Abstract
The methyltransferase RsmG methylates the N7 position of nucleotide G535 in 16S rRNA of Bacillus subtilis (corresponding to G527 in Escherichia coli). Disruption of rsmG resulted in low-level resistance to streptomycin. A growth competition assay revealed that there are no differences in fitness between the rsmG mutant and parent strains under the various culture conditions examined. B. subtilis rsmG mutants emerged spontaneously at a relatively high frequency, 10−6. Importantly, in the rsmG mutant background, high-level-streptomycin-resistant rpsL (encoding ribosomal protein S12) mutants emerged at a frequency 200 times greater than that seen for the wild-type strain. This elevated frequency in the emergence of high-level streptomycin resistance was facilitated by a mutation pattern in rpsL more varied than that obtained by selection of the wild-type strain.
Mutants resistant to streptomycin (Sm) can be classified into two distinct str phenotypes depending on whether they exhibit high- or low-level Sm resistance. The str mutations conferring high-level Sm resistance have been known for several decades to occur within rpsL, which encodes the ribosomal protein S12 (for reviews, see references 9 and 19). The mutations causing low-level resistance have been characterized only recently (22, 25). By use of comparative genome sequencing, we determined that low-level resistance in Streptomyces coelicolor is caused by mutations in rsmG (rRNA small subunit methyltransferase G), which encodes an S-adenosylmethionine (SAM)-dependent 16S rRNA methyltransferase (22). Analysis of the 16S rRNA by high-performance liquid chromatography (HPLC) showed that the ΔrsmG mutant lacked a 7-methylguanosine (m7G) modification. As the only naturally occurring m7G in 16S rRNA is at position G527 (Escherichia coli numbering system), this was assumed to be the site of RsmG methylation. Similar observations were made for rsmG-inactivated mutants of Escherichia coli, Staphylococcus aureus, Mycobacterium tuberculosis, and Mycobacterium smegmatis and led us to conclude that loss of a conserved m7G modification in 16S rRNA confers low-level Sm resistance in bacteria (25). In clinical isolates of M. tuberculosis, mutations within the rsmG gene were indeed an important cause of Sm resistance (25). In addition to conferring low-level Sm resistance, the rsmG mutation in S. coelicolor also led to the overproduction of antibiotics and enhanced expression of the SAM synthetase gene (22, 24).
Bacterial rRNAs have many methylated nucleotides. In E. coli, for example, there are 10 methylations in 16S rRNA and 14 methylations in 23S rRNA (1). Although the collective importance of these rRNA modifications for protein synthesis has been demonstrated (10, 17, 18), the function of individual methylations is still unclear, since inactivation of the genes encoding their cognate methyltransferases does not affect the cell's viability (1, 3, 20). To study the function of RsmG further, we have chosen Bacillus subtilis strain 168; genomic information and numerous tools for genetic, biochemical, and physiological analyses are available for this well-characterized system (7, 27). In the present study, we determined the precise location of the methylation target of B. subtilis RsmG, and we report here the physiological effects of inactivating rsmG in this species.
Strain construction.
The coding region of the rsmG gene was disrupted by insertion of a neomycin resistance (neo) gene. First, a DNA fragment containing rsmG (914 bp) was amplified by PCR using primers rsmG-F (5′-GTGAAATATGAAGGATATATTG-3′) and rsmG-R (5′-GTATCACCATAATATTACGATC-3′) and was cloned into plasmid pCR2.1 (Invitrogen) to form pCR2.1-rsmG. A 1.3-kbp SmaI fragment of neo derived from pBEST501 (16) was inserted into the HincII site of pCR2.1-rsmG. The resulting plasmid, pCR2.1-rsmG::neo, was linearized with KpnI and was used to transform B. subtilis 168. Neomycin-resistant transformants were selected on LB agar plates (with 3 μg/ml neomycin), and one recombinant, KO-756, was used for further study.
Strains with disrupted rsmG were complemented with an active copy of rsmG using plasmid pAPNC213 (21). This vector integrates specifically into the aprE locus and allows regulated expression of the target gene from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter. The full length of the coding region for rsmG was amplified by PCR using primers rsmGcom-F (5′-GAGGATCCCCGAGTAGAAAGGATGACGGC-3′; BamHI site underlined) and rsmGcom-R (5′-CATCCCGGGATTTTGATGAAAATATGATG-3′; SmaI site underlined). A BamHI-SmaI fragment containing the rsmG gene was inserted into pAPNC213 that had been treated with the same enzymes, generating pAPNCrsmG. The resulting plasmid, pAPNCrsmG, was used to transform the B. subtilis rsmG disruptant KO-756. Transformants were selected for plasmid-encoded resistance using 100 μg/ml spectinomycin; one of the spectinomycin-resistant transformants was used for complementation testing of rsmG.
Disruption of rsmG results in low-level Sm resistance in B. subtilis.
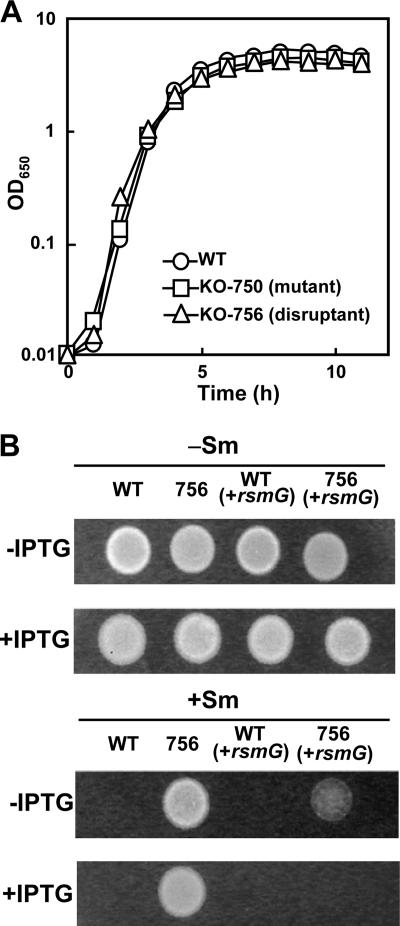
rsmG mutations have previously been shown to cause low-level Sm resistance in E. coli, M. tuberculosis, and S. coelicolor (22, 25). Similarly, disruption of rsmG in B. subtilis KO-756 (rsmG::neo) caused increased resistance to Sm (up to 100 μg/ml in LB medium). This resistance phenotype was eliminated by introduction of an active rsmG gene into KO-756 followed by induction with IPTG (Fig. 1B). In this complemented strain, the Sm MIC returned to the level for the wild-type strain (10 μg/ml in LB medium), unambiguously demonstrating a causal relationship between loss of rsmG activity and acquisition of low-level Sm resistance. We noted that disruption of rsmG conferred no resistance to any of the other antibiotics that we tested, including kanamycin, kasugamycin, spectinomycin, gentamicin, thiostrepton, lincomycin, chloramphenicol, erythromycin, and fusidic acid (the MICs in LB medium were 2, 1,500, 30, 0.3, 0.01, 10, 1, 0.05, and 0.02 μg/ml, respectively).
FIG. 1.
Growth and susceptibility to Sm of the B. subtilis wild-type (WT), rsmG frameshift (KO-750), and rsmG disrupted (KO-756) strains. (A) Strains were grown in LB medium at 37°C with shaking. (B) Wild-type and KO-756 (756) cells were grown to stationary phase, and 5-μl samples were spotted onto LB medium plates (with or without 30 μg/ml Sm and with or without 2 mM IPTG) and then incubated at 37°C for 12 h. OD650, optical density at 650 nm.
Although mutations that confer drug resistance often have a biological cost causing mutant bacteria to grow more slowly (2), the rsmG mutant KO-756 (and KO-750 [see Table 2]) grew as well as parent strain 168, both in LB medium (Fig. 1A) and in other media (not shown), and this result is consistent with earlier studies on E. coli and S. coelicolor (22, 25). Moreover, growth of the rsmG frameshift mutant KO-750 together with wild-type strain 168 in a competition assay (11) revealed no differences in the relative cell numbers (after five cycles of cultivation with reinoculation every 24 h or after 10 cycles of cultivation with reinoculation every 12 h) in various media, including LB medium (data not shown). Nor were there were differences in rsmG mutant or wild-type growth when the competition assay was performed using sterilized soil with four cycles of cultivation with reinoculation every 2 weeks. Finally, no differences between the parent and rsmG mutant strains were detected with respect to sensitivity of growth to high (55°C) or low (10°C) temperatures. These growth experiments demonstrate that the rsmG mutants are as fit as the wild-type strain under the various culture conditions tested. This finding contrasts with previous work on several other 16S rRNA methylases (1, 3, 20), which showed that knockout mutants were less fit than the wild-type strain.
TABLE 2.
Locations and identities of the mutations in the rpsL and rsmG genes
| Strain | Sm concn (μg/ml) used for selection | Position of mutation in rpsL or rsmGa | Amino acid substitution | Resistance to Sm (μg/ml)b |
|---|---|---|---|---|
| Wild-type 168 | —c | 10 | ||
| rpsL | ||||
| KO-670 | 1,000 | 167A→G | Lys56→Arg | 3,000 |
| KO-671 | 1,000 | 167A→C | Lys56→Thr | 5,000 |
| KO-672 | 1,000 | 166A→C | Lys56→Gln | 5,000 |
| rsmG | ||||
| KO-673 | 50 | 186,187TT→A | Frameshift (Val70→stop codon) | 100 |
| KO-674 | 50 | 499G-541C→Δ | Frameshift (Gly167→stop codon) | 100 |
| KO-675 | 50 | 326T→A | Frameshift (Leu109→stop codon) | 100 |
| KO-676 | 50 | 242G→T | Gly81→Val | 100 |
| KO-677 | 50 | 574G→Δ | Frameshift (Val214→stop codon) | 100 |
| KO-678 | 50 | 231 (AGCGGG) insertion | Gly-Ala insertion at position 77 | 100 |
| KO-679 | 50 | 313C→T | Arg105→Trp | 100 |
| KO-680 | 50 | 125C→A | Thr42→Asn | 100 |
| KO-681 | 50 | 360T→TT | Frameshift (Asp126→stop codon) | 100 |
| KO-682 | 50 | 231 (AGCGGG) insertion | Gly-Ala insertion at position 77 | 100 |
| KO-683 | 50 | 558A→Δ | Frameshift (Val214→stop codon) | 100 |
| KO-684 | 50 | 659A→Δ | Frameshift | 100 |
| KO-685 | 50 | 231A-236G→Δ | Ala78, Gly79→Δ | 100 |
| KO-707 | 50 | 558A→Δ | Frameshift (Val214→stop codon) | 100 |
| KO-708 | 50 | 310A→AAA | Frameshift (Glu110→stop codon) | 100 |
| KO-709 | 50 | 231A-236G→Δ | Ala78, Gly79→Δ | 100 |
| KO-710 | 50 | 606G-644T→Δ | Leu203-Ile215→Δ | 100 |
| KO-711 | 50 | 233C→A | Ala78→Glu | 100 |
| KO-712 | 50 | 528G-708A→Δ | Ala177-Pro236→Δ | 100 |
| KO-713 | 50 | NDd | 100 | |
| KO-714 | 50 | ND | 100 | |
| KO-750e | 50 | 100 | ||
| KO-756f | 100 |
Numbering from the start codon (ATG) of the open reading frame.
Determined 16 h after incubation on LB agar at 37°C.
Wild-type rpsL and rsmG genes.
ND, mutations were not detected within the rsmG gene.
KO-750 is the result of replacing the rsmG gene in strain 168 with the mutant copy from KO-673 by transformation.
Strain KO-756 is an rsmG disruptant (rsmG::neo) of strain 168. Selection was carried out with 100 μg/ml spectinomycin.
We previously reported that mutation of rsmG in E. coli did not lead to higher levels of accuracy in translation, and this contrasts with the results for most of the high-level-Sm-resistance mutations, such as those in rpsL, that have been characterized in E. coli. This was interpreted to indicate that E. coli ribosomes lacking RsmG methylation might have a reduced affinity for Sm (25). However, in the case of the rsmG mutations of B. subtilis, a detectable increase in translational accuracy was indeed observed in a readthrough induction assay, although the accuracy was not increased as much as that observed for an rpsL (K56N) mutant (Table 1).
TABLE 1.
Characterization of translation in mutant ribosomes in vivo
| Strain | In vivo readthrough induction ratios for Glu-105 codon of LacIa
|
|
|---|---|---|
| GAA (native) | UGA | |
| 168 (wild type) | 34 ± 9.9 | 6.0 ± 2.7 |
| WL2 (rpsL [K56N]) | 33 ± 8.9 | 0.99 ± 0.13 |
| KO-750 (rsmG) | 33 ± 10 | 2.2 ± 0.19 |
| KO-756 (rsmG::neo) | 27 ± 4.1 | 2.2 ± 0.36 |
The level of translational accuracy was estimated in vivo in a UGA readthrough system that measures the regulation of lacZ by LacI (13). Briefly, codon 105 of lacI was replaced by an opal codon (UGA); readthrough of the opal codon is required to generate the full-length LacI protein, which would then repress the expression of lacZ. For measurement of UGA readthrough, cells were grown to an optical density at 650 nm of 0.5 in Spizizen's salts minimum medium in the presence of IPTG (10 mM) or in the absence of IPTG prior to measurement of β-galactosidase activity (13). The UGA readthrough levels were expressed as the induction ratio (the β-galactosidase activity of the culture supplemented with IPTG divided by the activity of the culture without IPTG).
RsmG methylates the N7 position of G527 in 16S rRNA.
We recently showed that E. coli RsmG catalyzes a SAM-dependent m7G modification in E. coli 16S rRNA (25). This observation was confirmed here for B. subtilis using reversed-phase HPLC analysis of the rRNA nucleosides, which showed that RsmG also catalyzes an m7G modification within 16S rRNA (data not shown). Since only one m7G modification, at G527, is found in the 16S rRNA of E. coli, these findings suggest that RsmG modifies the same position in B. subtilis (nucleotide G535 in the B. subtilis sequence). We tested this possibility here by determining the exact target site of the RsmG methyltransferase.
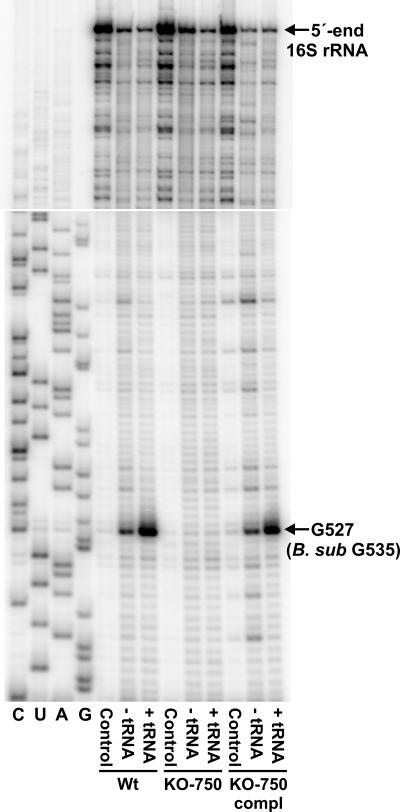
Total RNA was isolated as previously described (6) from the B. subtilis wild-type strain, the rsmG frameshift mutant KO-750, and the mutant strain complemented with an active rsmG gene. The RNAs were cleaved at N7-methylguanosine positions by reduction with NaBH4, followed by β-elimination with acetic acid-aniline (26, 33); a tRNA carrier, hypermodified at N7 of guanosines by dimethyl sulfate treatment, was added to enhance cleavage at the N7-methylated guanosines in the rRNA (34). The rRNAs were scanned using a series of primers by reverse transcriptase extension (32). The only difference seen was in the termination of reverse transcriptase immediately upstream of nucleotide G527 (G535 in B. subtilis) on the 16S rRNA templates (Fig. 2). The band formed in the wild-type sample indicates that there was N7 methylation at this guanosine; there was no such termination after loss of RsmG activity in the rsmG mutant KO-750, although the N7-methylation signal returned in the strain complemented with an active rsmG gene (Fig. 2). In combination, the HPLC and primer extension data conclusively demonstrate that RsmG is responsible for N7 methylation at position G527 in 16S rRNA.
FIG. 2.
Identification of the site of RsmG methylation: gel autoradiogram of primer extension of the rRNAs from B. subtilis wild-type strain 168 (Wt), rsmG frameshift mutant KO-750, and the mutant strain complemented with an active copy of rsmG (KO-750 compl). The oligodeoxynucleotide primer 5′-CCTGCGAGCCCTTTACGCC-3′ was hybridized to 16S rRNA nucleotides 567 to 585 (E. coli numbering) and extended with reverse transcriptase. Control, untreated samples; − tRNA, the rRNAs were treated with NaBH4 and aniline to break the phosphodiester backbone at m7G methylation sites (26, 33); + tRNA, rRNAs were treated with NaBH4 and aniline after addition of dimethyl sulfate-modified tRNA to enhance cleavage at m7G methylation sites (34). The position of the RsmG methylation site at nucleotide G527 (corresponding to G535 in the B. subtilis 16S rRNA sequence) is indicated. Wild-type rRNA was used as the template for the dideoxy sequencing reactions (lanes C, U, A, and G). The top panel shows the upper part of the gel autoradiogram extending to the 5′ end of the rRNA.
Nucleotide position 527 is invariably a guanosine in bacteria, and database searches of the available genome sequences further revealed that all bacteria have a homologue of rsmG. The identity of this nucleotide and its methylation are thus highly conserved and can be inferred to be functionally important. Nucleotide G527 is situated within a hairpin loop (the so-called 530 loop) that is one of the most highly conserved features of 16S rRNA, and mutations in this loop have been associated with resistance to Sm (29). This region of 16S rRNA is situated close to the ribosomal protein S12, and both of these ribosomal components play a major role in translational fidelity (4, 5, 23, 28, 29, 31). The previous studies coincide with the findings obtained here showing that loss of G527 methylation confers Sm resistance.
Emergence of high-level Sm resistance in B. subtilis rsmG mutants.
Spontaneous mutations that lead to high-level Sm resistance (an approximately 100-fold increase in the MIC) generally emerge at a low frequency in bacteria (10−11 to 10−9), with the majority of these mutations occurring within rpsL (8). Consistently, in B. subtilis wild-type strain 168 (MIC in LB medium, 10 μg/ml), spontaneous mutants conferring high-level Sm resistance (MIC, at least 1,000 μg/ml) arose at a low frequency, between 2 × 10−11 and 8 × 10−10. In contrast, mutants with low-level Sm resistance (MIC, 100 μg/ml) emerged at a much higher frequency, in the range from 3 × 10−7 to 4 × 10−6. Most of these mutants (19 out of 21) contained changes in rsmG, and these changes were in many cases frameshift mutations that resulted in a stop codon immediately downstream of the mutation site (Table 2). Strikingly, but consistent with previous observations for E. coli and S. coelicolor (22, 25), the B. subtilis rsmG mutants (and the rsmG disruption mutant KO-756) produced spontaneous mutants showing resistance to a high level of Sm (5,000 μg/ml) at a frequency on the order of 10−6 or 10−7. The data for rsmG mutant KO-750 are shown in Table 3 and show that there was a 500- to 2,000-fold-greater frequency of mutation to high-level Sm resistance than that observed for the wild-type strain. About one-third (68 out of 190) of the high-level-Sm-resistant rsmG mutants were found to have a mutation in rpsL (Table 3), but a majority (122 out of 190) had no mutation in either the rpsL, rpsD (encoding ribosomal protein S4), or rpsE (encoding ribosomal protein S5) gene. Importantly, the rsmG rpsL double mutants displayed a pattern of rpsL mutations more varied than the patterns derived directly by selection of the wild-type strain. For example, the mutations Lys101→Glu, Pro104→Thr, and Pro104→Arg are mutations that are novel or are found only rarely in high-level-Sm-resistant isolates. From a clinical microbiology viewpoint, the increased frequency and variety of these high-level-Sm-resistance mutations in the rsmG strains are significant; however, the underlying physiological mechanism by which they occur remains to be clarified. We can rule out the possibility that RsmG functions as an antimutator-like protein, since the rsmG mutation in E. coli does not affect the frequency at which mutants resistant to antibiotics other than Sm emerge (25).
TABLE 3.
Effect of rsmG mutation on the emergence of high-level-Sm-resistant mutantsa
| Strain | Frequency of high-level-Sm-resistant mutantsb | Position of mutation detected in rpsL gene | Amino acid exchange | No. of mutants |
|---|---|---|---|---|
| Wild-type 168 | 3 × 10−10-4 × 10−9 | 166A→C | Lys56→Gln | 13 |
| 167A→C | Lys56→Thr | 10 | ||
| 167A→G | Lys56→Arg | 15 | ||
| 167A→T | Lys56→Ile | 9 | ||
| 168A→C | Lys56→Asn | 14 | ||
| 311C→T | Pro104→Leu | 15 | ||
| Total | 76 | |||
| KO-750 (rsmG) | 5 × 10−7-2 × 10−6 | 166A→C | Lys56→Gln | 10 |
| 167A→C | Lys56→Thr | 5 | ||
| 167A→G | Lys56→Arg | 9 | ||
| 167A→T | Lys56→Ile | 2 | ||
| 168A→C | Lys56→Asn | 5 | ||
| 168A→T | Lys56→Asn | 2 | ||
| 301A→G | Lys101→Glu | 2 | ||
| 310C→A | Pro104→Thr | 1 | ||
| 311C→T | Pro104→Leu | 12 | ||
| 311C→A | Pro104→Gln | 6 | ||
| 311C→G | Pro104→Argc | 1 | ||
| 313G→A | Gly105→Arg | 12 | ||
| 313G→C | Gly105→Arg | 1 | ||
| Total | 68 |
Selection for high-level resistance was performed with Sm at 5,000 μg/ml (for the rsmG mutant) or at 3,000 μg/ml (for the wild type).
To measure the frequency of resistant mutants, single colonies were isolated, and cells originating from each of about 10 clones were examined separately.
The mutant with this mutation required 1,000 μg/ml of Sm for growth.
In S. coelicolor, rsmG mutations conferring low-level Sm resistance result in overproduction of the antibiotic actinorhodin (22, 24, 30). The S. coelicolor rsmG mutants exhibit enhanced expression of SAM synthetase, accompanied by increased protein synthesis activity at late growth phase, which eventually leads to overproduction of antibiotics (actinorhodin, undecylprodigiosin, and calcium-dependent antibiotics) (22). It is believed that the increases in SAM synthetase activity and protein synthesis activity caused by the rsmG mutation are both linked to the activation of secondary metabolism. In the B. subtilis rsmG frameshift mutant KO-750, however, there was no increase in production of either bacilysin or neotrehalosadiamine (data not shown), which are antibiotics that this organism produces at late growth phase (14, 15). Consistent with these results, KO-750 showed neither an increase in protein synthesis at late growth phase nor an increase in SAM synthetase activity (data not shown). Thus, in contrast to the situation in Streptomyces, secondary metabolism in B. subtilis is not activated by mutation of rsmG. Furthermore, the rsmG mutation did not affect sporulation, competence, or protease production, at least under usual culture conditions (data not shown), which again contrasts in part with the reduced ability to sporulate that was exhibited by the S. coelicolor rsmG mutants (22).
Concluding remarks.
In the present study we determined the exact location of the rRNA methylation target for RsmG and thereby further clarified one molecular mechanism underlying low-level Sm resistance. Sm is still an important drug for the treatment of tuberculosis, and our findings provide new insight into the role of rRNA modification in the acquisition of antibiotic resistance. The phylogenetic conservation of RsmG and of the 16S rRNA sequence in the 530 loop suggests that methylation at this rRNA site should confer some selective advantage. Nevertheless, the apparent lack of a disadvantage in cells that no longer can methylate the G527 position clearly prompts a question concerning the biological importance of this modification.
Concerning secondary metabolite production, we previously reported that certain B. subtilis mutants possess low-level Sm resistance and exhibit a 10- to 50-fold increase in antibiotic production (12). These mutants have been reevaluated, and consistent with the findings presented here, none of these strains had a mutation in the rsmG gene. This indicates that another type of mechanism, fundamentally different from that involving rsmG mutation, can be acquired by B. subtilis to confer low-level Sm resistance. In contrast to what happens in the rsmG mutants, acquisition of low-level resistance to Sm by this unidentified mechanism may be linked with activation of secondary metabolism.
Acknowledgments
This work was supported by grants to K.O. from the Organized Research Combination System and the Effective Promotion of Joint Research of Special Coordination Funds (Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government). Support of S.D. by the Danish Research Agency (FNU grant 21-04-0520) and the Nucleic Acid Center of the Danish Grundforskningsfond is also gratefully acknowledged.
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Andersen, N. M., and S. Douthwaite. 2006. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 359:777-786. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Basturea, G. N., K. E. Rudd, and M. P. Deutscher. 2006. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA 12:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 5.Cundliffe, F. 1990. Recognition sites for antibiotics within rRNA, p. 479-490. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology., Washington, DC.
- 6.Douthwaite, S., T. Powers, J. Y. Lee, and H. F. Noller. 1989. Defining the structural requirements for a helix in 23S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 209:655-665. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin, J., and R. Losick. 2001. Linking nutritional status to gene activation and development. Genes Dev. 15:1051-1054. [DOI] [PubMed] [Google Scholar]
- 8.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Böttger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9:1239-1246. [DOI] [PubMed] [Google Scholar]
- 9.Gorini, L. 1974. Streptomycin and misreading of the genetic code, p. 791-803. In M. Nomura, A. Tissières, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 10.Green, R., and H. F. Noller. 1999. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry 38:1772-1779. [DOI] [PubMed] [Google Scholar]
- 11.Gutgsell, N., N. Englund, L. Niu, Y. Kaya, B. G. Lane, and J. Ofengand. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin resistant(str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inaoka, T., K. Kasai, and K. Ochi. 2001. Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis. J. Bacteriol. 183:4958-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 15.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 279:3885-3892. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khaitovich, P., T. Tenson, P. Kloss, and A. S. Mankin. 1999. Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry 38:1780-1788. [DOI] [PubMed] [Google Scholar]
- 18.Krzyzosiak, W., R. Denman, K. Nurse, W. Hellmann, M. Boublik, C. W. Gehrke, P. F. Agris, and J. Ofengand. 1987. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into functional 30S ribosome. Biochemistry 26:2353-2364. [DOI] [PubMed] [Google Scholar]
- 19.Kurland, C. G., D. Hughes, and M. Ehrenberg. 1996. Limitations of translational accuracy, p. 979-1004. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 20.Lesnyak, D. V., J. Osipiuk, T. Skarina, P. V. Sergiev, A. A. Bogdanov, A. Edwards, A. Savchenko, A. Joachimiak, and O. A. Dontsova. 2007. Methyltransferase that modifies guanine 966 of the 16S rRNA: functional identification and tertiary structure. J. Biol. Chem. 282:5880-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539-3552. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura, K., T. Hosaka, S. Tokuyama, S. Okamoto, and K. Ochi. 2007. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 189:3876-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogle, J. M., and V. Ramakrishnan. 2005. Structural insights into translational fidelity. Annu. Rev. Biochem. 74:129-177. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto, S., A. Lezhava, T. Hosaka, Y. Okamoto-Hosoya, and K. Ochi. 2003. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2). J. Bacteriol. 185:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto, S., A. Tamaru, C. Nakajima, K. Nishimura, Y. Tanaka, S. Tokuyama, Y. Suzuki, and K. Ochi. 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63:1096-1106. [DOI] [PubMed] [Google Scholar]
- 26.Peattie, D. A. 1979. Direct chemical method for sequencing RNA. Proc. Natl. Acad. Sci. USA 76:1760-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 28.Poehlsgaard, J., and S. Douthwaite. 2005. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3:870-881. [DOI] [PubMed] [Google Scholar]
- 29.Powers, T., and H. F. Noller. 1991. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 10:2203-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Springer, B., Y. G. Kidan, T. Prammananan, K. Ellrott, E. C. Böttger, and P. Sander. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 33.Wintermeyer, W., and H. G. Zachau. 1975. Tertiary structure interactions of 7-methylguanosine in yeast tRNAPhe as studied by borohydride reduction. FEBS Lett. 58:306-309. [DOI] [PubMed] [Google Scholar]
- 34.Zueva, V. S., A. S. Mankin, A. A. Bogdanov, and L. A. Baratova. 1985. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur. J. Biochem. 146:679-687. [DOI] [PubMed] [Google Scholar]