Abstract
Biosynthesis of cephamycin C in Streptomyces clavuligerus involves the initial conversion of lysine to α-aminoadipic acid. Lysine-6-aminotransferase and piperideine-6-carboxylate dehydrogenase carry out this two-step reaction, and genes encoding each of these enzymes are found within the cephamycin C gene cluster. However, while mutation of the lat gene causes complete loss of cephamycin production, pcd mutants still produce cephamycin at 30% to 70% of wild-type levels. Cephamycin production by pcd mutants could be restored to wild-type levels either by supplementation of the growth medium with α-aminoadipic acid or by complementation of the mutation with an intact copy of the pcd gene. Neither heterologous PCR nor Southern analyses showed any evidence for the presence of a second pcd gene. Furthermore, cell extracts from pcd mutants lack detectable PCD activity. Cephamycin production in the absence of detectable PCD activity suggests that S. clavuligerus must have some alternate means of producing the aminoadipyl-cysteinyl-valine needed for cephamycin biosynthesis.
Biosynthesis of the penicillin-cephalosporin-cephamycin class of antibiotics originates from three amino acids, l-α-aminoadipic acid, l-cysteine, and l-valine. In bacterial cephamycin producers, α-aminoadipic acid in turn arises from lysine via a two-step process catalyzed by lysine-6-aminotransferase (LAT) and piperideine-6-carboxylate dehydrogenase (PCD), and so LAT and PCD activities are considered the first two steps of cephamycin biosynthesis. While lat genes have been found in the cephamycin gene clusters of all actinomycetes that have been studied (5, 24), the pcd gene has been more elusive. No pcd gene was identified in the cephamycin gene cluster of Nocardia lactamdurans (13). However, the possibility that piperideine-6-carboxylate (P6C) might convert nonenzymatically into α-aminoadipate was discounted by the finding that N. lactamdurans cell extracts contain PCD activity (7, 13). Therefore, in N. lactamdurans, a PCD enzyme encoded by a gene located outside the cluster regions examined to date is presumed to provide α-aminoadipate for cephamycin biosynthesis.
In contrast, both lat and pcd genes were found within the cephamycin gene cluster in Streptomyces clavuligerus. This represents the first and only example to date of a pcd gene found within a cephamycin gene cluster (3, 20). The pcd gene in S. clavuligerus is adjacent to, and transcriptionally coupled with, cefD and cefE. The latter genes encode isopenicillin N epimerase and desacetoxycephalosporin C synthase, enzymes from the middle of the cephamycin C biosynthetic pathway. The genes lat, pcbAB, and pcbC, which encode enzymes for the biosynthetic steps immediately before and after PCD at the beginning of the pathway, also form an operon but are located well apart from the pcd-cefD-cefE genes. Although the grouping of pcd together with genes from the middle rather than the early part of the pathway seems unusual, control of expression of the entire cluster by the pathway-specific regulator CcaR and other regulatory elements could mean that coordinate expression of individual genes within the cluster can be accomplished regardless of the physical location of the genes.
The PCD enzyme has been purified to homogeneity from S. clavuligerus, and N-terminal amino acid sequence information is consistent with that deduced from the 5′ end of the pcd gene (7, 20). pcd was also expressed at a high level in Escherichia coli, and the recombinant protein exhibited PCD activity when assayed in vitro. Furthermore, pcd from S. clavuligerus has been shown to cross-hybridize with other Streptomyces spp. known to produce β-lactam antibiotics but not with nonproducer species (3, 7), and cell extracts prepared from β-lactam-producing species of Streptomyces show PCD activity whereas those from nonproducer species do not.
In addition to their roles in cephamycin biosynthesis, PCD and LAT activities can be found in some non-antibiotic-producing bacteria, where they function in lysine catabolism. Lysine catabolism in bacteria is characterized by its diversity. Pseudomonas spp. are reported to have at least four separate pathways for lysine catabolism, including one that involves the intermediacy of α-aminoadipate (14). Although cephamycin-producing actinomycetes must convert lysine to α-aminoadipate, it is not clear whether the α-aminoadipate pathway is also a route for lysine catabolism in these organisms. Madduri et al. reported that catabolism of lysine via cadaverine is the primary pathway for lysine catabolism in both cephamycin-producing and non-cephamycin-producing actinomycetes and supported this claim by showing that S. clavuligerus cannot grow on α-aminoadipate as a sole source of nitrogen (14). Furthermore, complete genome sequences are now available for two non-cephamycin-producing species of Streptomyces, and no orthologues of genes encoding enzymes required for α-aminoadipate production are present in those sequences. However, Leitao et al. indicated that lysine is not significantly degraded to cadaverine in S. clavuligerus and N. lactamdurans (12), leaving open the question of how lysine catabolism is accomplished in cephamycin-producing species.
Recently, a second example of a pcd gene was characterized from Flavobacterium lutescens, a species known to produce α-aminoadipate but not β-lactam antibiotics, although β-lactam-producing members of that genus are known (8). The F. lutescens pcd gene shows 53% sequence identity to pcd from S. clavuligerus, and it was also expressed in E. coli to reveal levels of PCD activity. Examination of sequence databases shows a number of other genes discovered during genome sequencing projects that are annotated as pcd genes and that show strong similarity to the pcd genes from S. clavuligerus and F. lutescens, although their products have not been characterized. Many of these database entries are from Pseudomonas spp. and are not associated with antibiotic production. In addition, apparent pcd genes are present in Mycobacterium spp., providing examples of the presence of pcd genes in non-β-lactam-producing actinomycetes.
Against this backdrop, we were interested in investigating the role of the pcd gene in cephamycin production in S. clavuligerus, and so pcd disruption mutants were prepared and characterized.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Streptomyces clavuligerus NRRL 3585 was obtained from the Northern Regional Research Laboratories, Peoria, IL. Streptomyces lividans TK24 was generously provided by D. Hopwood, John Innes Institute, Norwich, United Kingdom. Escherichia coli DH5α (New England Biolabs, Mississauga, Ontario, Canada) was used as the general cloning host for DNA manipulations. pBluescript II SK/KS+ (Stratagene) are generalized E. coli cloning vectors providing a blue-white selection. pDA109 is a pBluescript-derived vector carrying a 3.2-kb BamHI fragment of S. clavuligerus genomic DNA encompassing all of the pcd and cefE genes and part of the cefD gene from the cephamycin gene cluster (3). pDA172 is a pUC119-based vector carrying a 1.8-kb EcoRI/KpnI fragment of S. clavuligerus DNA encompassing the lat gene. pDA517 (3) and pSK-Neo (16) are pBluescript-derived vectors carrying apramycin and neomycin resistance gene cassettes, respectively, useful for selection with both E. coli and Streptomyces strains. pJOE829 is a Streptomyces high-copy-number plasmid vector generously provided by J. Altenbuchner, University of Stuttgart. pSET152, an E. coli-Streptomyces shuttle vector that is a high-copy-number plasmid in E. coli but an integrative plasmid in Streptomyces spp., was obtained from NRRL. pOW309 is an E. coli-Streptomyces shuttle cosmid containing most of the cephamycin gene cluster from rhs to midway through pcbAB (21) generously provided by P. Skatrud, Eli Lilly Co. pMAL-c2 (New England BioLabs) is an expression vector designed to produce fusion proteins where the protein of interest is fused to the C terminus of the maltose binding protein from E. coli.
Cultures of S. clavuligerus and S. lividans were maintained as described previously (19). All fermentation studies to monitor cephamycin production by S. clavuligerus were conducted using glycerol spore stocks to inoculate Trypticase soy broth-1% soluble starch medium seed cultures. After 40 h of incubation the seed cultures were transferred at 2% (vol/vol) into soy (19), starch asparagine (SA) (19), or SA-plus-lysine (SAL) medium unless otherwise indicated, and incubation was continued for up to 96 h. SAL medium is a modification of SA medium in which l-asparagine is reduced to 1 g/liter and l-lysine is added to 1g/liter. All cultures were incubated at 28°C on a rotary shaker at 250 rpm.
Cultures of E. coli were grown and maintained on LB liquid or solid media, according to the method of Sambrook et al. (23). Where appropriate, media were supplemented with ampicillin (100 μg/ml), apramycin (25 μg/ml), hygromycin (200 μg/ml), or neomycin (50 μg/ml).
DNA manipulations and Southern analyses.
Routine DNA manipulations used for E. coli, such as plasmid isolation, digestion, ligation, and analysis by agarose gel electrophoresis, were carried out as described by Sambrook et al. (23). Routine DNA manipulations for Streptomyces spp. were performed according to the method of Kieser et al. (11).
Southern hybridization was carried out as described previously (23) using high-stringency hybridization and washing conditions. Hybridization was carried out for 16 h at 65°C, and the nylon blots were washed three times at 10 min each time at 21°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS), once for 15 min at 65°C in 1× SSC-0.1% SDS, and finally twice for 15 min each time at 65°C in 0.1× SSC-0.1% SDS. Oligonucleotide primers hetero-PCDfor (5′-SGG CGA RGT SCA GGA RAT GAT CGA; see http://bioinformatics.org/sms2/iupac.html for nucleic acid IUPAC code) and RT-PCDrev (5′-GCG CCC GAG GTG CCG ATG T) were used in a PCR with plasmid pDA109 as the template (96°C for 5 min; 96°C for 30 s, 65°C for 30 s, and 68°C for 1 min for 29 cycles; and 68°C for 5 min) to generate an approximately 1-kb DNA fragment internal to the pcd gene for use as a probe in Southern analysis. Oligonucleotide primers hetero-PCDfor and hetero-PCDrev (5′-CAD ACS GCG SCC GGG AAG TTG AA) were used in PCR mixtures (96°C for 2 min; 96°C for 30 s, 60°C for 30 s, and 68°C for 2 min for 29 cycles; and 68°C for 5 min) containing 6% dimethyl sulfoxide with genomic DNA from the Δpcd2 mutant as the template to attempt to amplify DNA fragments from heterologous pcd genes that might be present in S. clavuligerus.
Disruption of the pcd gene.
The pcd::apr mutant was obtained by subcloning the pcd gene from cosmid pOW309 (3) as a 3.2-kb BamHI fragment (also containing cefE and part of cefD) into pBluescript to give pDA109. The pcd gene was then disrupted by insertion of the 1.5-kb apramycin resistance gene cassette (Aprr cassette) from pDA517 into a BclI site located 480 bp from the start codon of pcd after both fragments were made blunt by treatment with the Klenow fragment of DNA polymerase I. Clones with the Aprr cassette fragment in both orientations relative to the pcd gene were obtained. Plasmids bearing the disrupted pcd gene were then fused to pJOE829 via their HindIII sites to create E. coli/Streptomyces shuttle vectors. The shuttle vectors were introduced into S. clavuligerus by transformation after passage through S. lividans as described previously (1), and gene disruption mutants arose via double recombination. Apramycin-resistant, hygromycin-sensitive clones were selected as potential pcd::apr mutants and then confirmed by Southern analysis. Mutants with the disrupting Aprr cassette in both orientations relative to the pcd gene were obtained.
Four independent pcd deletion mutants (Δpcd1-4) were obtained using the REDIRECT PCR-targeted mutagenesis procedure (9). Briefly, 2B8, a recombinant cosmid carrying an S. clavuligerus genomic DNA fragment spanning the cephamycin gene cluster cloned into the cosmid vector pWE15 (Stratagene) (10), was used as the basis for the PCR targeting. Oligonucleotide primers Pcd-fwd (5′-CGC CGC ACA TCC AAG GCA TAG AGA GCA CAC ACC GTC ATG ATT CCG GGG ATC CGT CGA CC) and Pcd-rev (5′-CAC GGG ACG GCG CCC GGC GCC GCG GGC CGG GGC GGA CTA TGT AGG CTG GAG CTG CTT C) were used to amplify a disruption cassette carrying oriT and the aac(3)IV gene flanked by sequences homologous to the 5′ and 3′ ends of the pcd gene by use of plasmid pIJ773 as the template. The pcd gene was then exchanged for the disruption cassette within cosmid 2B8 by means of the λRED recombination functions carried within E. coli BW25113, and the recombinant cosmid lacking pcd was conjugated into S. clavuligerus as described previously (25), where the entire native pcd gene was replaced by the disruption cassette by homologous recombination.
Complementation of pcd::apr mutants.
In order to use pSET152 to prepare a complementation construct for the pcd::apr mutant, it was first necessary to replace the Aprr cassette determinant. pSET152 was digested with ApaI-SacI to release a 0.7-kb fragment that comprised most of the Aprr cassette gene, and it was replaced by an approximately 1-kb ApaI-SacI fragment from pSK-Neo carrying the Neor gene cassette. The resulting vector, pSET-Neo, was digested with EcoRV, and the 3.2-kb pcd-containing BamHI fragment from pDA109 was inserted after it had been made blunt by treatment with the Klenow fragment of DNA polymerase I. The resulting construct, pSET-Neo-pcd, was then introduced into the pcd::apr mutant by conjugation using established techniques (25).
HPLC and mass spectroscopy.
Culture filtrates were analyzed for the presence of cephamycin C and 7-desmethoxycephamycin C by reversed-phase high-pressure liquid chromatography (HPLC) using a 5-μm packing Hypercarb column (Phenomenex, Torrance, CA) (2.1 by 100 mm) and operating under isocratic conditions, with 8% acetonitrile-92% 10 mM ammonium formate (pH 4.0) as the mobile phase, and detection at 268 nm. Mass spectra were obtained using a ZMD-2 single quadropole instrument from Waters Scientific (Milford, MA).
Purification of cephamycin C and 7-desmethoxycephamycin C.
Cephamycin C and 7-desmethoxycephamycin C were partially purified from lyophilized culture filtrates from soy-medium-grown cultures of S. clavuligerus by extraction with 70% ethanol followed by open-column chromatography on AG-50W resin (Bio-Rad Laboratories, Mississauga, Ontario, Canada) (X4; H+ form; 200/400 mesh) as described by O'Sullivan et al. (18). Purification to homogeneity, as judged by HPLC-mass spectrometry analysis, was subsequently accomplished by semipreparative HPLC using a μ-Bondapak column (RadPak A radial compression cartridge; Waters Scientific) as the stationary phase under isocratic conditions with 5% methanol-95% 0.04 M ammonium acetate (pH 4.45) as the mobile phase. Purified material was lyophilized, and concentrations of standard samples were determined based on their absorbance values at 264 nm by use of a molar extinction coefficient of ɛ264 in water of 6,900 for cephamycin C (17).
PCD enzyme assays.
Cell extracts were prepared from wild-type, pcd::aprB3, and Δpcd2 cultures grown in soy, SA, and SAL media for 48 h and used to measure PCD activity essentially as described by de la Fuente et al. (7). PCD activity in cell extracts (desalted into buffer A) was measured using coupled radiometric assays containing 0.2 μCi l-[U-14C]lysine (312 mCi/mmol) (Amersham Biosciences), 10 mM 2-ketoglutarate, 7 mM NAD+, 7.4 μM pyridoxal phosphate, 3 mU recombinant LAT, and S. clavuligerus PCD (50 to 200 μg protein) in 0.1 M MOPS (morpholinepropanesulfonic acid)-NaOH buffer (pH 8.0) (final volume, 250 μl). Reaction mixtures were incubated for 1 h at 30°C and then stopped by the addition of 250 μl of methanol. Unlabeled lysine and α-aminoadipic acid were also added to stopped reaction mixtures to achieve a final concentration of 2 mM. After 30 min at 4°C, precipitated protein was removed by centrifugation and 10-μl amounts of supernatant were applied to silica gel thin-layer chromatography plates (Whatman) and developed in ethanol-1 M ammonium acetate-glacial acetic acid (6:1:1 by volume). Labeled products were visualized using a Fuji FLA-500 imager, and thin-layer chromatography plates were also sprayed with ninhydrin (0.2% in ethanol) to locate unlabeled standards.
Production of S. clavuligerus LAT in E. coli.
LAT enzyme used to carry out the initial conversion of lysine to piperideine-6-carboxylate was produced at a high level in E. coli as a fusion protein bound to maltose binding protein. Oligonucleotide primers Dyl9 (5′-GCC GGA TCC ATG GGC GAA GCA GCA CGC CAC CC) and Dyl10 (5′-GCC GAA TTC GCG TCA GAC GCT CTC GGC GAC CGC) were used in a PCR with plasmid pDA172 as the template to amplify the lat gene from S. clavuligerus and introduce BamHI and EcoRI (underlined) sites into the 3′ and 5′ ends of the PCR product. These restriction sites allowed lat to be inserted into plasmid pMAL-c2 (New England BioLabs), creating a translational fusion with the maltose binding protein. The maltose binding protein-LAT fusion was produced in E. coli and purified using amylose resin (New England BioLabs) according to the manufacturer's instructions. A 1-IU quantity of LAT activity converts 1 μM of lysine to P6C in 1 min.
Chemical and bioassays.
DNA was analyzed as a measure of growth using the method of Burton (4). Bioassays for cephamycin C and 7-desmethoxycephamycin C were carried out as described previously (19) using pure cephamycin C as a standard.
RESULTS
Preparation of pcd::apr gene disruption mutants.
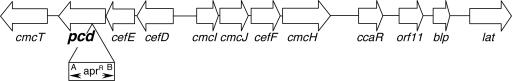
The pcd gene in S. clavuligerus is located adjacent to cefD and cefE on one flank and to cmcT on the other (Fig. 1). A subcloned copy of the pcd gene was disrupted by insertion of an apramycin resistance gene cassette (Aprr cassette) into the centrally located BclI site and was then introduced into S. clavuligerus, where it replaced the chromosomal copy via double recombination. pcd::apr disruption mutants were identified initially as apramycin-resistant, hygromycin-sensitive clones, and then the mutations were confirmed by Southern analysis. The pcd::apr mutants showed no obvious growth or morphological changes relative to the wild-type strain.
FIG. 1.
Organization of the cephamycin gene cluster of Streptomyces clavuligerus in the region of the pcd and lat genes. The Aprr gene cassette used to disrupt pcd is represented as an open box, with arrowheads marked A and B indicating the two possible orientations of the Aprr cassette.
Effect of pcd disruption on cephamycin production.
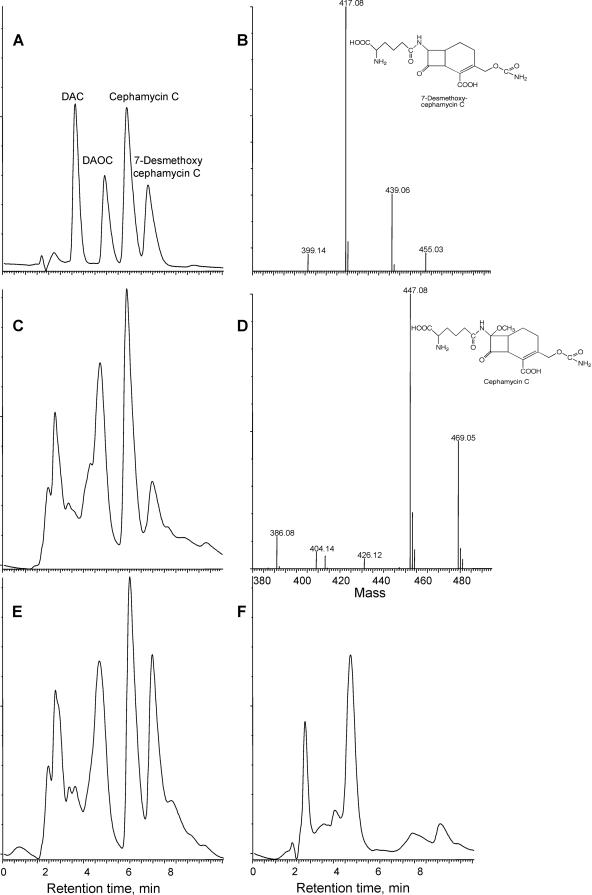
Eight pcd::apr mutants, four with an Aprr cassette in the same orientation as pcd and four with an Aprr cassette in the opposite orientation, were surveyed for their cephamycin-producing ability. Cultures were grown in soy medium, and samples were removed at 48, 72, and 96 h of incubation and assayed for cephamycin production (Table 1). Wild-type cultures showed good cephamycin production levels, with a high of 167 μg/ml at 96 h, but unexpectedly, all of the pcd::apr mutants also showed considerable levels of bioactivity. The pcd::apr mutants produced bioactive material at about 29% to 37% of wild-type levels at all three time points. Culture filtrates were also subjected to HPLC with mass spectrometric analysis to ensure that the bioactive material produced was cephamycin C. Cephamycin C, isolated from culture supernatants of wild-type S. clavuligerus by preparative HPLC, was used as an authentic standard. Although the HPLC chromatograms of soy-medium-grown culture supernatants are complex in the region of cephamycin C, it was clear that all of the mutants produced cephamycin C (Fig. 2). In addition to cephamycin C, both the mutants and the wild type also produced 7-desmethoxycephamycin C (Fig. 2), as has been reported previously (15).
TABLE 1.
Effect of mutation of the pcd gene on cephamycin production in S. clavuligerus
| Culturea | Cephamycin production (μg/ml) atb:
|
||
|---|---|---|---|
| 48 h | 72 h | 96 h | |
| Wild type 1 | 153 | 176 | 160 |
| Wild type 2 | 93.0 | 134 | 174 |
| pcd::apr-A1 | 30.8 | 57.5 | 55.0 |
| pcd::apr-A3 | 52.5 | 50.0 | 71.5 |
| pcd::apr-A5 | 21.5 | 65.8 | 50.0 |
| pcd::apr-A7 | 17.5 | 32.0 | 60.0 |
| pcd::apr-B1 | 40.2 | 57.5 | 55.0 |
| pcd::apr-B2 | 46.0 | 50.0 | 65.5 |
| pcd::apr-B3 | 40.2 | 57.5 | 65.8 |
| pcd::apr-B4 | 35.0 | 38.5 | 65.8 |
Mutant cultures designated A have the Aprr cassette in the same orientation as the pcd gene; cultures designated B have the Aprr cassette in the opposite orientation.
Single cultures were assayed in duplicate; data presented represent the average values for the results of the two assays.
FIG. 2.
HPLC-mass spectrometry analyses of culture filtrates from wild-type and pcd::apr mutant strains of S. clavuligerus. (A) HPLC profile of standard cephalosporin and cephamycin compounds, with detection by UV absorption at 268 nm. DAC, desacetylcephalosporin C; DAOC, desacetoxycephalosporin C. (B) Mass spectrum confirming the identity of 7-desmethoxycephamycin C from culture filtrates of a 96-h culture of pcd::apr-B3 mutant of S. clavuligerus grown on soy medium. (C) HPLC profile of culture filtrate from 96-h culture of pcd::apr-B3 mutant of S. clavuligerus grown on soy medium. (D) Mass spectrum confirming the identity of cephamycin C from culture filtrates of a 96-h culture of a pcd::apr-B3 mutant of S. clavuligerus grown on soy medium. (E) HPLC profile of culture filtrate from 96-h culture of wild-type S. clavuligerus grown on soy medium. (F) HPLC profile of uninoculated soy medium.
The pcd::apr mutants still contained the entire pcd gene, but the gene was disrupted by insertion of an Aprr cassette into its middle, leaving open the possibility that the cephamycin production seen in the mutants might have been due to residual PCD activity expressed from one or the other of the pcd gene fragments. To eliminate this possibility, a second collection of pcd deletion mutants, Δpcd1-Δpcd4, in which the entire pcd gene was deleted and replaced by the Aprr cassette, was prepared by PCR-targeted mutagenesis. The results seen with the Δpcd1-Δpcd4 mutants were similar to those seen with the pcd::apr mutants. Cephamycin production in the Δpcd1-Δpcd4 mutants ranged from a low of 37% to as much as 70% of the levels seen with the wild-type cultures.
Complementation of the pcd mutation.
To confirm that the decrease in cephamycin production in pcd mutants was due to the loss of the ability to make α-aminoadipate, two types of complementation experiments were conducted. In the first case, a representative pcd mutant, pcd::apr-B3, was chosen for examination in more detail and was grown in soy medium with or without supplementation with α-aminoadipate to 5 mM. Cultures were sampled at 48, 72, and 96 h and analyzed for antibiotic production and growth (Table 2). Bioassays showed that supplementation of the pcd::apr-B3 mutant with α-aminoadipate restored it to wild-type levels of cephamycin production. Supplementation with α-aminoadipate also stimulated cephamycin production by the wild-type culture slightly compared to the results seen with unsupplemented soy medium, indicating that under these growth conditions, the level of α-aminoadipate available limits cephamycin production. These results showed that the decreased cephamycin production in the pcd::apr mutants was due to a lack of α-aminoadipate.
TABLE 2.
Effect of supplementation with α-aminoadipate on cephamycin production by wild-type and pcd::apr mutant strains of S. clavuligerus grown on soy medium
| Culture | Cephamycin productiona [μg/ml (μg/mg DNA)]
|
|
|---|---|---|
| No α-aa | +5 mM α-aa | |
| Wild type | ||
| 72 h | 188 (903) | 240 (1,030) |
| 96 h | 140 (645) | 181 (939) |
| pcd::apr-B3 | ||
| 72 h | 80 (334) | 256 (998) |
| 96 h | 75 (396) | 155 (787) |
Cultures were grown in triplicate for each time point. Each sample was assayed in duplicate; data represent averages. α-aa, α-aminoadipate.
Similarly, the pcd::apr-B3 mutant was complemented genetically by introduction of an intact copy of the pcd gene. The pcd gene was cloned into a modified version of pSET152 in which the Aprr cassette was replaced by a neomycin resistance cassette. The resulting pSETneo-pcd plasmid was introduced into pcd::apr-B3 by conjugation, and the cephamycin production by exconjugants was assessed after fermentation on soy medium. Integration of an intact copy of the pcd gene restored cephamycin production to wild-type levels in the pcd::apr-B3 mutant, again indicating that the decreased production of cephamycin by pcd::apr mutants was due to the loss of PCD activity (data not shown).
Effect of lysine supplementation on cephamycin production by the pcd::apr mutant.
Complementation of the pcd::apr-B3 mutant with an intact pcd gene indicates that the decreased cephamycin production in the mutant was due to an insufficiency of PCD. However, the fact that the mutant was still able to produce cephamycin implies that it must have some other means of producing α-aminoadipate, possibly a second PCD activity associated with lysine catabolism rather than with cephamycin production. To investigate this possibility, wild-type S. clavuligerus and the pcd::apr-B3 mutant were grown on a defined SA medium that contained asparagine as the sole N source and on a modified SA medium in which half of the asparagine is replaced by an equal amount of lysine (SAL). Cultures were harvested at 72 and 96 h to determine growth and cephamycin production levels (Table 3). Cephamycin production was lower in SA than in soy medium, but the wild-type culture still produced 277 μg cephamycin/mg DNA in SA medium. Lysine stimulated cephamycin production somewhat in wild-type cultures, as has been reported previously (22), but a more pronounced effect was seen with the pcd::apr-B3 mutant. The mutant produced very little cephamycin when grown in SA medium (about 13% of wild-type levels), but when grown in SAL medium, cephamycin production was restored to about 50% of wild-type levels.
TABLE 3.
Effect of supplementation with lysine on cephamycin production by wild-type and pcd::apr mutant strains of S. clavuligerus grown on SA medium
| Culture | Cephamycin productiona [μg/ml(μg/mg DNA)]
|
|
|---|---|---|
| SA | SAL | |
| Wild type | ||
| 72 h | 31.6 (277) | 67.7 (594) |
| 96 h | 28.2 (157) | 47.5 (302) |
| pcd::apr-B3 | ||
| 72 h | 4.1 (31.4) | 33.7 (284) |
| 96 h | 5.0 (27.8) | 26.0 (167) |
When lysine was supplemented, the amount of asparagine in SA medium was reduced to 1.0 g/liter and lysine was added to 1.0 g/liter. Cultures were grown in triplicate for each time point. Each sample was assayed in duplicate; data represent averages.
To ensure that cephamycin production by the pcd::apr-B3 mutant was not just an artifact caused by the presence of preexisting α-aminoadipate in the media, cephamycin production by a lat deletion mutant, Δlat (2), was also examined. If exogenous α-aminoadipate was present in these media, then lat mutants should have shown a level of cephamycin production similar to that seen with the pcd mutants. However, when a Δlat mutant was grown on soy, SA, or SAL medium, no cephamycin production was evident unless α-aminoadipate was provided (Table 4).
TABLE 4.
Production of cephamycin by a Δlat mutant of S. clavuligerus
| Growth medium | Cephamycin productiona
|
|
|---|---|---|
| μg/ml | μg/mg DNA | |
| Soy | 4.6b | 11.2b |
| SA | 0 | 0 |
| SAL | 0 | 0 |
| SA + 5 mM α-aminoadipate | 131.8 | 653.0 |
When lysine was supplemented, the amount of asparagine in SA medium was reduced to 1.0 g/liter and lysine was added to 1.0 g/liter. Duplicate cultures were grown for 72 h. Each culture was assayed in duplicate; data represent averages.
Traces of bioactivity seen in the Δlat mutant grown on soy medium were due to non-β-lactam antibiotics produced under these growth conditions.
Searching for a pcd paralogue in S. clavuligerus.
The increase in cephamycin production by pcd mutants when grown in SAL versus SA medium could indicate the presence of a second PCD activity associated with lysine catabolism. To explore this possibility, degenerate PCR primers were designed to amplify a region of the pcd gene that is highly conserved in all known or putative pcd genes listed in GenBank. Primers hetero-PCDfor and hetero-PCDrev amplified a single band, the expected 171-bp DNA fragment from pcd, when wild-type S. clavuligerus genomic DNA was used as the template. However, no DNA fragment was amplified when genomic DNA from one of the deletion mutants, Δpcd3, was used as the template (data not shown).
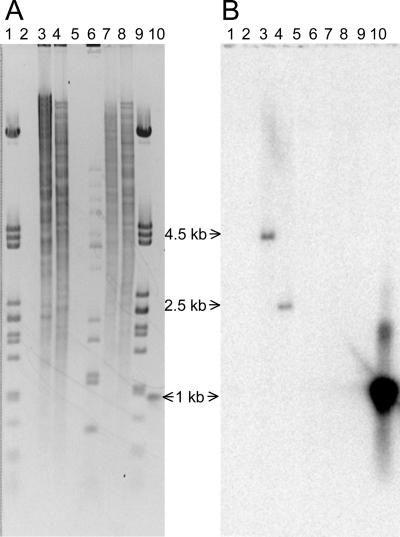
In an alternative approach to search for evidence indicating the presence of a second pcd gene, a larger ∼1-kb probe fragment (still internal to pcd) was amplified by PCR and used in Southern analyses to examine digests of wild-type and Δpcd3 genomic DNA. Restriction endonuclease digests of wild-type genomic DNA clearly showed hybridizing bands of the appropriate size when probed with the PCR fragment, but no hybridizing bands of any size were seen in digests of DNA from the Δpcd3 mutant (Fig. 3). Furthermore, no weakly hybridizing bands that might indicate the presence of a cross-reacting pcd gene were seen in either the wild-type or the Δpcd3 mutant strains.
FIG. 3.
Southern analysis of wild-type and Δpcd3 mutant strains of S. clavuligerus to search for paralogues of pcd. Samples of genomic DNA (2 μg) from wild-type and Δpcd3 mutant strains of S. clavuligerus were digested with either BbuI or NruI and separated on a 0.8% agarose gel. DNA fragments were transferred to a nylon sheet and hybridized with an approximately 1-kb radioactively labeled fragment internal to pcd. (A) Digested DNA fragments after electrophoresis and staining with ethidium bromide. (B) Nylon blot after hybridization with the labeled probe. Lanes 1 and 9, lambda DNA digested with PstI; lane 6, lambda DNA digested with BstEII; lanes 3 and 4, genomic DNA from wild-type cells digested with BbuI and NruI, respectively; lanes 7 and 8, genomic DNA from Δpcd3 mutant cells digested with BbuI and NruI, respectively; lane 10, probe DNA. Lanes 2 and 5 are empty.
PCD activity in cell extracts from wild-type and pcd mutant strains of S. clavuligerus.
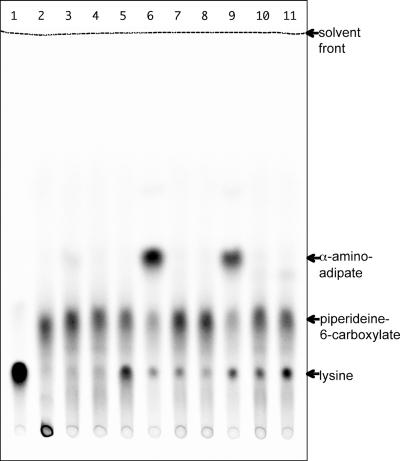
Despite the residual ability of pcd mutants of S. clavuligerus to synthesize cephamycin, no evidence for the existence of a second pcd gene was obtained. Therefore, cell extracts prepared from wild-type, pcd::apr-B3 mutant, and Δpcd2 mutant strains of S. clavuligerus were assayed for PCD activity directly. Δpcd2 was chosen from among the pcd deletion mutants because it showed the highest level of residual cephamycin production (∼70% of wild type) despite deletion of the pcd gene. Cell extracts were prepared from 48-h cultures of each strain grown on soy, SA, and SAL media and assayed in a coupled radiometric assay using [14C]lysine together with recombinant LAT from S. clavuligerus to form [14C]P6C (Fig. 4). Cell extracts from wild-type S. clavuligerus grown on any of the three media showed PCD activity, although levels were much higher in SA and SAL extracts than in soy extracts. In contrast, neither of the pcd mutants showed any PCD activity on any of the growth media tested despite the fact that the cell extracts were prepared from cultures that were actively synthesizing cephamycin.
FIG. 4.
PCD enzyme assays of cell extracts from wild-type and pcd mutant strains of S. clavuligerus. Cultures were grown on soy, SA, or SAL medium, and PCD activity was measured in a radiometric assay system using reaction mixtures containing l-[14C]lysine with recombinant LAT to provide labeled P6C as the substrate. Reaction mixtures were separated by silica gel thin-layer chromatography in ethanol-1 M ammonium acetate-glacial acetic acid (3:1:1), and then radioactivity was located on chromatograms by use of a phosphorimager. Lane 1, no-LAT control; lane 2, no-cell-extract control; lanes 3, 6, and 9, wild-type cell extracts from soy-, SA-, and SAL-grown cultures; lanes 4, 7, and 10, pcd::apr B3 cell extracts from soy-, SA-, and SAL-grown cultures; lanes 5, 8, and 11, Δpcd2 cell extracts from soy-, SA-, and SAL-grown cultures.
Wild-type cell extracts from soy-grown cultures showed unexpectedly low levels of PCD activity compared to extracts from SA- or SAL-grown cultures even though cephamycin production levels were much higher in soy-grown cultures. To ensure that extracts from soy-grown cultures did not contain some inhibitory factor giving artificially low values for PCD activity, PCD was purified from cell extracts of wild-type cultures grown on both SA and soy media by use of Hi-Trap Blue HP cartridges (GE Healthcare) as described previously (20). In each case, the PCD activity recovered after chromatography was comparable to that applied to the column, and so the PCD activity measured in crude cell extracts appears to reflect accurately the PCD content of the cells (data not shown).
DISCUSSION
S. clavuligerus contains a pcd gene located within the cephamycin gene cluster and encoding an active PCD enzyme. Database searches using the deduced PCD protein sequence show that many genes encoding similar proteins exist, although only one other protein has been characterized and confirmed to be a PCD enzyme. The protein with the highest similarity to S. clavuligerus PCD is a putative PCD from Mycobacterium avium subsp. paratuberculosis, with end-to-end identity of 69%. Several other mycobacterial species contain pcd genes encoding PCD proteins with similar levels of identity, and PCD from Pseudomonas syringae pv. tomato strain DC3000 also shows high identity at 65%, whereas the only other characterized PCD, that from F. lutescens, ranks somewhat lower at 53%. The corresponding pcd genes from these species show end-to-end identity ranging from 65% to 70% at the nucleotide level. None of these other species are known to produce cephamycin, although the possibility exists that F. lutescens may contain elements of a cryptic cephamycin biosynthetic gene cluster, since cephamycin-producing species of Flavobacterium are known. Nonetheless, it seems clear that the PCD activity associated with cephamycin production in S. clavuligerus resembles that of enzymes associated with lysine catabolism found in other genera. On the other hand, cross-hybridization studies have shown that genes similar to pcd exist in other cephamycin-producing species of Streptomyces but not in nonproducers. This suggests that, at least within the genus Streptomyces, pcd-type genes are usually associated with cephamycin production rather than with lysine catabolism. Certainly, those Streptomyces spp. for which whole-genome sequences are available (none are cephamycin producers) do not contain close homologues of pcd.
α-Aminoadipate is a precursor of cephamycin, and since it is produced in S. clavuligerus from lysine by the action of the LAT and PCD enzymes, pcd was expected to be an essential gene for cephamycin production. However, when pcd was disrupted, cephamycin production continued in mutants at about 30% to 70% of wild-type levels. Chemical complementation using either exogenous α-aminoadipate or genetic complementation by provision of an intact copy of the pcd gene restored the pcd::apr mutants to wild-type levels of cephamycin productivity. This showed that pcd must contribute at least part of the α-aminoadipate for cephamycin production and also ruled out the possibility that the deleterious effect of the pcd mutation on cephamycin production was an indirect result of polar effects on downstream genes. However, it remains clear that S. clavuligerus pcd mutants must have alternative means of producing the aminoadipyl-cysteinyl-valine needed for cephamycin production.
A simple explanation for the residual cephamycin-producing ability of pcd mutants would be that S. clavuligerus contains an additional PCD enzyme. However, Southern analyses conducted to search for pcd paralogues did not show any indication of cross-hybridizing bands attributable to the presence of a second pcd gene in S. clavuligerus. Similarly, PCR carried out using primers designed to amplify a region highly conserved in known and putative pcd genes yielded only a single product arising from the known pcd. When a larger PCR product internal to pcd was used as a probe for Southern analyses, only single hybridizing bands attributable to pcd were seen with the wild type, and no hybridization was seen with the pcd deletion mutant. Therefore, if S. clavuligerus does contain a second pcd gene, it must be sufficiently different in sequence from the known pcd gene to escape detection by heterologous PCR or Southern analyses using a pcd-specific probe.
The inability to detect a second pcd gene correlates with the lack of detectable PCD activity in pcd mutants. No PCD activity was detected in any of the pcd mutant cell extracts, including those made from lysine-supplemented cultures, suggesting that pcd mutants lack the ability to convert P6C into α-aminoadipate. Therefore, the stimulatory effect of lysine on cephamycin production by both wild-type and pcd mutant strains must just reflect an increase in levels of cephamycin precursors or biosynthetic enzymes, as has been noted previously upon lysine supplementation (22). However, the tightly blocked phenotype of lat mutants indicates that the only route to production of α-aminoadipate in S. clavuligerus is via LAT with P6C as an intermediate (2), leaving it unclear how pcd mutants are able to produce cephamycin. In this regard, Coque et al. (6) have purified ACV synthetase from Nocardia lactamdurans (expressed in S. lividans) and have shown that the purified enzyme would accept only α-aminoadipate or its lactam, 6-oxopiperidine-2-carboxylic acid, but not P6C as a substrate for synthesis of ACV. However, in view of the ability of pcd mutants from S. clavuligerus to form cephamycin despite the lack of detectable means to synthesize α-aminoadipic acid, the substrate specificity of ACV synthetase from S. clavuligerus should be examined to determine whether both P6C and α-aminoadipate may serve as substrates. An alternative possibility is that P6C converts spontaneously to α-aminoadipate at some low rate in S. clavuligerus and that PCD activity serves only to improve the efficiency of α-aminoadipate formation.
Acknowledgments
This study was supported by grants from the Natural Sciences and Engineering Research Council of Canada. D.C.A. was supported by a studentship from the Alberta Heritage Foundation for Medical Research.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Aidoo, K. A., A. Wong, D. C. Alexander, R. A. Rittammer, and S. E. Jensen. 1994. Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene 147:41-46. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. C., M. J. Brumlik, L. Lee, and S. E. Jensen. 2000. Early cephamycin biosynthetic genes are expressed from a polycistronic transcript in Streptomyces clavuligerus. J. Bacteriol. 182:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, D. C., and S. E. Jensen. 1998. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J. Bacteriol. 180:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, K. 1957. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem. J. 62:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coque, J. J., P. Liras, L. Laiz, and J. F. Martin. 1991. A gene encoding lysine 6-aminotransferase, which forms the β-lactam precursor α-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J. Bacteriol. 173:6258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coque, J. J., J. L. de la Fuente, P. Liras, and J. F. Martin. 1996. Overexpression of the Nocardia lactamdurans alpha-aminoadipyl-cysteinyl-valine synthetase in Streptomyces lividans. The purified multienzyme uses cystathionine and 6-oxopiperidine 2-carboxylate as substrates for synthesis of the tripeptide. Eur. J. Biochem. 242:264-270. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, J. L., A. Rumbero, J. F. Martin, and P. Liras. 1997. δ-1-Piperideine-6-carboxylate dehydrogenase, a new enzyme that forms α-aminoadipate in Streptomyces clavuligerus and other cephamycin C-producing actinomycetes. Biochem. J. 327:59-64. [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii, T., T. Narita, H. Agematu, N. Agata, and K. Isshiki. 2000. Cloning and characterization of pcd encoding Δ1-piperideine-6-carboxylate dehydrogenase from Flavobacterium lutescens IFO3084. J. Biochem. 128:975-982. [DOI] [PubMed] [Google Scholar]
- 9.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin, W., Y. G. Ryu, S. G. Kang, S. K. Kim, N. Saito, K. Ochi, S. H. Lee, and K. J. Lee. 2004. Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology 150:1485-1493. [DOI] [PubMed] [Google Scholar]
- 11.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. John Innes Foundation, Norwich, England.
- 12.Leitao, A. L., F. J. Enguita, A. de la Fuente, P. Liras, and J. F. Martin. 1999. Inducing effects of diamines on transcription of the cephamycin C genes from the lat and pcbAB promoters in Nocardia lactamdurans. J. Bacteriol. 181:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liras, P. 1999. Biosynthesis and molecular genetics of cephamycins. Antonie Leeuwenhoek 75:109-124. [DOI] [PubMed] [Google Scholar]
- 14.Madduri, K., C. Stuttard, and L. C. Vining. 1989. Lysine catabolism in Streptomyces spp. is primarily through cadaverine: β-lactam producers also make α-aminoadipate. J. Bacteriol. 171:299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malmberg, L. H., W. S. Hu, and D. H. Sherman. 1993. Precursor flux control through targeted chromosomal insertion of the lysine ɛ-aminotransferase (lat) gene in cephamycin C biosynthesis. J. Bacteriol. 175:6916-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosher, R. H., A. S. Paradkar, C. Anders, B. Barton, and S. E. Jensen. 1999. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 43:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagarajan, R., and D. O. Spry. 1971. The 3-cephem chromophore. J. Am. Chem. Soc. 93:2310-2312. [DOI] [PubMed] [Google Scholar]
- 18.O'Sullivan, J., R. T. Aplin, C. M. Stevens, and E. P. Abraham. 1979. Biosynthesis of a 7-α-methoxycephalosporin. Incorporation of molecular oxygen. Biochem. J. 179:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paradkar, A. S., and S. E. Jensen. 1995. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J. Bacteriol. 177:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Llarena, F. J., A. Rodriguez-Garcia, F. J. Enguita, J. F. Martin, and P. Liras. 1998. The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of α-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J. Bacteriol. 180:4753-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao, R. N., M. A. Richardson, and S. Kuhstoss. 1987. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 153:166-198. [DOI] [PubMed] [Google Scholar]
- 22.Rius, N., K. Maeda, and A. L. Demain. 1996. Induction of l-lysine epsilon-aminotransferase by l-lysine in Streptomyces clavuligerus, producer of cephalosporins. FEMS Microbiol. Lett. 144:207-211. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Tobin, M. B., S. Kovacevic, K. Madduri, J. A. Hoskins, P. L. Skatrud, L. C. Vining, C. Stuttard, and J. R. Miller. 1991. Localization of the lysine ɛ-aminotransferase (lat) and δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (pcbAB) genes from Streptomyces clavuligerus and production of lysine ɛ-aminotransferase activity in Escherichia coli. J. Bacteriol. 173:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, L., K. Tahlan, T. L. Kaziuk, D. C. Alexander, and S. E. Jensen. 2004. Transcriptional and translational analysis of the ccaR gene from Streptomyces clavuligerus. Microbiology 150:4137-4145. [DOI] [PubMed] [Google Scholar]