Abstract
Transcription of DNA transfer genes is a prerequisite for conjugative DNA transfer of F-like plasmids. Transfer gene expression is sensed by the donor cell and is regulated by a complex network of plasmid- and host-encoded factors. In this study we analyzed the effect of induction of the heat shock regulon on transfer gene expression and DNA transfer in Escherichia coli. Raising the growth temperature from 22°C to 43°C transiently reduced transfer gene expression to undetectable levels and reduced conjugative transfer by 2 to 3 orders of magnitude. In contrast, when host cells carried the temperature-sensitive groEL44 allele, heat shock-mediated repression was alleviated. These data implied that the chaperonin GroEL was involved in negative regulation after heat shock. Investigation of the role of GroEL in this regulatory process revealed that, in groEL(Ts) cells, TraJ, the plasmid-encoded master activator of type IV secretion (T4S) system genes, was less susceptible to proteolysis and had a prolonged half-life compared to isogenic wild-type E. coli cells. This result suggested a direct role for GroEL in proteolysis of TraJ, down-regulation of T4S system gene expression, and conjugation after heat shock. Strong support for this novel role for GroEL in regulation of bacterial conjugation was the finding that GroEL specifically interacted with TraJ in vivo. Our results further suggested that in wild-type cells this interaction was followed by rapid degradation of TraJ whereas in groEL(Ts) cells TraJ remained trapped in the temperature-sensitive GroEL protein and thus was not amenable to proteolysis.
Bacterial conjugation is a cell-to-cell-contact-dependent DNA transfer mechanism which represents one of the major routes for horizontal gene transfer among bacteria. Conjugative plasmids or other self-transmissible elements such as integrative conjugative elements carrying DNA transfer (tra) genes are highly relevant for human health problems because they can mediate the rapid spread of antibiotic resistance genes (29, 50) and can confer the capacity to form biofilms (11, 32). The bacterial conjugation machinery is composed of a nucleoprotein complex required for conjugative DNA replication, termed the relaxosome (24), and a dedicated DNA transporter, which is a cell envelope-spanning protein complex. It is assumed that this DNA transporter also secretes a filamentous surface appendage, the sex pilus, which is required for initial contact with recipient cells. This DNA and protein translocation machinery required for bacterial conjugation is ancestrally related to protein secretion machineries of pathogenic gram-negative bacteria and represents a subsystem of type IV secretion (T4S) systems (for recent reviews see references 7 and 35).
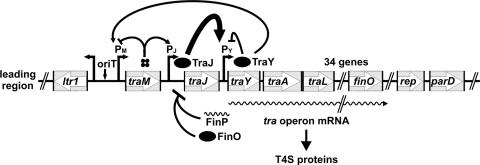
The expression of plasmid-encoded T4S system genes which are required for DNA transfer is controlled at various levels both by plasmid and host factors. In repressed F-like plasmids of enterobacterial origin (R1, R100, and pSLT) plasmid- and chromosome-encoded regulators act together to form a complex regulatory circuit (Fig. 1). The regulatory system of plasmid R1 is dominated by repression of traJ mRNA translation via the FinOP repressor system (19, 20). This repressor system is very efficient, and only a small fraction of a given cell population (approximately 0.1% in logarithmically growing cells) can escape from FinOP control. This leads to synthesis of TraJ, which then reaches an (unknown) threshold concentration required for the transcriptional activation of transfer genes. The TraJ protein stimulates transcription of the tra operon from the PY promoter, a process for which the host-encoded ArcA response regulator is necessary (39, 44, 45). It has been shown that ArcA-P, the phosphorylated form of this response regulator, binds to the PY promoter region in vitro (44), suggesting that ArcA-P can activate transcription from this promoter in vivo. The molecular mechanism underlying PY promoter activation by TraJ and ArcA, however, is unclear. The TraY protein, encoded by the first gene of the tra operon, is a sequence-specific DNA binding protein with binding sites in cognate oriT and PY promoter regions (15, 28). TraY participates in DNA processing for transfer (17, 27) and has been identified as a positive regulator of traM transcription (30, 42). Furthermore, it has been proposed that TraY is involved in transcriptional regulation of the tra operon, where it presumably acts as a repressor as reported for plasmid R100 (45). TraY of F binds with fivefold-higher affinity to its binding site at oriT (28); thus it seems likely that TraY first pushes the system to the “on” state, and only when TraY molecules accumulate would TraY then serve to turn the system off (i.e., the default state in repressed plasmids) by binding to the PY promoter. Another protein involved in regulation of tra gene expression is TraM. The TraM protein promotes contact formation between the DNA to be transferred and the T4S machinery via interaction with the coupling protein TraD (1, 9, 26). In addition, by binding as a tetramer to several sites in the oriT region overlapping with the PM promoters (31, 49) it acts as a relaxosomal component (22) and a transcriptional autorepressor (36).
FIG. 1.
Schematic representation of the R1 tra region and its regulation by plasmid-encoded factors. Positive regulation is indicated by arrows and negative regulation by black bars. Further details are outlined in the text.
Besides ArcA, several other host factors that have the potential to regulate the expression of the tra genes have been identified. IHF has been reported to be required for TraI-catalyzed nicking at oriT (17, 27) and transfer gene expression (8, 10). H-NS (40, 52), Lrp (2, 40), and cyclic AMP receptor protein (41) are believed to act in response to the physiology of the host cell and primarily affect conjugation by regulating transcription from the PJ promoter. In addition, the methylation state of the DNA can influence the expression of traJ and finP, as shown in case of the pSLT virulence plasmid (2-4, 48). Hfq, an RNA chaperone, can destabilize traM and traJ mRNAs and decrease the synthesis of the corresponding proteins (51). Last but not least, it has been demonstrated that protein TraJ is unstable in a cpxA* background, in which the sensor kinase of the CpxAR system constitutively activates the response regulator CpxR (13). However, the mechanism responsible for TraJ destabilization initiated by constitutive activation of CpxR remained unclear. Thus, as outlined above, multiple regulatory inputs by host factors ultimately determine whether in repressed plasmids PY promoter activation by the plasmid-encoded activator protein TraJ and transfer gene expression will eventually occur. This complex regulation of conjugative functions by host factors is thought to optimally adjust the expression level of the transfer genes to the physiological condition of the bacterial host in order to minimize the metabolic burden and to maximize the success of horizontal DNA spread (55).
Recent studies in our laboratory revealed that the expression and assembly of the R1-encoded T4S machinery elicit extracytoplasmic and cytoplasmic stress responses (54). The cytoplasmic stress response mediated by the expression/assembly of the T4S machinery is highly similar to the classical heat shock response, including increased levels of the heat shock sigma factor σ32 and concomitant transcriptional induction of the heat shock regulon. Among the activated genes in this regulon are the dnaJK and groESL operons, encoding the two main chaperone machineries in Escherichia coli. Based on these observations that sex factor expression induces stress in bacterial cells, we suggested a model in which the elicited stress response may down-regulate or fine-tune transfer gene expression in order to limit potentially detrimental effects to the cell (54). Here we test the validity of this model. By using heat shock as a tool to induce the expression of heat shock proteins (HSPs) we show that heat shock severely reduces transfer of plasmid R1-16, a derepressed variant of plasmid R1. We also show that this process depends on the presence of a functional GroEL protein. We describe that the master regulator of tra operon transcription, TraJ, becomes unstable after heat shock and that GroEL plays a key role in its destabilization.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1. The traJ gene was amplified using the oligonucleotide pair traJ-N-flag (TAGAATTCCATGGACTACAAAGACGACGATGACAAGTGTGCGCTGGACCGTAGA) and traJ-C (CGTCTAGATTATTACTTAACACCATAAAATTCACG) for addition of an N-terminal FLAG tag and the oligonucleotide pair traJ-N (TAGAATTCCATGGCGCTGGACCGTAGAG) and traJ-C-flag (CGTCTAGATTATTACTTGTCATCGTCGTCTTTGTAGTCCTTAACACCATAAAATTCACG) for addition of a C-terminal FLAG tag. Underlined letters in the oligonucleotide sequences indicate the DNA sequence of the FLAG peptide (DYKDDDDK). The PCR fragments were restricted with EcoRI and XbaI (recognition sequences are indicated by italics in the oligonucleotide sequences) and ligated into pGZ119EH, yielding pTGNF4 (FLAG-TraJ expression vector) and pTGCF1 (TraJ-FLAG expression vector). The inserted sequences were verified by DNA sequencing. The GST-TraY expression plasmid pGEX-traY was kindly provided by E. Zechner.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Source and/or reference |
|---|---|---|
| Bacterial strains | ||
| MC4100 | F−araD139 Δ(argF-lac)V169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 5 |
| J5 | F−pro met lac+ λ+ | IMB collection |
| NRK117 | MC4100 groEL44(Ts) zje::Tn10 Tcr | K. Ito; 23 |
| Plasmids | ||
| R1-16 | Derepressed variant of IncFII plasmid R1, Kmr | 12 |
| pAR183 | Variant of IncFII plasmid R1, Kmr | A. Reisner; 33 |
| pGZ119EH | ColD replicon, IPTG-inducible expression vector, Cmr | 25 |
| pTGNF4 | N-terminally FLAG-tagged traJ cloned into pGZ119EH | This study |
| pTGCF1 | C-terminally FLAG-tagged traJ cloned into pGZ119EH | This study |
| pGEX-traY | N-terminal GST-tagged traY cloned into pGEX-6P-1 (Pharmacia Biotech) | E. Zechner |
| pExtraM | Temp-inducible TraM expression vector, Apr | 49 |
Media and growth conditions.
For all experiments 2× TY medium (16 g tryptone liter−1, 10 g yeast extract liter−1, and 5 g NaCl liter−1; prewarmed to the respective incubation temperature) was used. Cultures were grown in Erlenmeyer flasks (300-ml total volume for stability experiments and pull down assays and 100-ml total volume for all other experiments) in a shaking water bath to provide aeration. E. coli precultures (4- to 10-ml culture volumes) grown to stationary phase at 22°C or 37°C were diluted 1:22 or 1:50 (for protein stability and pull down experiments) into prewarmed medium (50 ml for stability experiments and pull down assays and 18 ml for all other experiments) and incubated at 37°C or 43°C. For RNA and protein isolation at various time points after heat shock or dilution into fresh medium 5 × 109 CFU of the cultures (between 0.5 and 4 ml) were harvested by centrifugation at 2,600 × g for 4 min at 4°C and the cell pellets were quickly frozen in liquid nitrogen and stored at −70°C until further processing steps were applied. When necessary, antibiotics were added to the following concentrations: 100 μg ampicillin ml−1, 20 μg chloramphenicol ml−1, 40 μg kanamycin ml−1, 15 μg tetracycline ml−1, and 200 μg rifampin ml−1.
Preparation of RNA and Northern blot analyses.
Total RNA was isolated from E. coli cells harvested as described above using the RNeasy minikit (QIAGEN), by following the manufacturer's protocol (available from http://www1.qiagen.com/literature/). RNA concentration was determined by measuring the absorption at 260 nm. Northern blot analysis was performed according to reference 34. Briefly, 5 μg of total RNA from each sample was denatured and separated in a 1.2% agarose gel containing 1.2 M formaldehyde. RNA was transferred onto a nylon membrane (Hybond-N; Amersham Biosciences) by capillary transfer and cross-linked by UV radiation. For hybridization and detection, reagents of the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science) were used according to the manufacturer's protocol (http://www.roche-applied-science.com/pack-insert/1585614a.pdf). The traA and traJ DNA probes were PCR amplified from plasmid R1-16 using the oligonucleotide pairs traA_fw (CGTCTGAATATGCTTCGCC) and traA_rev (AGAACCGCTGCACCAATAC) and traJ-N and traJ-C (see above). The probes were labeled with digoxigenin-11-2′-deoxyuridine-5′-triphosphate (DIG-11-dUTP; Roche Applied Science). For detection an anti-DIG-11-dUTP alkaline phosphatase conjugate and disodium 3-(4-methoxyspiro-[1,2-dioxetane-3,2-{5-chloro}-tricyclo-{3.3.1.13,7}-decan-4-yl] phenyl phosphate) (CSPD) were used. The chemiluminescent bands were visualized on X-ray films. For quantification of the signals the blots were scanned densitometrically using a Personal densitometer (Molecular Dynamics) and ImageQuant 5.1 software (Molecular Dynamics).
Determination of TraM levels after heat shock.
For determination of steady-state TraM levels E. coli MC4100(R1-16) and E. coli NRK117(R1-16) cultures were grown and harvested as described above. The cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA) and lysed by sonication four times for 30 seconds each with a Branson sonifier 250 using a microtip. To avoid heat denaturation of proteins, samples were cooled on ice. After removal of cell debris by centrifugation for 10 min at 5,000 × g and 4°C the protein concentration of each supernatant was determined using the Bio-Rad protein assay. Proteins were precipitated with 10% trichloroacetic acid for 30 min on ice. After centrifugation for 10 min at 23,000 × g and 4°C the protein pellets were washed three times with 1 ml ice-cold double-distilled water. Protein pellets were dissolved in final sample buffer, and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting were essentially performed as described previously (31). Briefly, 20 μg of total protein per lane was separated on a 15% SDS-polyacrylamide gel (PAG). After electrophoresis, proteins were electrotransferred onto nitrocellulose membranes (Immobilon-P; Millipore). Subsequently the membranes were blocked for 15 min with TST buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 80) containing 3% dry milk (Bio-Rad). The immunological detection of TraM was performed using an affinity-purified polyclonal antiserum raised against purified TraM (31) diluted 1:2,000 in TST buffer containing 1% dry milk, and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (diluted 1:15,000 in TST buffer containing 1% dry milk; Sigma-Aldrich). For immunological detection of GroEL a horseradish peroxidase-conjugated polyclonal antibody (Sigma-Aldrich; diluted 1:15,000 in TST buffer containing 1% dry milk) was used by following the manufacturer's instructions. For photochemical detection the ECL system (Amersham Biosciences) was used. Quantification was performed as described above.
Mating assays.
Overnight cultures of the donor strains E. coli MC4100(R1-16) and E. coli NRK117(R1-16) grown at 22°C (for subsequent heat shock) or 37°C (no heat shock) in 2× TY medium containing the appropriate antibiotics were diluted 1:22 into 18 ml of fresh, prewarmed medium and further incubated at 43°C (heat shock) or 37°C (no heat shock). Directly after dilution as well as 15 min before the time points, indicated in the figure legends, after heat shock or dilution, 450 μl of the cultures was transferred into prewarmed 1.5-ml reaction tubes and 50 μl of the recipient strain E. coli J5 from an overnight culture grown at 37°C was added. To allow mating, the suspensions were further incubated at 37°C or 43°C for 15 min. DNA transfer was interrupted by vigorously mixing for 1 min. Serial dilutions prepared in 0.9% NaCl were plated on MacConkey agar containing kanamycin and incubated at 37°C to allow formation of colonies. The preincubation time at 37°C or 43°C plus 15 min of mating represent the total time in minutes after dilution or heat shock. The conjugation frequency is expressed as the number of transconjugants (red colonies) per donor cell (white colonies). Heat shock mating assays with concomitant expression of TraJ-FLAG were performed as described above with wild-type and groEL(Ts) cells both harboring the repressed conjugative plasmid pAR183 (a kind gift from A. Reisner) and plasmid pTGCF1. Expression of TraJ-FLAG was induced by addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). For Western blot analyses of TraJ-FLAG and maltose binding protein (MBP) 1-ml aliquots of the donor cultures were harvested as described above after heat shock. The cell pellets were resuspended in phosphate-buffered saline buffer, and proteins corresponding to 0.2 unit of optical density at 600 nm were separated electrophoretically in 12.5% SDS-PAG. Following transfer of proteins onto a nitrocellulose membrane TraJ-FLAG was detected immunologically using an anti-FLAG antibody (Sigma; diluted 1:4,000 in TST buffer containing 1% dry milk powder) and an anti-mouse horseradish peroxidase conjugate (Amersham Pharmacia; diluted 1:15,000 in TST buffer containing 1% dry milk powder). MBP was detected as described previously (53).
Stability of GST-TraY, FLAG-TraJ, and TraJ-FLAG.
Overnight cultures of E. coli MC4100 and E. coli NRK117 cells harboring either pGEX-traY or pTGNF4 or pTGCF1 grown to stationary phase at 37°C in 2× TY medium containing the appropriate antibiotics were diluted 1:50 into 50 ml of fresh medium and incubated for 60 min at 37°C in a shaking water bath. The expression of fusion proteins was induced by addition of IPTG (0.075 mM for GST-TraY expression and 0.5 mM for FLAG-TraJ and TraJ-FLAG expression). After 60 min 1-ml aliquots of the cultures were harvested by centrifugation at 2,600 × g for 4 min at 4°C as an expression control. The cultures were then centrifuged at 2,600 × g for 5 min at 27°C, and the cell pellets were resuspended in 40 ml of 2× TY medium containing 0.2% glucose prewarmed to 43°C. After a 15-min incubation at 43°C 1-ml aliquots were harvested as described above. Subsequently, rifampin was added to the cultures to inhibit further transcription. Fifteen, 30, and 45 min after addition of rifampin 1-ml aliquots were harvested as described above. The cell pellets were resuspended in phosphate-buffered saline buffer, and proteins corresponding to 0.2 unit of optical density at 600 nm were separated electrophoretically in 12.5% SDS-PAG. After proteins were transferred onto a nitrocellulose membrane, GST-TraY was detected with a glutathione S-transferase (GST)-specific antibody (Sigma; diluted 1:16,000 in TST buffer containing 1% dry milk) and horseradish-peroxidase-conjugated anti-rabbit immunoglobulin G. FLAG-TraJ, TraJ-FLAG, and MBP were detected as described above. Bands on X-ray films were quantified as described above. Half-lives of proteins were calculated using the Prism software, version 3.03 (Graph Pad Software Inc.).
Stability of TraM.
Overnight cultures of E. coli MC4100 and E. coli NRK117 harboring pExtraM (49) were grown to stationary phase at 30°C, diluted 1:50 into 50 ml of fresh medium, and incubated for 60 min at 30°C. The expression of TraM was induced by shifting the cultures to 37.8°C. After 60 min 1-ml aliquots of the cultures were harvested by centrifugation at 2,600 × g for 4 min at 4°C as an expression control. All subsequent steps were performed as described for GST-TraY and FLAG-tagged TraJ.
Pull down assay.
Overnight cultures of E. coli MC4100 and E. coli NRK117 cells both harboring either pGZ119EH (vector control) or pTGNF4 were diluted 1:50 into 100 ml of 2× TY medium containing chloramphenicol and incubated for 60 min at 37°C. Then the cultures were shifted to 43°C, and 1 mM IPTG was added for induction of FLAG-TraJ expression. After 45 min cells were harvested by centrifugation for 5 min at 2,600 × g and 4°C. After a washing, the cells were resuspended in 1.5 ml PD buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing complete protease inhibitor (Roche Diagnostics) and lysed by sonication as described above. After addition of Triton X-100 (0.1% final concentration) and incubation on ice for 10 min cell debris was removed by centrifugation as described above and the crude extract was transferred into a fresh 1.5-ml reaction tube. Five hundred microliters of the crude extract was incubated with 40 μl of anti-FLAG M2 affinity gel (Sigma; equilibrated with PD buffer containing Triton X-100) on ice for 90 min. The affinity gel matrix was then sedimented by centrifugation at 10,000 rpm for 10 seconds in a microcentrifuge. The supernatant was removed, and the affinity gel matrix was washed three times with 350 μl of cold PD buffer containing Triton X-100. For elution of proteins the affinity gel matrix was incubated with 100 μl 0.1 M glycine-HCl, pH 3.5, at room temperature for 5 min and then sedimented as described above. The supernatant was transferred into a fresh 1.5-ml reaction tube containing 10 μl neutralization buffer (0.5 M Tris-HCl, 1.5 M NaCl, pH 7.5). Proteins of the crude extracts and in the pull down eluates were separated electrophoretically on 12.5% SDS-PAG and FLAG-TraJ, GroEL, and MBP were detected by immunoblotting as described above.
RESULTS
A functional GroEL protein is essential for the efficient repression of conjugative transfer and transfer gene expression of plasmid R1-16 after heat shock.
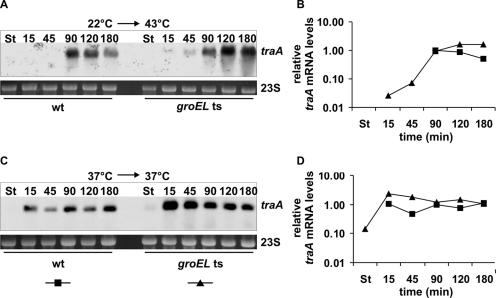
We previously found that expression and assembly of the T4S machinery encoded by plasmid R1 elicited extracytoplasmic and cytoplasmic stress responses in E. coli. Based on our findings we proposed a model in which a negative-feedback loop existed to ensure proper fine-tuning and to limit transfer gene expression (54). Our model predicted that the induction of HSPs negatively affects conjugative transfer and transfer (tra) gene expression. To test the validity of our model, we used heat shock as a tool to induce the expression of HSPs in cells harboring R1-16. To investigate a possible role of the heat shock chaperonin GroEL, we compared DNA transfer and tra transcript levels in wild-type and groEL(Ts) cells carrying the well-characterized E191G mutation in GroEL, which is encoded by the groEL44 allele (6). Our experimental setup was designed to allow induction of HSPs by heat shock and concomitant shift of groEL(Ts) cells to the restrictive temperature of 43°C (for details see Materials and Methods). For the investigation of the tra transcript levels we used a probe specific for the stable region of the tra transcript, which comprises the traA coding sequence and the 5′ region of the traL coding region (18, 21). At 22°C no conjugative DNA transfer was detectable in wild-type and groEL(Ts) cells and no traA transcripts were detectable (Fig. 2A and 3A and B). It is important that, at 22°C in E. coli cells harboring the derepressed plasmid R1-16, at no time during growth is transfer gene expression detectable nor does any DNA transfer occur (data not shown), suggesting a suppression of tra expression at low temperatures. Compared to what was found for non-heat-shocked cells (Fig. 2B), heat shock had severe effects on transfer of R1-16 in wild-type cells (Fig. 2A). Fifteen minutes after shift from 22°C to 43°C conjugative DNA transfer was below 1 × 10−5 transconjugant per wild-type donor cell and increased to a still very low level of 1 × 10−4 after 130 min of incubation at 43°C. Full transfer competence (0.1 transconjugant per donor cell) of wild-type cells was gained 180 min after heat shock, indicating that the increased temperature per se does not inhibit DNA transfer. In line with the observation of severely reduced DNA transfer, no traA transcripts were detectable in wild-type cells 15 and 45 min after heat shock (Fig. 3A and B). Ninety minutes after heat shock the traA transcript level increased dramatically (about 100-fold) but slightly decreased over further incubation at 43°C. In comparison, non-heat-shocked wild-type cells already resumed tra transcription 15 min after dilution into fresh medium (Fig. 3C and D).
FIG. 2.
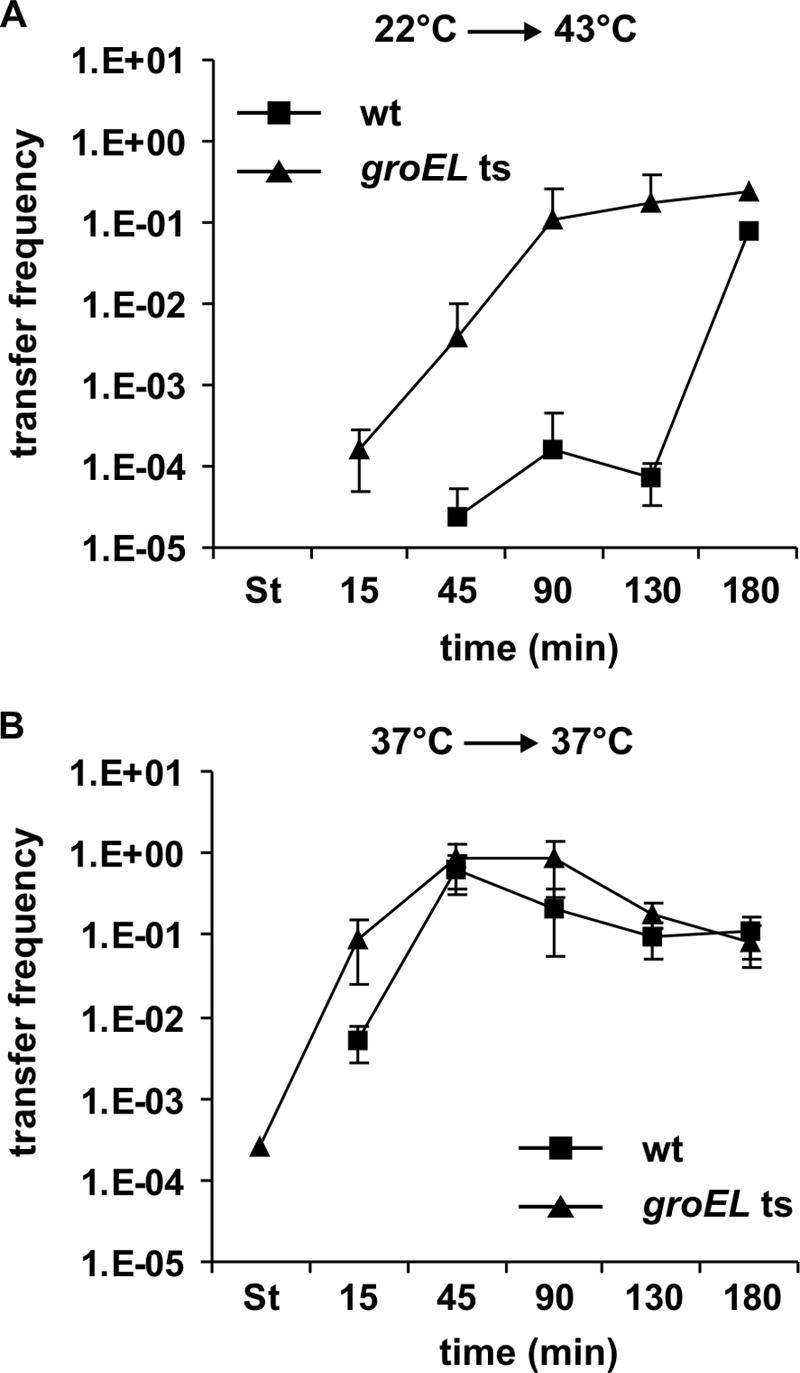
Heat shock represses transfer of R1-16 in a GroEL-dependent way. Mating assays with E. coli MC4100 cells (wild type [wt]) and NRK117 cells [groEL(Ts)] harboring R1-16 were performed as described in Materials and Methods. (A) Heat shock mating assays. Cells carrying the groEL(Ts) allele gained full transfer competence (>0.1 transconjugant per donor cell) 90 min after heat shock, whereas wild-type cells gained full transfer competence 180 min after heat shock. (B) Mating assays without heat shock. Both strains gained full transfer competence 45 min after dilution. The transfer frequencies are expressed as transconjugants per donor cell. Shown are means ± standard deviations of at least three independent experiments. Data points that are not depicted indicate that DNA transfer was not detectable at these time points (transfer frequency < 10−7 transconjugant per donor cell). St, stationary phase.
FIG. 3.
Heat shock represses tra operon transcription in a GroEL-dependent way. Northern blot analyses of traA mRNA levels in E. coli MC4100(R1-16) (wild type [wt]) and NRK117(R1-16) [groEL(Ts)] with (A) and without (C) heat shock. The 23S rRNA is shown as a loading control. (B and D) Quantification of traA signals. The traA signal intensities were normalized against the amount of RNA loaded on the gel, and the intensities of the traA signals in wild-type cells 90 min after heat shock or dilution were set as 1. Data points that are not depicted indicate that traA mRNA was not detectable at these time points. Experiments were repeated twice; one representative result is shown. St, stationary phase at 22°C/37°C.
In sharp contrast to that from wild-type cells, DNA transfer from cells carrying the groEL(Ts) allele was clearly detectable 15 and 45 min after heat shock (Fig. 2A; 1.6 × 10−4 transconjugant per donor cell after 15 min and 3.8 × 10−3 transconjugant per donor cell after 45 min). These cells gained full transfer competence 90 min after heat shock. It is important that incubation of the groEL(Ts) strain at the restrictive temperature arrested cell division but did not lead to cell death during the duration of the experiment (data not shown). Therefore, the increased transfer frequencies of groEL(Ts) cells compared to wild-type cells after heat shock were not an artifact caused by death of the temperature-sensitive donor strain. In compliance with the results of the mating assays groEL(Ts) cells showed weak but detectable levels of the traA transcripts 15 and 45 min after heat shock. At 90 min after heat shock the traA transcript level was similar to the that of the wild-type strain. In contrast to those of the wild-type strain, groEL(Ts) strain traA transcript levels further increased 120 and 180 min after heat shock, indicating deregulation of tra operon transcription (Fig. 3A and B).
Northern blot analyses of traA transcript levels and mating assays without heat shock showed that after stationary-phase repression wild-type and groEL(Ts) cells both resumed conjugation and tra operon transcription 15 min after dilution into fresh medium and gained full transfer competence after 45 min (Fig. 2B and 3C and D), demonstrating highly similar regulatory kinetics of conjugative DNA transfer at the permissive temperature. However, the typical repression of DNA transfer in stationary-growth phase (52, 54) was not as pronounced in groEL(Ts) cells as in wild-type cells. Upon prolonged incubation at 37°C a slight decrease in transfer frequency of R1-16 could be observed in both cases, reflecting the reentry of the strains into stationary phase (Fig. 2B).
Taken together our data strongly suggested that heat shock transiently repressed tra gene expression and conjugative transfer of plasmid R1-16 in wild-type cells. Results obtained with the groEL(Ts) strain suggested that the heat shock chaperonin GroEL (HSP60) is involved in this repression process. The following experiments were designed to elucidate the role of GroEL in regulation of tra gene expression and conjugative DNA transfer.
In groEL(Ts) cells steady-state levels of the essential transfer protein TraM are increased after heat shock.
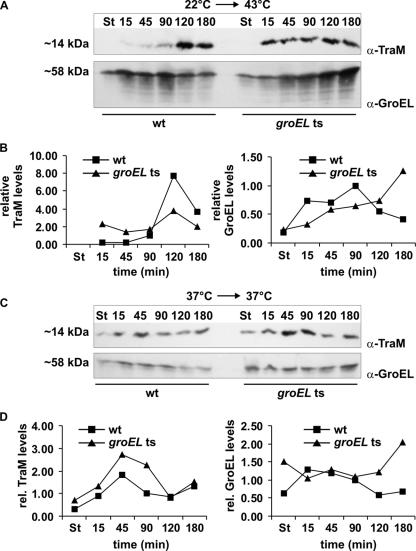
The traM gene is transcribed separately from the tra operon, and its expression is positively affected by TraY, encoded by the first gene of the tra operon (Fig. 1). Therefore we reasoned that steady-state levels of protein TraM should be affected by heat shock. Wild-type and groEL(Ts) cells both harboring R1-16 were either subjected to heat shock or incubated at 37°C, total proteins were separated by SDS-polyacrylamide gel electrophoresis, and Western blotting was performed with anti-TraM and anti-GroEL antisera. GroEL levels were analyzed in order to monitor the heat shock response. In stationary phase at 37°C the TraM protein levels in wild-type cells were fourfold lower than they were 15 min after dilution into fresh medium (Fig. 4C and D). Compared to wild-type cells no significant differences in TraM steady-state levels were detectable in groEL(Ts) cells grown at the permissive temperature. At 22°C no TraM was detectable in both wild-type and groEL(Ts) cells (Fig. 4A and B). Similar to heat shock-mediated repression of conjugative DNA transfer and tra operon transcription in wild-type cells—and in sharp contrast to non-heat-shocked cells—the steady-state levels of TraM were significantly reduced in wild-type cells until 90 min after heat shock. Maximum TraM levels were reached 120 min after heat shock, and again a slight decrease could be observed after 180 min. The results obtained by Northern blot analyses of tra transcript levels and Western blot analyses of TraM levels after heat shock imply that in a first step transcription of the tra operon resumes at about 90 min after heat shock and TraY and other Tra proteins are synthesized. As a consequence of increased TraY levels TraM synthesis becomes fully activated.
FIG. 4.
After heat shock TraM steady-state levels are transiently reduced dependent on a functional GroEL protein. Western blot analyses of TraM and GroEL steady-state levels in E. coli MC4100(R1-16) (wild type [wt]) and NRK117(R1-16) [groEL(Ts)] were performed with (A) and without (C) heat shock. (B and D) Quantification of TraM and GroEL signals. The intensity of the TraM or the GroEL signals in wild-type cells 90 min after heat shock or dilution was set as 1. Data points that are not depicted indicate that no protein was detectable at these time points. St, stationary phase at 22°C/37°C.
In groEL(Ts) cells no reduction of the TraM levels was detectable after heat shock (Fig. 4A and B). Therefore, similar to its role in DNA transfer and tra operon transcription, GroEL plays a key role in the heat shock-mediated reduction of TraM steady-state levels. We considered two possible explanations for these observations: (i) indirectly, GroEL could be involved in transcriptional regulation of the plasmid-encoded key regulatory genes traM, traY, and traJ and (ii) GroEL could be involved in stabilization or destabilization of TraM, TraY, and/or TraJ. Further experiments were designed to test these hypotheses.
Identification of protein TraJ as a target of GroEL-mediated proteolysis.
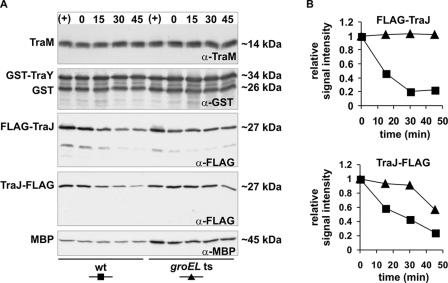
To investigate if GroEL is directly involved in the regulation of the traM promoter (PM), the traY promoter (PY), or the traJ promoter (PJ) after heat shock, we performed β-galactosidase assays with wild-type and groEL(Ts) cells carrying a traM-lacZ fusion (31), a traY-lacZ fusion (44), or a traJ-lacZ fusion (20). No differences in PM, PY, and PJ promoter activities in wild-type and groEL(Ts) cells could be observed after heat shock (data not shown), demonstrating that GroEL is not involved in regulating these promoters. Therefore we tested whether the induction of HSPs affects the stability of plasmid-encoded key regulators as outlined above and a possible involvement of GroEL in this process. To address this question, we compared the stabilities of TraM, of a functional GST-TraY fusion, and of functional either N- or C-terminally FLAG-tagged TraJ (named FLAG-TraJ or TraJ-FLAG, respectively) in wild-type and groEL(Ts) cells after heat shock (for details see Materials and Methods). These analyses revealed that both TraM and TraY were highly stable after heat shock and no differences in stabilities in wild-type and groEL(Ts) cells were observed (Fig. 5A). In control experiments we determined the stabilities of FLAG-TraJ and TraJ-FLAG in wild-type cells which had not been subjected to heat shock (Fig. 6A). Even without heat shock FLAG-TraJ and TraJ-FLAG proteins were less stable than TraM and TraY, implying rapid turnover and high susceptibility of TraJ to proteases.
FIG. 5.
GroEL is involved in destabilization of FLAG-TraJ and TraJ-FLAG after heat shock. Stabilities of TraM, GST-TraY, FLAG-TraJ, TraJ-FLAG, and MBP in E. coli MC4100 cells (wild type [wt]) and NRK117 cells [groEL(Ts)] after heat shock were determined as described in Materials and Methods. (A) Western blot analyses were performed with total cell lysates after induction of TraM, GST-TraY, FLAG-TraJ, or TraJ-FLAG expression (+), immediately before (0) and 15, 30, and 45 min after addition of rifampin using antibodies specific for TraM, for GST, for the FLAG peptide, and for MBP. α, anti. (B) Quantification of the FLAG-TraJ and TraJ-FLAG signals shown in panel A. The time zero signals were set as 1. Shown are the results of one representative experiment.
FIG. 6.
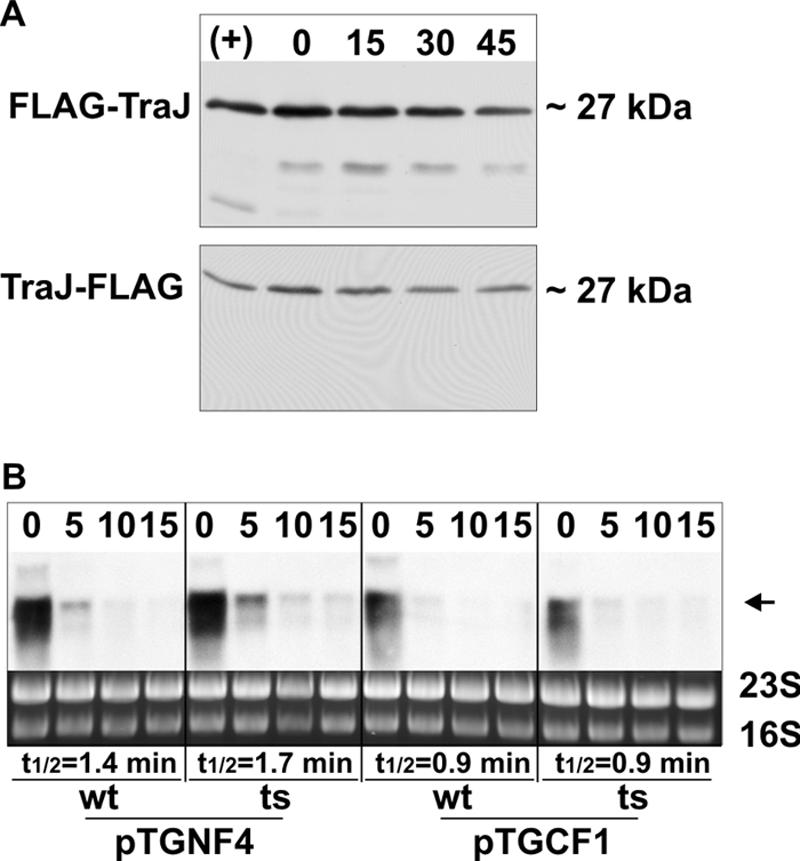
(A) FLAG-TraJ and TraJ-FLAG are more stable without heat shock. Stability of FLAG-TraJ and TraJ-FLAG in E. coli MC4100 cells harboring either pTGNF4 or pTGCF1 was determined as described in Materials and Methods. Western blot analyses were performed with total cell lysates after induction of FLAG-TraJ and TraJ-FLAG expression (+) immediately before (0) and 15, 30, and 45 min after addition of rifampin using an antibody specific for the FLAG peptide. (B) GroEL is not involved in regulating traJ and mRNA stability. Heat-shocked E. coli MC4100 (wild type [wt]) and NRK117 cells [groEL(Ts)] harboring either pTGNF4 or pTGCF1 were harvested immediately before (0) and at the indicated time points (minutes) after addition of rifampin to the cultures. Total RNA was isolated, and Northern blot analyses were performed using a traJ-specific probe. The arrow indicates the signal corresponding to the traJ transcripts. 23S and 16S rRNAs are shown as a loading control.
In contrast to TraM and GST-TraY and further underscoring the high protease susceptibility, both FLAG-TraJ and TraJ-FLAG were highly destabilized in wild-type cells after heat shock and half-lives decreased to less than 20 min (Fig. 5A and B). However, in groEL(Ts) cells the half-lives of these proteins were significantly longer, exceeding 40 min in both cases. As a control we determined the half-life of MBP after a shift to 43°C. No difference in the half-lives of MBP was detectable in wild-type and groEL(Ts) cells, excluding the possibility of general protein destabilization after heat shock. To exclude the possibility that the differences in FLAG-TraJ and TraJ-FLAG stability are the result of altered mRNA degradation, we isolated total RNA of wild-type and groEL(Ts) cells expressing these proteins immediately before and at different time points after addition of rifampin. Subsequent Northern blot analyses were performed using a traJ-specific probe. The traJ transcript levels before addition of rifampin were similar in wild-type and groEL(Ts) cells (Fig. 6B), indicating that RNA degradation was only marginally affected by the groEL(Ts) mutation. Thirty minutes after addition of rifampin no traJ transcripts were detectable in both cases (not shown). Therefore, the increased half-lives of FLAG-TraJ and TraJ-FLAG in groEL(Ts) cells after heat shock can be attributed to protein stabilization.
GroEL interacts with TraJ in vivo.
If GroEL was indeed involved in proteolysis, we hypothesized that it then should interact, albeit transiently, with TraJ. Thus, we tested experimentally if GroEL could be pulled down with FLAG-TraJ expressed in wild-type and groEL(Ts) cells using anti-FLAG agarose (for details see Materials and Methods). As shown in Fig. 7, a weak GroEL signal was visible in the eluate when wild-type cells expressing FLAG-TraJ were used. In contrast, a strong GroEL signal was present in the eluate in the case of groEL(Ts) cells expressing FLAG-TraJ. These data can be explained by the trapping of TraJ in the GroEL44 mutant protein at the nonpermissive temperature of 43°C. It is conceivable that, in this case, FLAG-TraJ cannot be prepared for proteolytic degradation by an as yet unknown GroEL-dependent protease and remains stable (Fig. 5). In the case of wild-type GroEL, FLAG-TraJ is rapidly degraded, as reflected by a lower stability and by its absence in the eluate (Fig. 7). These data corroborate the finding that proteolysis of TraJ, at least in part, proceeds via GroEL and show that GroEL directly interacts with TraJ in vivo.
FIG. 7.
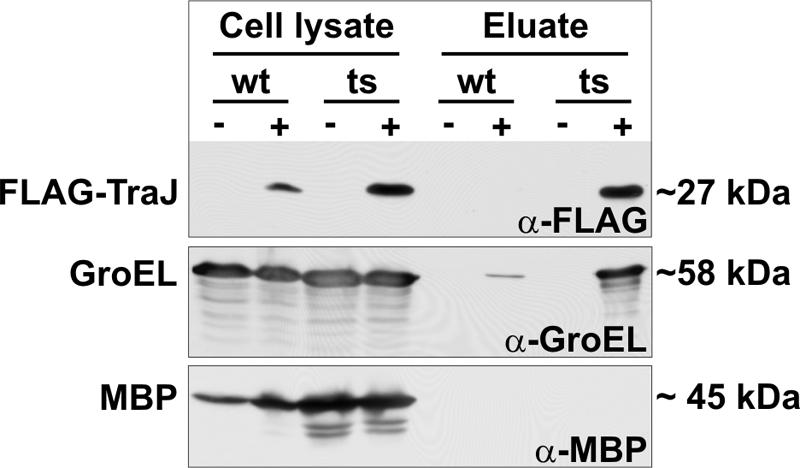
The GroEL temperature-sensitive (ts) protein traps FLAG-TraJ. Pull down experiments with lysates of E. coli MC4100 (wild-type [wt]) and NRK117 [groEL(Ts)] cells harboring either pGZ119EH (vector control; −) or pTGNF4 (FLAG-TraJ expression vector, +) shifted to 43°C were performed as described in Materials and Methods. Proteins in the cell lysates and in the pull down eluates were separated electrophoretically and analyzed by immunoblotting with antibodies specific for the FLAG peptide, for GroEL, and for MBP as a control. Shown are the results of one representative experiment.
GroEL-dependent TraJ degradation is physiologically relevant.
To validate the physiological relevance of the observed GroEL-mediated destabilization of TraJ in wild-type cells after heat shock, we exploited the fact that FLAG-tagged TraJ expressed in trans can induce a repressed plasmid R1 variant with a functional FinOP system (pAR183; transfer frequency approximately 10−4 transconjugant per donor cell) to full transfer competence. We hypothesized that the increased half-life of TraJ in groEL(Ts) cells after heat shock should result in higher steady-state levels of this protein compared to that in wild-type cells. As a consequence, induction of transfer of pAR183 should be detectable earlier after heat shock in groEL(Ts) cells than in wild-type cells.
At 22°C no transfer of pAR183 was detectable in wild-type and groEL(Ts) cells although cells expressed TraJ-FLAG (data not shown). These results imply that TraJ provided in trans cannot overcome repression of DNA transfer at low temperatures. Fifteen minutes after heat shock transfer of pAR183 was still below the detection limit in wild-type cells irrespective of induction of TraJ-FLAG expression with IPTG (Fig. 8A). However, when cells carried the groEL(Ts) allele, conjugative transfer was clearly detectable 15 min after heat shock. These results are similar to those obtained with plasmid R1-16 and demonstrate that the TraJ-FLAG fusion protein provided in trans can functionally substitute for the TraJ protein, which operates in the context of the natural conjugation system. In line with this finding Western blot analyses showed that the steady-state levels of TraJ-FLAG were significantly higher in groEL(Ts) cells than in wild-type cells (Fig. 8B). As already seen with R1-16 (Fig. 2A), transfer of pAR183 from wild-type and groEL(Ts) donors reached similar frequencies at later time points. As predicted, and corroborating our hypothesis that degradation of TraJ is dependent on GroEL, prolonged incubation of groEL(Ts) cells at 43°C resulted in accumulation of TraJ-FLAG (Fig. 8B).
FIG. 8.
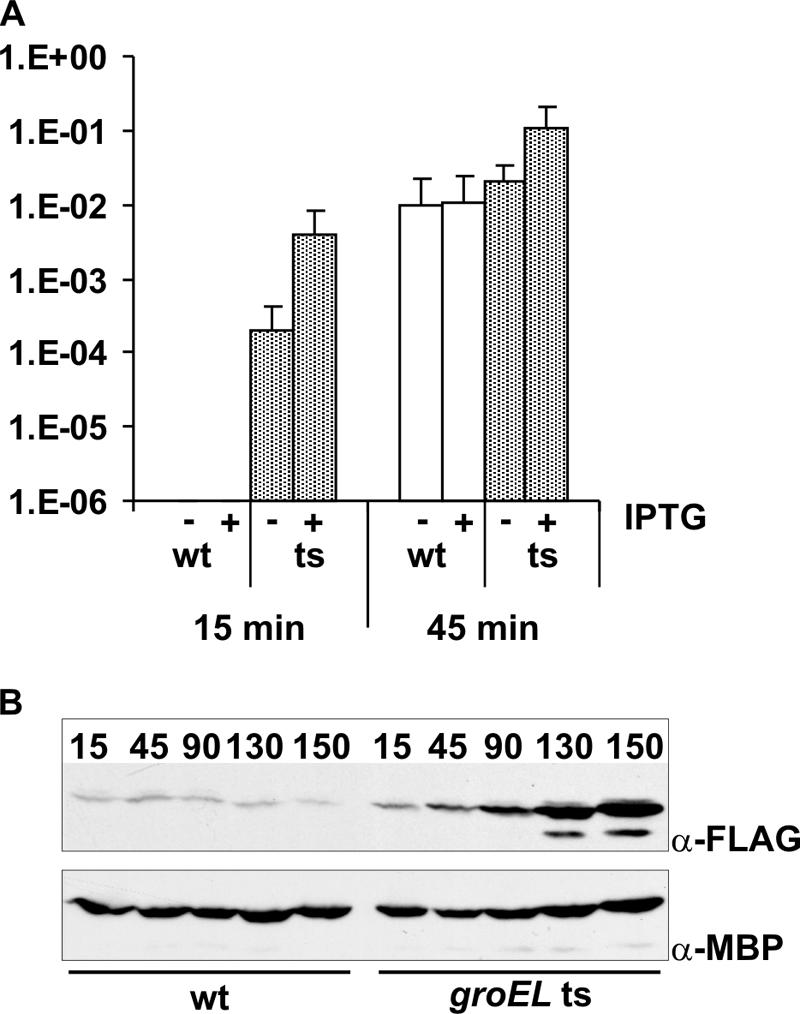
Derepression of pAR183 transfer after heat shock in wild-type and groEL(Ts) (ts) cells. (A) Heat shock mating assays with E. coli MC4100 cells (wild type [wt]) and NRK117 cells [groEL(Ts)] harboring both the repressed plasmid pAR183 and the TraJ-FLAG expression plasmid pTGCF1 were performed as described in Materials and Methods. Fifteen minutes after heat shock conjugative transfer was clearly detectable in the case of groEL(Ts) cells whereas no transfer was detectable in the case of wild-type cells. (B) Western blot analyses of TraJ-FLAG steady-state levels after heat shock in wild type and groEL(Ts) cells harboring plasmid pAR183 and the TraJ-FLAG expression plasmid. TraJ-FLAG accumulated in groEL(Ts) cells, in contrast to wild-type cells, after heat shock. Steady-state levels of MBP are shown as a loading control.
DISCUSSION
We here report that the heat shock chaperonin GroEL is involved in destabilization of TraJ, the master activator of tra operon transcription in F-like conjugative plasmids. As evidenced by the results of our experiments, turnover of protein TraJ is highly reduced in a groEL(Ts) mutant at the restrictive temperature. In line with these observations we found that overexpression of GroESL reduced DNA transfer 10-fold (our unpublished results). We propose that GroEL triggers degradation of TraJ by binding and partially unfolding the protein, thereby facilitating access of the as yet unknown heat shock protease(s) responsible for subsequent proteolysis. This is a likely explanation for our observations. It has been proposed that, besides correct folding of newly synthesized or misfolded proteins during cellular stress, chaperone complexes like DnaJK/GrpE or GroESL also stimulate degradation of proteins by ATP-dependent proteases such as Lon or Clp (16, 37, 38). Why is TraJ stabilized in the groEL(Ts) strain at the restrictive temperature? It has been suggested that the E191G mutation in GroEL, which is encoded by the groEL44(Ts) allele, leads to weakened interaction between GroEL and its cochaperone GroES (6). Therefore, it seems likely that TraJ is not or only very slowly released from the mutant GroEL. We suggest that this prolonged interaction results in the “clogging” of the GroEL-dependent degradation pathway and that therefore TraJ is stabilized in groEL(Ts) cells. Supporting this hypothesis is the fact that GroEL interaction with FLAG-TraJ at the nonpermissive temperature can be readily shown in the temperature-sensitive strain. TraJ is the master activator of tra operon transcription, and our data strongly suggest that it is the key target of triggered proteolysis under stressful conditions. We could demonstrate the physiological relevance of this process since the increased stability of TraJ in groEL(Ts) cells led to earlier induction of plasmid pAR183 transfer after heat shock compared to that in wild-type cells. We suggest that in wild-type cells rapid destabilization of TraJ, not only in response to heat shock but also in response to other stress conditions which induce the σ32 regulon (see below), results in fast and highly efficient down-regulation of Tra protein synthesis. In E. coli, an analogous regulatory mechanism ensures the function of the heat shock regulon itself: the paradigm for regulation by triggered proteolysis in bacteria is the rapid destabilization of the transiently stabilized σ32 factor, which leads to a rapid shutoff of the bacterial stress response. It has been shown that FtsH (HflB) protease-dependent degradation of σ32 (14, 47) is triggered by the DnaJK/GrpE chaperone complex (43, 46). Current work performed by our group aims to elucidate which GroEL-dependent protease(s) is involved in the degradation of TraJ.
As consequence of TraJ destabilization after heat shock no components of the T4S machineries are synthesized, presumably until the heat shock response is shut down and steady-state levels of heat shock proteins decrease. We suggest that the observed transient repression of DNA transfer after heat shock serves to ensure that the damages resulting from heat shock are adjusted without piling up additional stress caused by the expression and the assembly of the T4S machinery. This hypothesis is corroborated by the finding that at the restrictive temperature the concomitant expression of the T4S system components and TraJ-FLAG leads to death of groEL(Ts) cells, whereas in wild-type cells or in groEL(Ts) cells expressing only FLAG-tagged TraJ no such effects were observed (our unpublished results). Furthermore, we recently found that the expression and assembly of the R1-encoded T4S machinery elicit extracytoplasmic and cytoplasmic stress responses under physiological conditions (54). The extracytoplasmic stress response is triggered by the activation of the host-encoded CpxAR two-component system and results in activation or repression of the downstream targets of the CpxR response regulator. One of the activated downstream targets of CpxR is the rpoH gene, encoding the heat shock sigma factor σ32. The cytoplasmic stress response is highly similar to the classical heat shock response, resulting in transcriptional activation of the heat shock regulon, including the groESL operon. These findings suggested that stress proteins including the GroESL chaperone complex exert regulatory and/or accessory functions in conjugative DNA transfer not only after stress-inducing conditions but also under physiological conditions (54). The results presented here are in accordance with this model in which GroEL is part of a regulatory feedback loop that serves to limit tra gene expression and thus the stress that is created by expression and assembly of the T4S system.
Acknowledgments
We thank Koreaki Ito for the kind gift of the E. coli strain NRK117, Andreas Reisner for providing plasmid pAR183, and Ellen Zechner for providing plasmid pGEX-traY. We are indebted to Thomas Wilfinger and Christian Wels for construction of TraJ expression plasmids and functionality tests.
Research in our laboratory is supported by the Fonds zur Förderung der Wissenschaftlichen Forschung, grant no. P17857-B12.
Footnotes
Published ahead of print on 22 June 2007.
REFERENCES
- 1.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 3.Camacho, E. M., and J. Casadesus. 2005. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 57:1700-1718. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, E. M., A. Serna, C. Madrid, S. Marques, R. Fernandez, F. de la Cruz, A. Juarez, and J. Casadesus. 2005. Regulation of finP transcription by DNA adenine methylation in the virulence plasmid of Salmonella enterica. J. Bacteriol. 187:5691-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 6.Chatellier, J., F. Hill, N. W. Foster, P. Goloubinoff, and A. R. Fersht. 2000. From minichaperone to GroEL 3: properties of an active single-ring mutant of GroEL. J. Mol. Biol. 304:897-910. [DOI] [PubMed] [Google Scholar]
- 7.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dempsey, W. B., and B. E. Fee. 1990. Integration host factor affects expression of two genes at the conjugal transfer origin of plasmid R100. Mol. Microbiol. 4:1019-1028. [DOI] [PubMed] [Google Scholar]
- 9.Disqué-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamas, P., L. Caro, D. Galas, and M. Chandler. 1987. Expression of F transfer functions depends on the Escherichia coli integration host factor. Mol. Gen. Genet. 207:302-305. [DOI] [PubMed] [Google Scholar]
- 11.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 12.Goebel, W., W. Lindenmaier, H. Schrempf, R. Kollek, and D. Blohm. 1977. Dissociation and recombination of fragments with defined functions of the antibiotic resistance factor R1, p. 261-275. In J. Drews and G. Högenauer (ed.), Topics in infectious diseases, vol. 2. Springer Verlag, Wien, Austria. [Google Scholar]
- 13.Gubbins, M. J., I. Lau, W. R. Will, J. M. Manchak, T. L. Raivio, and L. S. Frost. 2002. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J. Bacteriol. 184:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamoto, S., and E. Ohtsubo. 1990. Specific binding of the TraY protein to oriT and the promoter region for the traY gene of plasmid R100. J. Biol. Chem. 265:6461-6466. [PubMed] [Google Scholar]
- 16.Kandror, O., M. Sherman, and A. Goldberg. 1999. Rapid degradation of an abnormal protein in Escherichia coli proceeds through repeated cycles of association with GroEL. J. Biol. Chem. 274:37743-37749. [DOI] [PubMed] [Google Scholar]
- 17.Karl, W., M. Bamberger, and E. L. Zechner. 2001. Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J. Bacteriol. 183:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koraimann, G., and G. Högenauer. 1989. A stable core region of the tra operon mRNA of plasmid R1-19. Nucleic Acids Res. 17:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koraimann, G., C. Koraimann, V. Koronakis, S. Schlager, and G. Högenauer. 1991. Repression and derepression of conjugation of plasmid R1 by wild-type and mutated finP antisense RNA. Mol. Microbiol. 5:77-87. [DOI] [PubMed] [Google Scholar]
- 20.Koraimann, G., K. Teferle, G. Markolin, W. Woger, and G. Högenauer. 1996. The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol. Microbiol. 21:811-821. [DOI] [PubMed] [Google Scholar]
- 21.Koraimann, G., K. Teferle, R. Mitteregger, S. Wagner, and G. Högenauer. 1996. Differential mRNA decay within the transfer operon of plasmid R1: identification and analysis of an intracistronic mRNA stabilizer. Mol. Gen. Genet. 250:466-476. [DOI] [PubMed] [Google Scholar]
- 22.Kupelwieser, G., M. Schwab, G. Hogenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 23.Kusukawa, N., T. Yura, C. Ueguchi, Y. Akiyama, and K. Ito. 1989. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 8:3517-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 25.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, J., and L. S. Frost. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J. Bacteriol. 187:4767-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 28.Nelson, W. C., B. S. Morton, E. E. Lahue, and S. W. Matson. 1993. Characterization of the Escherichia coli F factor traY gene product and its binding sites. J. Bacteriol. 175:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Normark, B. H., and S. Normark. 2002. Evolution and spread of antibiotic resistance. J. Intern. Med. 252:91-106. [DOI] [PubMed] [Google Scholar]
- 30.Penfold, S. S., J. Simon, and L. S. Frost. 1996. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol. Microbiol. 20:549-558. [DOI] [PubMed] [Google Scholar]
- 31.Pölzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Högenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 32.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 33.Reisner, A., B. M. Holler, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schröder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 36.Schwab, M., H. Reisenzein, and G. Högenauer. 1993. TraM of plasmid R1 regulates its own expression. Mol. Microbiol. 7:795-803. [DOI] [PubMed] [Google Scholar]
- 37.Sherman, M., and A. L. Goldberg. 1992. Heat shock in Escherichia coli alters the protein-binding properties of the chaperonin groEL by inducing its phosphorylation. Nature 357:167-169. [DOI] [PubMed] [Google Scholar]
- 38.Sherman, M., and A. L. Goldberg. 1992. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 11:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman, P. M., E. Wickersham, and R. Harris. 1991. Regulation of the F plasmid traY promoter in Escherichia coli by host and plasmid factors. J. Mol. Biol. 218:119-128. [DOI] [PubMed] [Google Scholar]
- 40.Starcic-Erjavec, M., J. P. van Putten, W. Gaastra, B. J. Jordi, M. Grabnar, and D. Zgur-Bertok. 2003. H-NS and Lrp serve as positive modulators of traJ expression from the Escherichia coli plasmid pRK100. Mol. Gen. Genomics 270:94-102. [DOI] [PubMed] [Google Scholar]
- 41.Starcic, M., D. Zgur-Bertok, B. J. Jordi, M. M. Wosten, W. Gaastra, and J. P. van Putten. 2003. The cyclic AMP-cyclic AMP receptor protein complex regulates activity of the traJ promoter of the Escherichia coli conjugative plasmid pRK100. J. Bacteriol. 185:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stockwell, D., V. Lelianova, T. Thompson, and W. B. Dempsey. 2000. Transcription of the transfer genes traY and traM of the antibiotic resistance plasmid R100-1 is linked. Plasmid 43:35-48. [DOI] [PubMed] [Google Scholar]
- 43.Straus, D., W. Walter, and C. A. Gross. 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev. 4:2202-2209. [DOI] [PubMed] [Google Scholar]
- 44.Strohmaier, H., R. Noiges, S. Kotschan, G. Sawers, G. Högenauer, E. L. Zechner, and G. Koraimann. 1998. Signal transduction and bacterial conjugation: characterization of the role of ArcA in regulating conjugative transfer of the resistance plasmid R1. J. Mol. Biol. 277:309-316. [DOI] [PubMed] [Google Scholar]
- 45.Taki, K., T. Abo, and E. Ohtsubo. 1998. Regulatory mechanisms in expression of the traY-I operon of sex factor plasmid R100: involvement of traJ and traY gene products. Genes Cells 3:331-345. [DOI] [PubMed] [Google Scholar]
- 46.Tilly, K., N. McKittrick, M. Zylicz, and C. Georgopoulos. 1983. The dnaK protein modulates the heat-shock response of Escherichia coli. Cell 34:641-646. [DOI] [PubMed] [Google Scholar]
- 47.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, et al. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torreblanca, J., S. Marques, and J. Casadesus. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdino, P., W. Keller, H. Strohmaier, K. Bischof, H. Lindner, and G. Koraimann. 1999. The essential transfer protein TraM binds to DNA as a tetramer. J. Biol. Chem. 274:37421-37428. [DOI] [PubMed] [Google Scholar]
- 50.Waldor, M. K. 2006. Disarming pathogens—a new approach for antibiotic development. N. Engl. J. Med. 354:296-297. [DOI] [PubMed] [Google Scholar]
- 51.Will, W. R., and L. S. Frost. 2006. Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J. Bacteriol. 188:124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Will, W. R., J. Lu, and L. S. Frost. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54:769-782. [DOI] [PubMed] [Google Scholar]
- 53.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]
- 54.Zahrl, D., M. Wagner, K. Bischof, and G. Koraimann. 2006. Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J. Bacteriol. 188:6611-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zechner, E. L., F. de la Cruz, R. Eisenbrand, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 1999. Conjugative DNA transfer processes, p. 87-173. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.