose isomerases. J. Bacteriol. 128:587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamionka, A., S. Parche, H. Nothaft, J. Siepelmeyer, K. Jahreis, and F. Titgemeyer. 2002. The phosphotransferase system of Streptomyces coelicolor. Eur. J. Biochem. 269:2143-2150. [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi, H., A. A. Vertes, S. Okino, M. Inui, and H. Yukawa. 2006. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch, R. 1882. Die Aetiologie der Tuberculose. Berl. Klin. Wochenschr. 19:221-230. [Google Scholar]
- 33.Lawlis, V. B., M. S. Dennis, E. Y. Chen, D. H. Smith, and D. J. Henner. 1984. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 47:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiden, M. C., E. O. Davis, S. A. Baldwin, D. C. Moore, and P. J. Henderson. 1987. Mammalian and bacterial sugar transport proteins are homologous. Nature 325:641-643. [DOI] [PubMed] [Google Scholar]
- 35.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10:204-219. [DOI] [PubMed] [Google Scholar]
- 36.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 37.Monedero, V., A. Maze, G. Boel, M. Zuniga, S. Beaufils, A. Hartke, and J. Deutscher. 2007. The phosphotransferase system of Lactobacillus casei: regulation of carbon metabolism and connection to cold shock response. J. Mol. Microbiol. Biotechnol. 12:20-32. [DOI] [PubMed] [Google Scholar]
- 38.Montero-Moran, G. M., S. Lara-Gonzalez, L. I. Alvarez-Anorve, J. A. Plumbridge, and M. L. Calcagno. 2001. On the multiple functional roles of the active site histidine in catalysis and allosteric regulation of Escherichia coli glucosamine 6-phosphate deaminase. Biochemistry 40:10187-10196. [DOI] [PubMed] [Google Scholar]
- 39.Moon, M. W., S. Y. Park, S. K. Choi, and J. K. Lee. 2007. The phosphotransferase system of Corynebacterium glutamicum: features of sugar transport and carbon regulation. J. Mol. Microbiol. Biotechnol. 12:43-50. [DOI] [PubMed] [Google Scholar]
- 40.Mota, L. J., P. Tavares, and I. Sa-Nogueira. 1999. Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 33:476-489. [DOI] [PubMed] [Google Scholar]
- 41.Munoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy, H. N., G. R. Stewart, V. V. Mischenko, A. S. Apt, R. Harris, M. S. McAlister, P. C. Driscoll, D. B. Young, and B. D. Robertson. 2005. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 280:14524-14529. [DOI] [PubMed] [Google Scholar]
- 43.Nobelmann, B., and J. W. Lengeler. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nothaft, H., D. Dresel, A. Willimek, K. Mahr, M. Niederweis, and F. Titgemeyer. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan, F., M. Jackson, Y. Ma, and M. McNeil. 2001. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parche, S., H. Nothaft, A. Kamionka, and F. Titgemeyer. 2000. Sugar uptake and utilisation in Streptomyces coelicolor: a PTS view to the genome. Antonie Leeuwenhoek 78:243-251. [DOI] [PubMed] [Google Scholar]
- 49.Park, J. H., and M. H. Saier, Jr. 1996. Phylogenetic characterization of the MIP family of transmembrane channel proteins. J. Membr. Biol. 153:171-180. [DOI] [PubMed] [Google Scholar]
- 50.Paula, S., A. G. Volkov, A. N. V. Hoek, T. H. Haines, and D. W. Deamer. 1996. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 70:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez-Rueda, E., and J. Collado-Vides. 2000. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28:1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakrishnan, T., P. S. Murthy, and K. P. Gopinathan. 1972. Intermediary metabolism of mycobacteria. Bacteriol. Rev. 36:65-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratledge, C., and J. Stanford. 1982. Nutrition, growth, and metabolism, p. 186-271. In C. Ratledge and J. Stanford (ed.), The biology of mycobacteria. Academic Press Inc. Ltd., London, United Kingdom.
- 55.Reizer, J., C. Hoischen, F. Titgemeyer, C. Rivolta, R. Rabus, J. Stülke, D. Karamata, M. H. Saier, Jr., and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 27:1157-1169. [DOI] [PubMed] [Google Scholar]
- 56.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1994. A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta 1197:133-166. [DOI] [PubMed] [Google Scholar]
- 57.Richardson, H. B., E. Shorr, and R. O. Loebel. 1931. Comparative studies in the respiratory metabolism of various acid-fast bacilli. Trans. Nat. Tuberc. Assoc. 27:205-210. [Google Scholar]
- 58.Rimmele, M., and W. Boos. 1994. Trehalose-6-phosphate hydrolase of Escherichia coli. J. Bacteriol. 176:5654-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romano, A. H., S. J. Eberhard, S. L. Dingle, and T. D. McDowell. 1970. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in bacteria. J. Bacteriol. 104:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sa-Nogueira, I., T. V. Nogueira, S. Soares, and H. de Lencastre. 1997. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology 143:957-969. [DOI] [PubMed] [Google Scholar]
- 61.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlösser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlösser, A., and H. Schrempf. 1996. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/cellotriose-transport system from the cellulose degrader Streptomyces reticuli. Eur. J. Biochem. 242:332-338. [DOI] [PubMed] [Google Scholar]
- 64.Schönert, S., T. Buder, and M. K. Dahl. 1999. Properties of maltose-inducible alpha-glucosidase MalL (sucrase-isomaltase-maltase) in Bacillus subtilis: evidence for its contribution to maltodextrin utilization. Res. Microbiol. 150:167-177. [DOI] [PubMed] [Google Scholar]
- 65.Smeulders, M. J., J. Keer, R. A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soerensen, K. I., and B. Hove-Jensen. 1996. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 178:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stahl, C., S. Kubetzko, I. Kaps, S. Seeber, H. Engelhardt, and M. Niederweis. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40:451-464. [DOI] [PubMed] [Google Scholar]
- 68.Stephan, J., J. G. Bail, F. Titgemeyer, and M. Niederweis. 2004. DNA-free RNA preparations from mycobacteria. BMC Microbiol. 4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailänder, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714-730. [DOI] [PubMed] [Google Scholar]
- 70.Sumiya, M., E. O. Davis, L. C. Packman, T. P. McDonald, and P. J. Henderson. 1995. Molecular genetics of a receptor protein for D-xylose, encoded by the gene xylF, in Escherichia coli. Receptors Channels 3:117-128. [PubMed] [Google Scholar]
- 71.Takayama, K., and J. O. Kilburn. 1989. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 33:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tobisch, S., P. Glaser, S. Krüger, and M. Hecker. 1997. Identification and characterization of a new β-glucoside utilization system in Bacillus subtilis. J. Bacteriol. 179:496-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Wezel, G. P., K. Mahr, M. Konig, B. A. Traag, E. F. Pimentel-Schmitt, A. Willimek, and F. Titgemeyer. 2005. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2). Mol. Microbiol. 55:624-636. [DOI] [PubMed] [Google Scholar]
- 74.Woodruff, P. J., B. L. Carlson, B. Siridechadilok, M. R. Pratt, R. H. Senaratne, J. D. Mougous, L. W. Riley, S. J. Williams, and C. R. Bertozzi. 2004. Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem. 279:28835-28843. [DOI] [PubMed] [Google Scholar]
- 75.Woodson, K., and K. M. Devine. 1994. Analysis of a ribose transport operon from Bacillus subtilis. Microbiology 140:1829-1838. [DOI] [PubMed] [Google Scholar]
- 76.Wu, L. F., J. M. Tomich, and M. H. Saier, Jr. 1990. Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J. Mol. Biol. 213:687-703. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, C. C., M. C. Durand, R. Jeanjean, and F. Joset. 1989. Molecular and genetical analysis of the fructose-glucose transport system in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 3:1221-1229. [DOI] [PubMed] [Google Scholar]