Abstract
RovA is a MarR/SlyA-type regulator that mediates the transcription of inv in Yersinia enterocolitica and Y. pseudotuberculosis. In Y. pseudotuberculosis, rovA transcription is controlled primarily by H-NS and RovA, which bind to similar regions within the rovA promoter. At 37°C, rovA transcription is repressed by H-NS. Transcription of rovA results when RovA relieves H-NS-mediated repression. The region of the rovA promoter that H-NS and RovA bind is not conserved in the Y. enterocolitica promoter. Using green fluorescent protein reporters, we determined that the Y. enterocolitica rovA (rovAYent) promoter is weaker than the Y. pseudotuberculosis promoter. However, despite the missing H-NS/RovA binding site in the rovAYent promoter, H-NS and RovA are still involved in the regulation of rovAYent. DNA binding studies suggest that H-NS and RovA bind with a higher affinity to the Y. pseudotuberculosis/Y. pestis rovA (rovAYpstb/Ypestis) promoter than to the rovAYent promoter. Furthermore, H-NS appears to bind to two regions in a cooperative fashion within the rovAYent promoter that is not observed with the rovAYpstb/Ypestis promoter. Finally, using a transposon mutagenesis approach, we identified a new positive regulator of rovA in Y. enterocolitica, LeuO. In Escherichia coli, LeuO regulates gene expression via changes in levels of RpoS and H-NS, but LeuO-mediated regulation of rovAYent appears to be independent of either of these two proteins. Together, these data demonstrate that while the rovA regulatory factors are conserved in Yersinia, divergence of Y. enterocolitica and Y. pseudotuberculosis/Y. pestis during evolution has resulted in modifications in the mechanisms that are responsible for controlling rovA transcription.
There are three pathogenic species of Yersinia that cause disease in humans. Y. pestis is a vector-borne pathogen that causes plague and has been responsible for three major pandemics, including an ongoing pandemic that began in the 1860s (45). Y. enterocolitica and Y. pseudotuberculosis are gastrointestinal pathogens that cause milder manifestations of disease and enter the host through consumption of contaminated food or water (2, 41). Sharing the same route of infection, Y. enterocolitica and Y. pseudotuberculosis utilize similar virulence factors that appear to be inactivated in Y. pestis, including the colonization factors invasin (Inv) and YadA. These adhesins are important for colonization by the oral pathogens but are no longer expressed in Y. pestis due to naturally occurring mutations (51, 52).
Invasin is the major adherence factor encoded in both Y. enterocolitica and Y. pseudotuberculosis (20, 36). Inv is believed to promote efficient entry into the Peyer's patches of the small intestine through interactions with β1 integrins on the surface of the M cells overlying these lymphoid tissues (4, 21, 31, 49). During in vitro culture, inv transcription is regulated by temperature and growth phase. The highest levels of Inv expression are observed in early-stationary-phase cultures incubated at 23 to 26°C. The pH of the culture medium has also been shown to alter the expression of inv in Y. enterocolitica cultures grown at 37°C (22, 43). Work to understand the regulation of inv has identified three proteins involved in modulating the levels of Inv (10, 12, 39, 40). H-NS and YmoA have been implicated in repression of inv transcription. H-NS is a histone-like protein that is important for the proper nucleoid packaging of the bacterial chromosome (1) and is involved in the regulation of multiple genes, including virulence factor genes, in response to temperature (1, 16, 38). YmoA is a member of the Hha/YmoA family of regulatory proteins (5, 29). Studies in Escherichia coli with the YmoA homolog Hha were the first studies to demonstrate that Hha/YmoA members interact with H-NS to enhance repression mediated by H-NS (42). Ellison and Miller extended these observations to Yersinia and demonstrated that YmoA interacts with H-NS during the regulation of inv transcription (10). The current model for inv repression proposes that H-NS recognizes and binds to the inv promoter, where YmoA interacts with H-NS to propagate the formation of a regulatory complex, blocking recognition of the promoter by RNA polymerase.
The third protein involved in inv regulation is RovA, which is necessary for the expression of Inv (40, 46). RovA is a member of the SlyA/MarR regulatory family, which contains homologs in several species of bacteria and archaea, including all three pathogenic species of Yersinia (11). SlyA/MarR family members have been shown to regulate a variety of functions, including resistance to antibiotics (15, 54), production of antimicrobial agents (55), and expression of virulence factors (19, 27, 46, 47). In Yersinia, microarray data suggest that RovA regulates the expression of multiple genes in addition to inv (3; J. S. Cathelyn and V. L. Miller, unpublished data). During inv regulation in Y. enterocolitica, RovA appears to act as a derepressor to relieve the negative regulation of the H-NS/YmoA complex. In vitro analysis of RovA and H-NS binding to the inv promoter indicates that RovA and H-NS bind to similar overlapping regions in both Y. pseudotuberculosis and Y. enterocolitica. Furthermore, RovA can displace H-NS bound to the inv promoter (10). Therefore, in vivo it is predicted that RovA binding either displaces H-NS/YmoA or relieves topological restraints imposed by H-NS/YmoA on the promoter to allow access to RNA polymerase, resulting in transcription of inv. This model is supported by work demonstrating that the RovA level relative to the H-NS/YmoA level within the cell is a key determinant of inv expression and that RovA is not required for inv expression in Y. enterocolitica if the inv promoter is truncated to remove the H-NS binding site (10). Thus, an understanding of how rovA is regulated is necessary to understand the expression of inv and other RovA-regulated genes.
The regulation of rovA in Y. pseudotuberculosis appears to use a mechanism similar to that observed for inv regulation in Y. enterocolitica. Heroven et al. have demonstrated that H-NS and RovA regulate the levels of rovA transcription in an E. coli surrogate system, suggesting that these proteins regulate the expression of rovA in Y. pseudotuberculosis as well (18). These authors reported that rovA transcription levels are low in wild-type E. coli but increase with either the addition of RovA or the inactivation of the E. coli hns gene. In Y. pseudotuberculosis, addition of a plasmid encoding a trans copy of hns also decreased the levels of RovA. In vitro, H-NS and RovA bind to similar regions within the Y. pseudotuberculosis rovA promoter, suggesting that RovA may use a conserved mechanism of H-NS derepression to regulate the expression of genes. Data suggest that RovA may have an additional negative role in autoregulation. As levels of RovA were increased in an E. coli strain carrying a rovA::lacZ reporter, a moderate decrease in rovA transcription (∼2-fold) was observed (18). In vitro data also suggest that RovA binds to a second region within the Y. pseudotuberculosis rovA promoter, although at a lower affinity than to the previously described binding site. Heroven et al. have suggested that as levels of RovA increase, RovA may bind to this second binding site, resulting in decreased transcription of the gene (18).
Recently, a third protein has been implicated in rovA regulation in Y. pseudotuberculosis. RovM (modulator of rovA expression) is a LysR-like protein that represses the expression of rovA in response to growth in minimal medium (17). Further, Heroven et al. demonstrated in vitro binding of recombinant RovM to the rovA promoter and suggested that RovM directly represses rovA expression through this interaction. RovM also autoregulates its own expression and affects the motility of Y. pseudotuberculosis independent of rovA. Interestingly, a rovM mutant is hypervirulent during oral infection of the mouse model, consistent with previous studies suggesting that there is a requirement for RovA in virulence (9, 46).
Comparisons of the DNA sequences upstream of rovA from Y. pseudotuberculosis, Y. pestis, and Y. enterocolitica reveal considerable divergence in the Y. enterocolitica rovA promoter. Notably, while the Y. pseudotuberculosis and Y. pestis promoter sequences are identical to each other (the rovA coding regions are also identical in these two species), there are two predicted open reading frames (ORFs) upstream of rovA in Y. enterocolitica that are not found in Y. pseudotuberculosis and Y. pestis. Furthermore, these ORFs appear to have replaced the predicted H-NS/RovA binding region reported in Y. pseudotuberculosis and Y. pestis. These significant differences in the Y. enterocolitica rovA (rovAYent) promoter suggest that the regulatory mechanism of rovA could differ in Y. enterocolitica. To understand how these differences in the rovA promoter affect the transcription of rovA in Y. enterocolitica, we defined the promoter of rovA in Y. enterocolitica, compared the expression of this promoter to the expression of the Y. pseudotuberculosis/Y. pestis promoter in different Yersinia backgrounds, and elucidated the effects of RovA, H-NS, and RovM on rovA transcription in Y. enterocolitica. Furthermore, we used transposon mutagenesis to identify a fourth regulator of rovA in Y. enterocolitica.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Y. enterocolitica and E. coli strains were grown in Luria-Bertani (LB) broth at 26°C or 37°C as indicated below. Y. pestis strains were grown in brain heart infusion (BHI) broth at 26°C. When appropriate, antibiotics were used at the following concentrations: nalidixic acid, 20 μg ml−1; tetracycline, 15 μg ml−1; kanamycin, 100 μg ml−1; carbenicillin, 100 μg ml−1; and spectinomycin, 100 μg ml−1. Primers used in plasmid construction in this study are listed in Table 2. To generate in-frame lacZ, rovA, leuO, and rpoS deletion strains, the regions flanking the genes were amplified by PCR, digested with SalI, NotI, and BamHI, and ligated into the suicide plasmid pSR47s (34). The suicide plasmids were introduced into Y. enterocolitica through conjugation, and mutants were selected as described previously (59). The YVM1251 rovA::lacZ reporter was generated using the system described by Maxson and Darwin (32). To construct rovA::green fluorescent protein (GFP) reporters, regions of the promoter were amplified by PCR, digested, and ligated into the low-copy-number vector pPROBE-gfp[LVA] (37). pLEUO was generated by amplifying leuO and 500 bp upstream of leuO by PCR and introducing the product into pCR2.1 by TOPO cloning (Invitrogen, Carlsbad, CA).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Y. enterocolitica strains | ||
| YVM150 | JB580 ΔyenR (R− M+) | 23 |
| YVM927 | JB580 ΔyenR (R− M+) ΔrovA | 10 |
| YVM1251 | JB580 ΔyenR (R− M+) pYV− ΔlacZ ΔaraGFB::[φ(rovAYent-lacZY)] | This study |
| YVM1252 | JB580 ΔyenR (R− M+) pYV− ΔlacZ ΔaraGFB::[φ(rovAYent-lacZY)] ΔleuO | This study |
| YVM1253 | JB580 ΔyenR (R− M+) ΔrpoS | This study |
| YVM1254 | JB580 ΔyenR (R− M+) ΔrovM | This study |
| TM102 | JB580 ΔyenR (R− M+) pYV− ΔlacZ ΔaraGFB::[φ(rovAYent-lacZY)] leuO::miniTn5 Knr | This study |
| Y. pestis strains | ||
| YP6 | CO92 pCD1− | 7 |
| YP12 | CO92 pCD1− ΔrovA | 3 |
| E. coli strains | ||
| MC4100 | F−araD139 Δ(lacIOPZYA)U169 rpsL thiA | 14 |
| VM1303 | MC4100 hns205::Tn10 Tetr | 10 |
| Plasmids | ||
| pPROBE-gfp[LVA] | Promoterless gfp with LVA tag to decrease GFP half-life, Knr | 37 |
| pYEL | pPROBE-gfp[LVA] containing rovAYent promoter fragment from nt −614 to 170 | This study |
| pYES | pPROBE-gfp[LVA] containing rovAYent promoter fragment from nt −445 to 170 | This study |
| pYPL | pPROBE-gfp[LVA] containing rovAYpstb/Ypestis promoter fragment from nt −622 to 170 | This study |
| pYPS | pPROBE-gfp[LVA] containing rovAYpstb/Ypestis promoter fragment from nt −443 to 170 | This study |
| pWKS30::StrSpec | pWKS30 with a Str/Spec cassette in the HindIII site of the polylinker, Ampr Strr Specr | 10 |
| pHNS | pWKS30::Str/Spec::hns with native promoter region | 10 |
| pLEUO | pCR2.1::leuO with native promoter region | This study |
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| Deletion constructs | |
| lacZ | |
| lacZ5′F | ACGCGTCGACGCCAGCAATACCCCATTTAGC |
| lacZ5′R | CGGGATCCAATTTCAGCCTTATCTTTTACGAAAGTTAGC |
| lacZ3′F | CGGGATCCGCAACAATATCAACACAGAATTTCTAATACGC |
| lacZ3′R | ATAAGAATGCGGCCGCTGCTGGGCTATAATCTGGTGC |
| leuO | |
| leuO5′F | ACGCGTCGACGGATTGGTTCATGCTTCTTATATTTTATGGCT |
| leuO5′R | CGGGATCCTACTAAGTTGTGTTCAAACATGCTTAACTCCAC |
| leuO3′F | CGGGATCCTAGTTATTATCATTAAGTCCTGCTGCTGAAGTTG |
| leuO3′R | ATAAGAATGCGGCCGCACGCTCTTGGATGGCAGC |
| rpoS | |
| rpoS5′F | ACGCGTCGACTGTCGCTACAACCGCACC |
| rpoS5′R | GGAAGATCTCATATGCTGCTCCTACCCGTG |
| rpoS3′F | GGAAGATCTTAGCGATACTCTCGCAAACAGTCTG |
| rpoS3′R | ATAAGAATGCGGCCGCCCGTATCAAAGCCATGACGCTA |
| Reporters | |
| rovA::lacZ | |
| rlacZF | TGCTCTAGAGGGTCAATGACAAATAATAAGCCTCCAGT |
| rlacZR | CGGGATCCACCAATCGCTTTCGCCAGTTG |
| rovA::gfp | |
| pYELF | CGGGATCCTGGTTATCATGAACTAATATTTTAACCAATCGGC |
| pYESF | CGGGATCCAGGGGGATTGCATATAATAATTCCACA |
| pYEL-2R | GGAATTCACCAATCGCTTTCGCCAGTTG |
| pYPLF | CGGGATCCTGCCGCCTTCCTGCAA |
| pYPSF | CGGGATCCTTTGAAATATTGATGATTCATATCAATTTACCCAAGTC |
| pYPL-2R | GGAATTCACCAATCGCTTTCGCCAGTTG |
| Complementation clone: pLEUO | |
| pLEUOF | ACGCGTCGACGGATTGGTTCATGCTTCTTATATTTTATGGCT |
| pLEUOR | GGGGTACCTTAAGAAGGAATATTAAGCTGGCTGAGTAATTC |
| Primer extension | |
| PE1 | CTAATCGTGCTAAATCAGATCC |
| PE2 | AAAATTATGTATTTACTAAAATTACCTCTTAAGGA |
| EMSA | |
| YP1-2F | TGC CGC CTT CCT GCA A |
| YP3-4F | TTTGAAATATTGATGATTCATATCAATTTACCCAAGTC |
| YP1-4R | ACCAATCGCTTTCGCCAGTTG |
| YP2-3R | GGACAATAGCAATAAATACGGGGAA |
| YE1-2F | TGGTTATCATGAACTAATATTTTAACCAATCGGC |
| YE3F | AGGGGGATTGCATATAATAATTCCACA |
| YE4F | ATTTAAGAGACTGATGATTCATATCAATTTACCAAC |
| YE5F | TTGGATGCCAGATATCACCC |
| YE6F | ACCTAGCATAACCGCCTTAAAAATT |
| YE1-4-5-6R | ACCAATCGCTTTCGCCAGTTG |
| YE2-3R | GGACAATAGCAATAAATACGGGGAA |
| Transposon sequencing | |
| TNF | CCCATGTCAGCCGTTAAGTGT |
Underlining indicates restriction sites.
Primer extension.
RNA was extracted from Y. enterocolitica using a RiboPure-Bacteria isolation kit (Ambion, Austin, TX) and treated with DNase I (Ambion) as described by the manufacturer. Primers PE1 and PE2 (Table 2) were labeled with 32P using T4 DNA polynucleotide kinase (New England Biolabs, Beverley, MA). One picomole of labeled primer was hybridized with 10 μg of total RNA and incubated with 20 U of Superscript III reverse transcriptase (Invitrogen). The reaction was terminated by addition of stop solution, and primer-extended products were separated on 8% polyacylamide-8 M urea gels (58).
GFP assays.
For Y. enterocolitica and E. coli, strains were grown in triplicate overnight in 2 ml of LB broth at 26°C with aeration and diluted to an optical density at 600 nm (OD600) of 0.1 in media containing the appropriate antibiotics. Then 0.7 ml of each diluted culture was inoculated into individual wells in a 48-well plate and grown with shaking for 10 h at 26°C in a Synergy HT kinetic plate reader (BioTek, Winooski, VT). The OD600 and fluorescence (measured at an excitation wavelength of 485 nm and an emission wavelength of 528 nm) of the cultures were determined at numerous intervals. For Y. pestis, bacteria were grown overnight in 2 ml of BHI broth at 26°C with aeration and diluted to an OD600 of 0.2 in 20 ml of BHI broth containing the appropriate antibiotics. The cultures were grown for 6 h with aeration, and the OD600 and fluorescence of the cultures were determined. Data for Y. pestis studies were normalized and expressed as fluorescence (in relative light units)/OD600.
EMSA.
Primers used to generate rovA promoter fragments used in an electrophoretic mobility gel shift assay (EMSA) are listed in Table 2. An approximately 500-bp fragment of the ysaE promoter, which is not regulated by RovA or H-NS, was used as a control for nonspecific binding. Protein purification and binding reactions were performed as described previously (10).
Transposon mutagenesis.
YVM1251 was mutagenized with the Tn5 transposon TnMod-RKm′ (6). TnMod-RKm′ contains an R6K origin of replication and a kanamycin resistance cassette which are integrated into the target DNA during successful transposition. Transposon mutants were selected on MacConkey plates with kanamycin containing 1% lactose incubated at 26°C and initially screened for effects on rovA::lacZ expression levels by determining colorimetric differences in colonies. Colonies with altered intensities of the red color compared to the majority of the colonies were purified, inoculated into 2-ml broth cultures, and grown overnight at 26°C, and β-galactosidase activities were determined (35). Mutants that demonstrated at least a 20% change in activity were stored at −80°C for future analysis.
To rule out false-negative mutants that affected the reporter and not the native rovA gene, anti-RovA Western blotting was performed as a secondary screen. Bacteria were grown in LB broth at 26°C to mid-logarithmic phase, harvested, and resuspended in Laemmli buffer containing 10% β-mercaptoethanol. A volume of each sample equivalent to an OD600 of 0.1 was separated on 15% polyacrylamide-sodium dodecyl sulfate gels and transferred to a polyvinylidene difluoride membrane. RovA was detected using rabbit polyclonal anti-RovA serum (1:5,000) as described previously (10). Levels of RovA from the transposon mutants were compared to levels of the protein from YVM1251. To verify that samples having an OD600 of 0.1 were equivalent, protein samples were also analyzed by Coomassie blue staining.
The R6K origin of replication within the transposon was used to reisolate the transposon along with flanking chromosomal DNA from the mutants as described below. Genomic DNA was isolated as previously described (33), and 50 μl was digested with EcoRV (New England Biolabs) overnight at 37°C. This restriction enzyme does not digest the DNA within the transposon, and therefore, the digested fragment containing the transposon had flanking ends that could be sequenced to identify the location of the transposon insertion within the Y. enterocolitica genome. The digested DNA was diluted 10-fold and ligated with T4 DNA ligase (New England Biolabs) overnight at 16°C. The ligated DNA was desalted and concentrated by ethanol precipitation to 30 μl. The entire ligation preparation was transformed into E. coli S17 λ pir by electroporation. Bacteria containing the religated transposon were selected on LB agar containing kanamycin. Three clones from each ligation were selected for sequencing using primer TNF. Sequence data were compared to the Y. enterocolitica genome from the Y. enterocolitica Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk/Projects/Y_enterocolitica/) using BLAST.
Western blot analysis.
Y. enterocolitica cultures were grown overnight in 2 ml of LB broth at 26°C with aeration, and volumes equivalent to 1.0 OD600 unit were harvested by centrifugation and resuspended in Laemmli buffer containing 10% β-mercaptoethanol. Whole-cell extracts equivalent to an OD600 of 0.1 were separated by polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. To confirm that protein samples were equivalent, total proteins were also analyzed by Coomassie blue staining prior to Western blot analysis. H-NS was detected with mouse anti-H-NS antibody kindly supplied by Yeong-Jae Seok, Seoul National University (50).
RESULTS
rovA promoter in Y. enterocolitica.
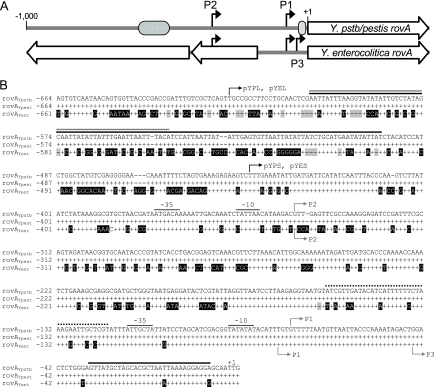
The DNA upstream of rovA in Y. enterocolitica differs greatly from the Y. pseudotuberculosis and Y. pestis DNA, with increasing divergence further 5′ of the gene (Fig. 1). In contrast, the Y. pseudotuberculosis and Y. pestis sequences are identical. The differences include dramatic changes in the region of the Y. enterocolitica promoter that corresponds to a predicted H-NS/RovA binding site of the Y. pseudotuberculosis rovA promoter. Furthermore, two putative ORFs upstream of rovA in Y. enterocolitica are absent in Y. pseudotuberculosis/Y. pestis. The ORFs overlap or replace regions that have been shown to be important for rovA regulation in Y. pseudotuberculosis (18). These variations in important regulatory domains of the Y. pseudotuberculosis promoter suggested that rovA regulation might differ in Y. enterocolitica.
FIG. 1.
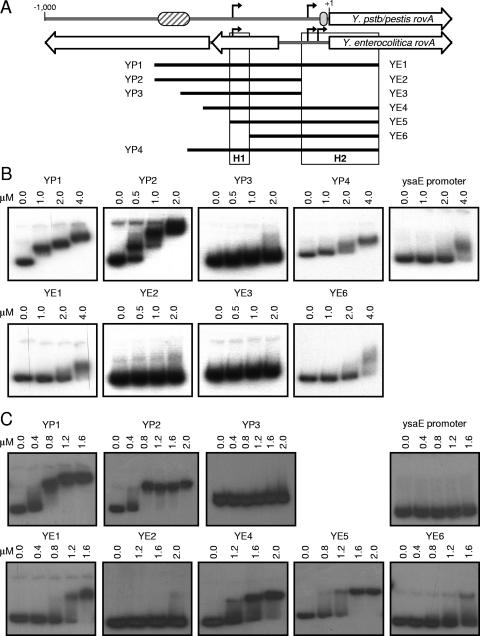
rovA promoters of Yersinia. (A) Linear representation of comparisons of the rovA regions from Y. pseudotuberculosis/Y. pestis (Y.pstb/pestis rovA) and Y. enterocolitica (Y. enterocolitica rovA). The black arrows indicate transcriptional initiation sites. The cross-hatched oval represents the predicted high-affinity RovA/H-NS binding site. The gray oval represents the predicted low-affinity RovA binding site. The large open arrows represent ORFs. (B) Sequence alignment of DNA upstream of rovA in Y. pseudotuberculosis (rovAYpstb) with similar regions in Y. pestis (rovAYpest) and Y. enterocolitica (rovAYent). Conserved nucleotides are represented by plus signs, and divergent nucleotides are indicated by a black background. Gaps are indicated by shaded dashes. The initiation codon for rovA is designated +1. The gray arrows indicate mRNA initiation sites for Y. pseudotuberculosis (above the sequences) or Y. enterocolitica (below the sequences). Predicted −35 and −10 regions for P1 and P2 in Y. pseudotuberculosis are also shown. Solid black, gray, and dotted lines above the sequences indicate predicted RovA, H-NS, and RovM binding sites, respectively, in Y. pseudotuberculosis (17, 18). The black arrows indicate the locations of the most 5′ nucleotides of reporter constructs.
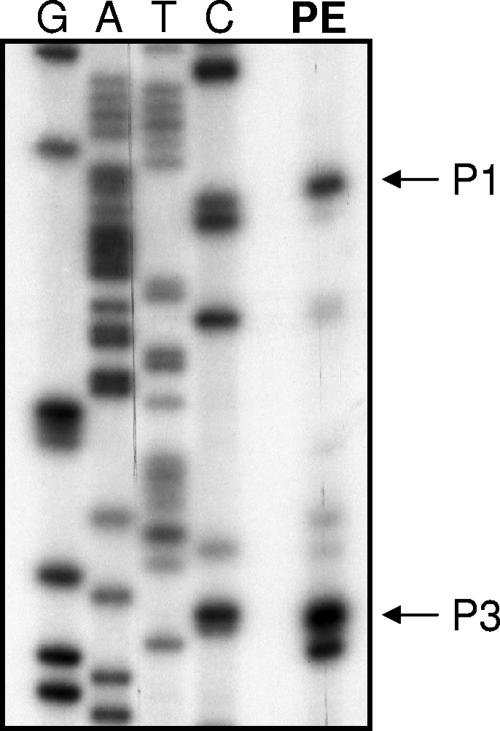
To begin our comparison of the rovA promoters (the rovAYent and Y. pseudotuberculosis/Y. pestis rovA [rovAYpstb/Ypestis] promoters), we first determined the transcriptional initiation sites of rovAYent using primer extension. In addition to the two initiation sites previously reported in Y. pseudotuberculosis (18), a third rovA putative transcript initiates downstream of P1 in Y. enterocolitica (Fig. 2). P2 begins at the same nucleotide in Y. enterocolitica that has been reported for Y. pseudotuberculosis and shares a very similar predicted −10/−35 region (one nucleotide difference in Y. enterocolitica) that is a strong σ70 consensus sequence. P1 of rovAYent appears to initiate approximately 3 nucleotides (nt) upstream of P1 in rovAYpstb/Ypestis but shares a conserved predicted −10/−35 region. P3 is predicted to begin ∼35 bp downstream of P1 and has a −10/−35 region with weaker similarity to the σ70 consensus sequence.
FIG. 2.
Mapping of rovA transcriptional initiation sites in Y. enterocolitica. Primer extension (lane PE) was performed to determine if transcription in Y. enterocolitica initiated from the same promoters that have been reported for Y. pseudotuberculosis. P3 indicates that there is a potential third transcript in Y. enterocolitica that initiates downstream of the conserved promoter P1. Sequencing ladders shown on left side of the gel.
A subset of environmental conditions that may affect the expression of rovA were tested, and only temperature influenced rovA transcription. As reported for Y. pseudotuberculosis, we observed an increase in the rovA transcript by Northern analysis in cultures grown at 26°C compared to cultures grown at 37°C (approximately fourfold higher at 26°C). This transcription profile correlated directly with protein levels in the cultures (10; data not shown). We did not observe a change in transcription in response to nutrient limitation, changes in the concentrations of iron or magnesium, increases in NaCl2 concentrations, or alterations in the pH of the medium (data not shown).
Transcription of rovA in Y. enterocolitica and the influence of expression by RovA.
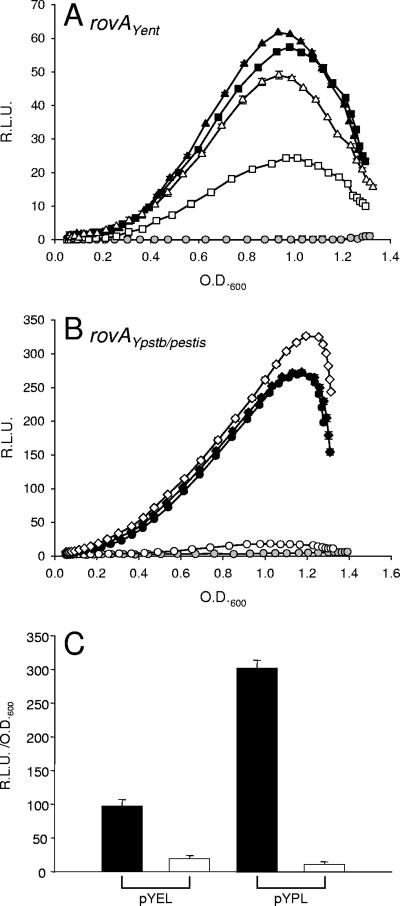
To further characterize the rovAYent promoter and compare the expression to that of the rovAYpstb/Ypestis promoter, we generated a series of GFP reporters fused to promoter regions from either rovAYent or rovAYpstb/Ypestis (Table 1 and Fig. 1). The first pair of reporters (pYEL from Y. enterocolitica and pYPL from Y. pseudotuberculosis/Y. pestis) includes the first 170 bp of rovA and approximately 620 bp of upstream DNA. pYPL includes the predicted H-NS and RovA binding sites of Y. pseudotuberculosis (Fig. 1) (18). The second pair of reporters (pYES and pYPS) also includes 170 bp of the rovA coding region but only approximately 445 bp of the upstream region, resulting in loss of the predicted H-NS/RovA binding site of Y. pseudotuberculosis. These reporters were transformed into wild-type and ΔrovA strains of Y. enterocolitica to compare the activities of the two promoters.
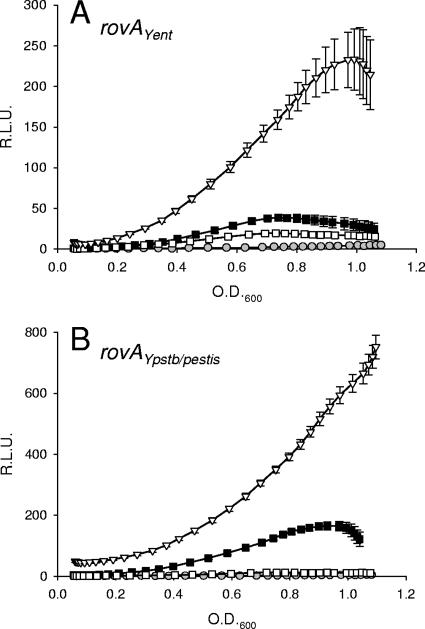
The pYPL reporter demonstrated approximately fivefold-higher activity than the equivalent Y. enterocolitica reporter, pYEL, suggesting that the rovAYpstb/Ypestis promoter is a stronger promoter than the rovAYent promoter in Y. enterocolitica (Fig. 3). pYPL also demonstrated a greater requirement for RovA than pYEL. The activity of pYPL decreased almost 20-fold when it was analyzed in a ΔrovA background. While a decrease in pYEL activity was also observed in the ΔrovA strain, it decreased by only 2.5-fold and was still expressed at levels above background levels. As expected, when we removed the H-NS/RovA binding site from the Y. pseudotuberculosis/Y. pestis promoter (pYPS), the requirement for RovA for expression was lost. Interestingly, when a similar amount of DNA was removed from the 5′ end of the Y. enterocolitica promoter (pYES), we observed a partial dependence on RovA. Similar differences in promoter strengths were observed when the reporters were transformed into Y. pestis (Fig. 3C). pYPL expression was more than threefold higher than pYEL expression during late stationary phase, and both reporters demonstrated negligible expression in the rovA mutant. These results indicate that the rovAYent promoter is less active than the rovAYpstb/Ypestis promoter, but regardless of the promoter activities, both promoters require RovA for maximal expression.
FIG. 3.
Influence of RovA on the expression of rovA. To determine the effect of RovA on the rovAYent (A) and rovAYpstb/Ypestis (B) promoters, expression patterns were determined in wild-type Y. enterocolitica (filled symbols) and a ΔrovA mutant (open symbols). Four GFP reporters were utilized: Y. enterocolitica rovA reporter fusions (squares, pYEL; triangles, pYES) and Y. pseudotuberculosis/Y. pestis rovA reporter fusions (circles, pYPL; diamonds, pYPS). Gray circles show the results for vector-only controls. The data are expressed in relative light units (R.L.U.) as a function of the OD600. (C) Expression of pYPL and pYEL in wild-type Y. pestis (filled bars) and a ΔrovA mutant (open bars). The data are expressed in relative light units (R.L.U.)/OD600. Experiments were performed in triplicate, and error bars were plotted but are typically too small to see clearly.
H-NS regulation of rovA.
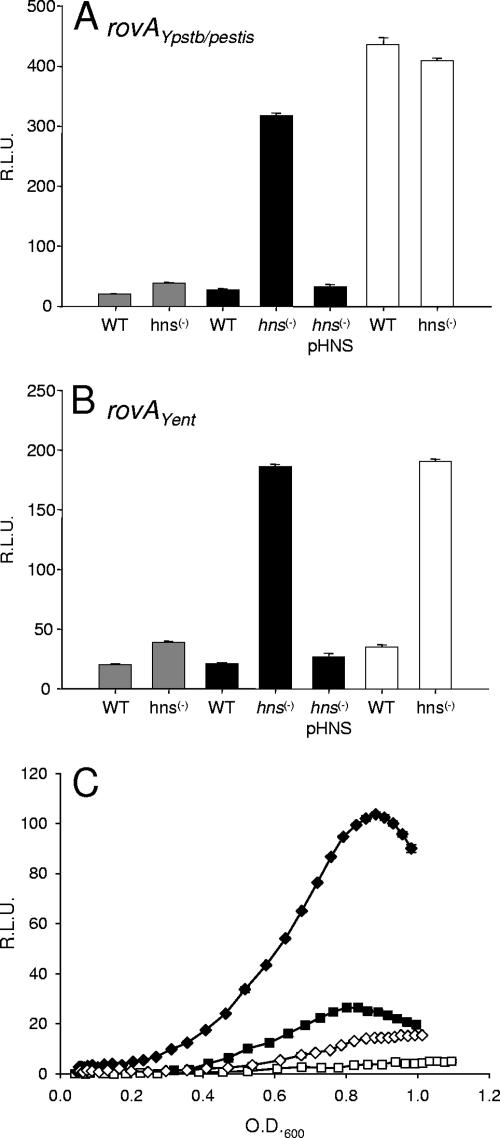
Heroven et al. previously demonstrated that H-NS is a negative regulator of rovA in Y. pseudotuberculosis, with repression achieved through direct interactions with a region of the promoter missing in the rovAYent promoter (Fig. 1) (18). Since rovAYent is temperature regulated and H-NS is a major mediator of temperature regulation in prokaryotes, we suspected that H-NS mediates rovA transcription even in the absence of the rovAYpstb/Ypestis H-NS binding site. hns is apparently required for growth of Yersinia as we and others have been unable to inactivate the gene in Y. enterocolitica or Y. pseudotuberculosis (17; M. B. Lawrenz, D. W. Ellison, C. Affolter, and V. L. Miller, unpublished data). To overcome this obstacle, two independent systems were utilized to provide insight into the role of H-NS regulation of rovAYent. Similar systems have been used previously to determine the impact of H-NS on Yersinia gene regulation (10, 18). We determined the expression of the rovA reporters in an E. coli hns mutant and compared the pattern to expression in wild-type E. coli or in the hns mutant complemented with hns from Y. enterocolitica expressed ectopically on a low-copy-number plasmid (pHNS). Activity of the promoters was determined as a function of fluorescence. For clarity, data from cultures at an OD600 of approximately 0.5 are displayed, but the trends were conserved throughout the growth curve. As reported previously, rovAYpstb/Ypestis transcription was dependent on the presence of H-NS (Fig. 4A). In the wild-type MC4100 background no fluorescence in the culture above the fluorescence of the control strain (vector only) was observed. Inactivation of hns resulted in dramatic increases in pYPL expression, which were more than 11-fold at this time point and 30-fold at the peak of the GFP expression (data not shown). rovAYent transcription followed a similar pattern (Fig. 4B). While hns inactivation also resulted in increased transcription of the rovAYent promoter, a more moderate increase was observed (the greatest increase observed was ninefold), and the levels of fluorescence from the pYEL reporter never reached those from the Y. pseudotuberculosis/Y. pestis reporter. Complementation with pHNS resulted in an expression pattern similar to that of wild-type MC4100 and expression of pYPL and pYEL at background levels. These results strongly indicate that H-NS negatively regulates rovAYent, at least in a surrogate E. coli background.
FIG. 4.
Influence of H-NS on the expression of rovA. To determine the effect of H-NS on the rovAYpstb/Ypestis (A) and rovAYent (B) promoters, expression patterns were determined in E. coli MC4100 (WT), an hns mutant [hns(−)], or an hns mutant complemented with Y. enterocolitica hns [hns(−) pHNS]. Filled bars, pYPL (A) or pYEL (B); open bars, pYPS (A) or pYES (B); gray bars, vector control. (C) Expression of pYPL (diamonds) and pYEL (squares) in wild-type Y. enterocolitica (filled symbols) and pHNS-complemented Y. enterocolitica (open symbols). The data are expressed in relative light units (R.L.U.) as a function of the OD600.
To support our E. coli data, we next investigated H-NS regulation of rovAYent in native Y. enterocolitica to determine if the regulation was conserved in the wild-type background. We transformed the reporters into a strain of Y. enterocolitica that also contained pHNS. The levels of H-NS in this strain are artificially high at 26°C due to the increased copy number of the gene (10; data not shown). The expression of both pYPL and pYEL was decreased in this strain by approximately 7.5-fold compared to the expression in wild-type Y. enterocolitica (Fig. 4C). Taken together with the data from E. coli, these results suggest that rovAYent is modulated by H-NS.
Deletion of 175 bp from the 5′ end of pYPL resulted in rovA transcription in wild-type E. coli (Fig. 4A), supporting the presence of an H-NS binding site within this region as predicted by in vitro DNA binding experiments of Heroven et al. (18). Deleting a similar region (based on distance from the rovA initiation codon) in Y. enterocolitica did not relieve H-NS-mediated repression (Fig. 4B). These results demonstrate that while H-NS represses rovA transcription in Y. enterocolitica, H-NS binds to a different region in the rovAYent promoter than in Y. pseudotuberculosis.
Binding of RovA and H-NS to the rovA promoter.
In vitro analysis of Y. pseudotuberculosis revealed that RovA and H-NS bind to a similar region in the rovA promoter (18). Despite the absence of this binding site in the Y. enterocolitica promoter, our in vivo data demonstrate that both proteins influence the expression of rovAYent. To determine whether these proteins directly regulate rovAYent through interactions with the Y. enterocolitica promoter, we compared binding of recombinant RovA and H-NS from Y. enterocolitica to fragments of the rovAYent and rovAYpstb/Ypestis promoters using an EMSA (Fig. 5). As predicted by Heroven et al. (18), RovA bound to fragments of the rovAYpstb/Ypestis promoter that contained the 5′ RovA binding site (YP1 and YP2) and bound more weakly (requiring at least twofold more protein to bind) to a fragment that contained only the 3′ RovA binding site (YP4) (Fig. 5B). The absence of both sites resulted in loss of RovA binding (YP3). Approximately fourfold-higher concentrations of RovA were required to initiate changes in the mobility of the Y. enterocolitica promoter (YE1), and these concentrations approached concentrations leading to nonspecific binding of the negative control (ysaE) (Fig. 5B).
FIG. 5.
Ability of RovA and H-NS to bind to the rovA promoters. (A) Schematic representation of the rovA promoters from Y. pseudotuberculosis/Y. pestis (Y.pstb/pestis rovA) and Y. enterocolitica and the PCR products used in EMSA analysis. The striped and gray ovals represent the predicted high-affinity RovA/H-NS and predicted low-affinity RovA binding sites, respectively, from Y. pseudotuberculosis. The black arrows represent transcriptional initiation sites. The black bars represent PCR products used for the EMSA from Y. pseudotuberculosis/Y. pestis (YP) and Y. enterocolitica (YE). H1 and H2 represent regions of H-NS binding in the Y. enterocolitica promoter. (B) EMSA performed with recombinant RovA-His. (C) EMSA performed with recombinant H-NS-His. The fragment used in each panel and the concentrations of the protein added to each reaction mixture are shown. The ysaE promoter is not regulated by either RovA or H-NS and was included as a negative control for binding.
H-NS bound to the Y. pseudotuberculosis/Y. pestis promoter at a concentration approximately threefold lower than the concentration at which it bound to the Y. enterocolitica promoter (YP1 and YE1), indicating that H-NS had a higher affinity for rovAYpstb/Ypestis (Fig. 5C). This interaction with the rovAYent promoter appears to be specific, as concentrations required for binding YE1 did not bind the ysaE control promoter. These results correlate directly with the degree of regulation by RovA and H-NS for these promoters. Both proteins bind to the Y. pseudotuberculosis/Y. pestis promoter with a greater affinity than they bind to the Y. enterocolitica promoter and have greater influence on the expression of rovAYpstb/Ypestis.
In vivo data demonstrated that truncation of the rovAYent promoter by 175 nt did not alter the effects of H-NS on transcription (Fig. 4A), indicating that the H-NS binding site is not present within this region. To further narrow the region within the Y. enterocolitica promoter that H-NS binds, we generated PCR fragments representing truncated regions of the promoter and analyzed the ability of H-NS to bind to these fragments. Deletion of 265 nt from the 5′ end of the promoter resulted in a dramatic decrease in H-NS binding (YE6), which was not restored until the fragment included the region 349 to 279 nt upstream of the rovA initiation codon (YE5). These results suggest the presence of an H-NS binding site in this region. Interestingly, H-NS did not bind to YE2, which contains this region but lacks sequence in the 3′ region of the promoter. This finding may indicate that a second H-NS binding site is present within the 3′ region of the promoter. A second low-affinity H-NS binding site within this region could also explain the weak binding observed at higher H-NS concentrations in YE6.
RovM regulation of rovA in Y. enterocolitica.
Recently, it was shown that RovM modulates the expression of rovA in Y. pseudotuberculosis (17). RovM is also present in Y. enterocolitica (ORF YE1343), and the region within the rovA promoter where RovM is predicted to bind is considerably more conserved between the species (76% identity) than the RovA/H-NS binding site (Fig. 1B). Therefore, we hypothesized that RovM is involved in the regulation of rovA in Y. enterocolitica. To determine the role of RovM in the regulation of rovAYent, we generated an in-frame deletion of rovM in Y. enterocolitica and transformed the pYPL and pYEL reporters into the strain. Cultures were grown as described above, and levels of rovA transcription in the rovM mutant were determined as a function of fluorescence and compared to levels in wild-type bacteria. As observed in Y. pseudotuberculosis, deletion of rovM in Y. enterocolitica resulted in an increase in rovA transcription (Fig. 6). Near peak rovAYpstb/Ypestis expression in wild-type Y. enterocolitica (OD600, ∼0.9), the levels of transcription in the rovM mutant increased approximately 3.2-fold. A moderately higher increase was observed for the rovAYent promoter (approximately 4.8-fold). These results support the hypothesis that RovM has a conserved role in the modulation of rovA expression in Yersinia.
FIG. 6.
Influence of RovM on the expression of rovA. To determine the effect of RovM on the rovAYent pYEL (A) and rovAYpstb/Ypestis pYPL (B) promoters, expression patterns were determined in wild-type (▪), ΔrovA mutant (□), and ΔrovM mutant (▿) Y. enterocolitica. The results for the vector-only control are indicated by gray circles. The data are expressed in relative light units (R.L.U.) as a function of the OD600.
LeuO is a positive regulator of rovA.
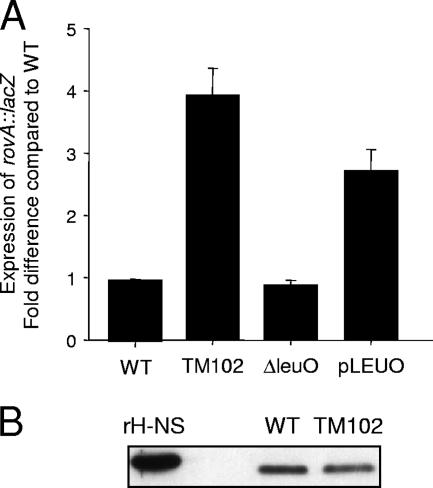
In order to identify novel factors involved in the regulation of rovA expression in Y. enterocolitica, we initiated a transposon mutagenesis screen in a strain of Y. enterocolitica with a second copy of the rovA promoter fused to a lacZ reporter integrated in the arabinose operon (YVM1251). Approximately 42,000 colonies from 21 independent conjugations were screened. There were 150 mutants that displayed at least a 20% variation in β-galactosidase activity compared to YVM1251 in an independent analysis in liquid culture. Eight of these mutants showed a reproducible >2-fold effect on rovA::lacZ expression. Upon Western blot analysis of native RovA levels, only four of the eight mutants demonstrated a difference in native RovA protein levels (data not shown). TM102 demonstrated the greatest activation of rovA of these four mutants (approximately threefold increase in rovA::lacZ expression) (Fig. 7) and was selected for further characterization.
FIG. 7.
LeuO regulation of rovA in Y. enterocolitica. (A) To determine the effect of LeuO on rovA transcription in Y. enterocolitica, the levels of β-galactosidase activity of the rovA::lacZ reporter were compared in the wild type (WT), a transposon mutant with a mutation in the promoter of leuO (TM102), a mutant with an in-frame deletion of leuO (ΔleuO), and a strain containing leuO on a multicopy plasmid (pLEUO). The expression is shown as the fold change compared with wild-type expression. (B) Whole-cell proteins (equivalent to 0.1 OD600 unit) from wild-type (WT) and TM102 cultures were harvested and analyzed by Western blotting with anti-H-NS antibody. Purified His-tagged H-NS (rH-NS) served as a positive control.
TM102 contains an insertion upstream of ORF YE0655 (56). YE0655 encodes a homolog of the LeuO regulator of E. coli (75% similarity and 62% identity; P = 9.9e−91). LeuO is a member of the LysR family of transcriptional regulators and was originally identified as a regulator of the cryptic bgl operon of E. coli (28). Subsequently, it was shown that leuO expression is ppGpp dependent and involved in the stringent response of bacteria (13, 30). It was unclear whether the location of the transposon insertion within the promoter of leuO resulted in inactivation of the gene or induction of leuO transcription. To determine which event occurred, we generated two Yersinia strains in the YVM1251 background: a strain with an in-frame deletion of leuO (YVM1252) and a strain containing an additional copy of leuO controlled by its native promoter on a multicopy plasmid (pLEUO). Deletion of the gene resulted in no changes in rovA::lacZ expression, while increased β-galactosidase levels were observed in the pLEUO-complemented strain (Fig. 7). These results indicate that LeuO is a positive regulator of rovA in Y. enterocolitica.
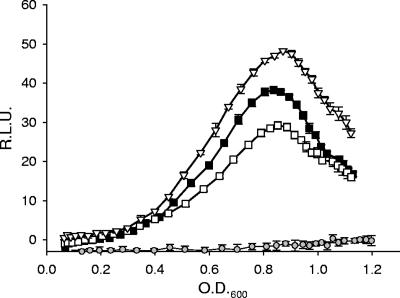
In addition to the bgl operon, LeuO has been shown to positively regulate osmY in E. coli (24). osmY expression is mediated by σS and induced during the transition from logarithmic to stationary phase and in response to osmotic stress (25, 60). Klauck et al. (24) demonstrated that LeuO indirectly regulated osmY by inducing the expression of a small regulatory RNA, dsrA, which altered the levels of σS in the bacterium. The changes in σS levels in the bacterium in turn influenced the expression of osmY. The similarities in osmY and rovA expression patterns (peak expression during the transition to stationary phase and regulation by LeuO) suggested that σS may also be a regulator of rovA in Y. enterocolitica. To address this possibility, we generated an in-frame deletion of rpoS in Y. enterocolitica and determined the effects on rovA using the pYEL reporter. Inactivation of rpoS did not result in a significant change in rovA::GFP expression compared to wild-type Y. enterocolitica (Fig. 8), demonstrating that σS is not a regulator of rovA. Furthermore, these results suggest that LeuO-mediated regulation of rovA is independent of σS.
FIG. 8.
Influence of σs on the expression of rovA. To determine the effect of σs on the expression of rovAYent (pYEL), expression patterns were determined in wild-type (▪), ΔrovA mutant (□), and ΔrpoS mutant (▿) Y. enterocolitica. The results for the vector-only control are indicated by gray circles. The data are expressed in relative light units (R.L.U.) as a function of the OD600.
LeuO has also been implicated in the regulation of H-NS translation in E. coli (24, 26). Since H-NS is a negative regulator of rovA, it is possible that the changes in rovA transcription in TM102 are a result of alterations in the steady-state levels of H-NS in the mutant. To determine if H-NS levels are affected by increased LeuO expression, total protein was harvested from wild-type and TM102 cultures and the levels of H-NS were compared by Western blot analysis (Fig. 7B). No significant differences in H-NS levels were observed, indicating that the increased expression of LeuO did not significantly alter the translation of H-NS in Y. enterocolitica. These data indicate that LeuO is unlikely to regulate rovAYent expression by altering the levels of H-NS in the cell.
DISCUSSION
Y. enterocolitica and Y. pseudotuberculosis share conserved virulence mechanisms to cause gastrointestinal infection. Invasin, the major adhesin for both species, interacts with β1-integrins on the surface of human cells (21). This interaction mediates invasion of host cells and may promote efficient translocation across the epithelial barrier during initial stages of colonization (20, 36, 44). Interestingly, inv has been inactivated in Y. pestis, which relies primarily on an arthropod vector to colonize a new host (51). The regulation of inv transcription is also conserved between Y. enterocolitica and Y. pseudotuberculosis. Interactions between H-NS and RovA have been shown to mediate the expression of inv in both species (10, 40). These proteins have also been implicated in the regulation of the rovA gene in Y. pseudotuberculosis (18). While rovA is highly conserved in all three species, inspection of the region upstream of rovA revealed that the promoter in Y. enterocolitica has greatly diverged from that in the other two pathogens. The differences in the promoters result in significantly lower levels of transcription in Y. enterocolitica from the rovA promoter. This decreased rovAYent activity may be due to transcription of the divergent ORFs upstream of rovAYent occurring at 26°C (Lawrenz and Miller, unpublished). However, we were interested in determining whether mechanisms reported for rovA regulation in Y. pseudotuberculosis are active in Y. enterocolitica.
Despite the divergence of the Y. enterocolitica rovA promoter from the promoter of the other two species and the absence of the rovAYpstb/Ypestis predicted H-NS/RovA binding site, we observed that rovA regulation in Y. enterocolitica remains responsive to H-NS and RovA. As reported for Y. pseudotuberculosis, H-NS represses rovAYent transcription; however, regulation is mediated through interactions with different regions within the promoter. rovA is also autoregulated in Y. enterocolitica, and RovA is required for maximal transcription. It should be noted that both regulators affect the level of transcription from the rovAYpstb/Ypestis promoter to a greater degree than that seen for rovAYent. These results support the hypothesis that H-NS and RovA mediate transcription in Y. enterocolitica. Interestingly, transcription of pYPL and pYEL is detectable in a rovA mutant, and the levels of activity from the reporters are similar. These data indicate that the basal levels of RovA in the repressed state are similar for both promoters. This low level of expression may be important to maintain a pool of RovA that it is available to quickly initiate rovA transcription in response to stimuli. However, the mechanism(s) that leads to derepression of rovA is not yet fully understood. Transcription may result from a combination of factors, including changes in affinity for the promoter by RovA or H-NS, alterations of the DNA structure within the promoter, stability of the regulators, and/or activity of other regulators.
The differences in transcriptional modulation by RovA and H-NS correlate directly with the ability of the proteins to bind to the promoters. Both proteins bind to the Y. pseudotuberculosis/Y. pestis promoter at lower concentrations and affect levels of transcription to higher degrees. The ability of the rovAYpstb/Ypestis promoter to bind the proteins at lower concentrations can be attributed mostly to the region identified by Heroven et al. that is absent in the rovAYent promoter (18). The current model for RovA/H-NS-mediated regulation in Y. pseudotuberculosis suggests that the two proteins compete for binding to this site, so that successful binding by RovA derepresses H-NS repression. In corroboration of this hypothesis, in vivo data presented here demonstrate that deletion of the RovA/H-NS binding site from the Y. pseudotuberculosis/Y. pestis promoter eliminated the requirement of RovA for expression of the pYPS reporter. We were also able to observe weak RovA binding to YP4, which includes a second proposed RovA binding site near the rovAYpstb/Ypestis coding region (Fig. 1) (18). Herevon et al. have suggested that this low-affinity site may be involved in negative auto-feedback regulation to repress rovA transcription when levels of RovA in the cell reach a certain value. This low-affinity site is conserved within the Y. enterocolitica promoter, with only three nucleotide changes.
The interactions between RovA and H-NS that mediate the transcription of rovAYent are less obvious due to the lack of observable RovA binding to the Y. enterocolitica promoter. While our in vivo data suggest that autoregulation occurs in Y. enterocolitica, our in vitro data indicate that RovA does not bind specifically to the rovAYent promoter. These in vitro results suggest that RovA regulation may occur through an indirect mechanism. However, we cannot eliminate the possibility that RovA derepresses H-NS directly in Y. enterocolitica and the level of specific binding of RovA to rovAYent is below the sensitivity of our assay. The possible limitation of our EMSA conditions to detect low-affinity RovA binding to rovAYent, and therefore direct regulation by RovA, is supported by the lack of specific RovA binding to the conserved RovA low-affinity binding site in the Y. enterocolitica promoter. Modifications in EMSA conditions have yet to demonstrate specific RovA binding, but future variations of in vitro binding conditions and/or in vivo binding assays, such as chromatin immunoprecipitation, may result in more sensitive assays that could aid in determining if RovA interacts with the Y. enterocolitica promoter.
Tran et al. have reported that RovA can directly activate transcription of rovAYpstb/Ypestis through interactions with RNA polymerase in vitro (57). Deletion of the high-affinity RovA/H-NS binding site did not alter expression of rovAYpstb/Ypestis in wild-type Y. enterocolitica. These results support the hypothesis that binding of RovA to the RovA/H-NS binding site in the Y. pseudotuberculosis/Y. pestis promoter primarily relieves negative regulation by H-NS. Furthermore, since expression of pYPS did not decrease in wild-type Y. enterocolitica, RovA binding to the high-affinity binding site in pYPL does not appear to significantly activate rovA transcription in vivo. If RovA actively induces transcription of the Y. pseudotuberculosis/Y. pestis promoter, these data suggest that it occurs through interactions with other regions of the promoter.
In vitro DNA binding assays indicate the presence of two H-NS binding sites in Y. enterocolitica: between nt −349 and −279 and between nt −128 and 171 (in relation to the initiation codon). H-NS demonstrates much lower affinity for either of these sites than for the reported Y. pseudotuberculosis site. H-NS also appears to cooperatively bind to these sites, as loss of the H2 site results in loss of binding to the H1 site (compare binding to YE2 and binding to YE4). The presence of multiple binding sites within a promoter is a common theme for H-NS-mediated repression and has been well described for the rrnB P1 and proU promoters (48, 53). Dorman and Deighan proposed that binding to two regions, in conjunction with protein oligomerization, leads to formation of a loop within these promoters that traps RNA polymerase and blocks initiation of transcription (8). A similar mechanism could occur within the rovAYent promoter. Binding to the H1 and H2 regions may result in loop formation that occludes all three promoters from interactions with RNA polymerase, leading to repression of transcription.
Based on sequence similarity alone, one would suspect that H-NS should also bind the same H2 region in Y. pseudotuberculosis/Y. pestis; however, binding to this region has not been observed by us or reported by others. The lack of data indicating a second binding site in Y. pseudotuberculosis may be due to weak binding of H-NS to the H2 site that is below the sensitivity of the footprinting analysis used to map interactions of H-NS with the rovAYpstb/Ypestis promoter (18). Alternatively, binding to the H2 region may have been missed because the probes used for footprinting the 5′ region of the promoter did not include both binding sites on the same fragment. In Y. enterocolitica, binding of H-NS to either the H1 or H2 site required the presence of the other site, indicating cooperative binding. The presence of two binding sites may also be necessary for H2 binding in Y. pseudotuberculosis. Finally, the strong binding of H-NS to the high-affinity site may mask binding to a second site. In Y. pseudotuberculosis/Y. pestis, the high-affinity site does not appear to require a second binding site. Therefore, promoter probes for EMSA that are generated without the H2 site would still interact with H-NS, masking the presence of this binding site in the promoter.
Heroven and Dersh previously demonstrated that RovM regulates the transcription of rovA in Y. pseudotuberculosis (17). Unlike the predicted H-NS/RovA binding site, the RovM predicted binding site is conserved in Y. enterocolitica. We demonstrated here that rovA transcription in Y. enterocolitica is also mediated by RovM. It seems likely that RovM binds to the same region in the Y. enterocolitica promoter and directly represses rovA transcription, as reported for Y. pseudotuberculosis.
In addition to H-NS, RovA, and RovM, we identified a fourth regulator involved in rovA regulation in Y. enterocolitica. leuO encodes a LysR-like regulator and is induced in the stringent response that occurs during amino acid starvation. Also, it is required to resume growth after starvation (30). The conditions for LeuO expression suggest that rovA responds to nutrient limitation or another stress signal. It has been shown that rovA transcription in Y. pseudotuberculosis is altered in cultures grown in minimal medium compared to cultures grown in rich medium (17, 40); however, we did not observe a similar pattern in Y. enterocolitica. The higher levels of rovA transcription from the Y. pseudotuberculosis promoter may allow detection of subtle changes in rovA expression that are not as easily observed for the weaker Y. enterocolitica promoter. The rovA response to starvation or decreased availability of nutrients could also explain why peak expression of rovA and the rovA-regulated gene inv occurs upon entry into late-logarithmic/early-stationary growth, when nutrient levels in the medium are starting to decline.
LeuO has been implicated in the regulation of several genes in E. coli, including the bgl operon and osmY (24, 28). In the case of osmY regulation, increased expression of LeuO represses the transcription of the small regulatory RNA dsrA, resulting in destabilization of the rpoS message and down regulation of osmY. Due to the difficulty in predicting small regulatory RNAs, it has yet to be determined if a dsrA-like homolog is present in Y. enterocolitica. However, we did not observe a requirement for rpoS for rovA regulation, indicating that LeuO affects the transcription of rovA by a mechanism that differs from that of osmY. It is unclear at this time whether regulation occurs through direct interaction between LeuO and the rovA promoter or if LeuO modulates an additional regulator that in turn acts on the rovA promoter. However, LeuO does not appear to affect rovA transcription by modulating H-NS steady-state levels in the bacterium.
In conclusion, we have demonstrated that conserved factors are active in the regulation of rovA in Y. enterocolitica and Y. pseudotuberculosis/Y. pestis despite considerable differences in the relative strengths and sequences of the rovA promoters. However, divergence between Yersinia species during evolution has resulted in modifications in the regulatory mechanisms. The identification of the stringent response protein LeuO as a rovA regulator strengthens the hypothesis that starvation or nutrient limitation may be an additional signal to Yersinia to induce rovA transcription, leading to induction of inv and other genes that prepare the bacterium to colonize its host. Future analysis of the mechanism by which LeuO regulates rovA may provide insight into whether a small regulatory RNA such as DsrA is also involved in rovA regulation.
Acknowledgments
We thank A. J. Darwin for providing the pAJD905 plasmid to generate the rovA::lacZ reporter strain and S. E. Lindow for providing the pPROBE-gfp[LVA] reporter plasmid. We also thank Damon Ellison for advice concerning EMSA protocols and for helpful discussions during the course of this study and Kim Walker for advice on primer extension.
This study was supported by NIH grants AI063746 and AI07172 (to M.B.L.) and AI052167 (to V.L.M.).
Footnotes
Published ahead of print on 15 June 2007.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cathelyn, J. S., S. D. Crosby, W. W. Lathem, W. E. Goldman, and V. L. Miller. 2006. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. USA 103:13514-13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. M-cell surface β1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M.-P. Sory, J.-C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 6.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doll, J. M., P. S. Zeitz, P. Ettestad, A. L. Bucholtz, T. Davis, and K. Gage. 1994. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 51:109-114. [DOI] [PubMed] [Google Scholar]
- 8.Dorman, C. J., and P. Deighan. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179-184. [DOI] [PubMed] [Google Scholar]
- 9.Dube, P. H., S. A. Handley, P. A. Revell, and V. L. Miller. 2003. The rovA mutant of Yersinia enterocolitica displays differential degrees of virulence depending on the route of infection. Infect. Immun. 71:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison, D. W., and V. L. Miller. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188:5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison, D. W., and V. L. Miller. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153-159. [DOI] [PubMed] [Google Scholar]
- 12.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, M., A. Majumder, K. J. Tsai, and H. Y. Wu. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276:64-70. [DOI] [PubMed] [Google Scholar]
- 14.Fleming, T. P., M. S. Nahlik, and M. A. McIntosh. 1983. Regulation of enterobactin iron transport in Escherichia coli: characterization of ent::Mu d(Apr lac) operon fusions. J. Bacteriol. 156:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 155:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, A., K. Paul, and R. Chowdhury. 2006. Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect. Immun. 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heroven, A. K., and P. Dersch. 2006. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 62:1469-1483. [DOI] [PubMed] [Google Scholar]
- 18.Heroven, A. K., G. Nagel, H. J. Tran, S. Parr, and P. Dersch. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53:871-888. [DOI] [PubMed] [Google Scholar]
- 19.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isberg, R. R., and S. Falkow. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262-264. [DOI] [PubMed] [Google Scholar]
- 21.Isberg, R. R., and J. M. Leong. 1990. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861-871. [DOI] [PubMed] [Google Scholar]
- 22.Isberg, R. R., A. Swain, and S. Falkow. 1988. Analysis of expression and thermoregulation of the Yersinia pseudotuberculosis inv gene with hybrid proteins. Infect. Immun. 56:2133-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R− M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 24.Klauck, E., J. Bohringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 25.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the sigma σ-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 27.Libby, S. J., A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhusudan, S., A. Paukner, Y. Klingen, and K. Schnetz. 2005. Independent regulation of H-NS-mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology 151:3349-3359. [DOI] [PubMed] [Google Scholar]
- 29.Madrid, C., J. M. Nieto, and A. Juarez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425-432. [DOI] [PubMed] [Google Scholar]
- 30.Majumder, A., M. Fang, K. J. Tsai, C. Ueguchi, T. Mizuno, and H. Y. Wu. 2001. LeuO expression in response to starvation for branched-chain amino acids. J. Biol. Chem. 276:19046-19051. [DOI] [PubMed] [Google Scholar]
- 31.Marra, A., and R. R. Isberg. 1997. Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer's patch intestinal epithelium. Infect. Immun. 65:3412-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxson, M. E., and A. J. Darwin. 2005. Improved system for construction and analysis of single-copy beta-galactosidase operon fusions in Yersinia enterocolitica. Appl. Environ. Microbiol. 71:5614-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 34.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci from Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 38.Muller, C. M., U. Dobrindt, G. Nagy, L. Emody, B. E. Uhlin, and J. Hacker. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagel, G., A. K. Heroven, J. Eitel, and P. Dersch. 2003. Function and regulation of the transcriptional activator RovA of Yersinia pseudotuberculosis. Adv. Exp. Med. Biol. 529:285-287. [DOI] [PubMed] [Google Scholar]
- 40.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 41.Naktin, J., and K. G. Beavis. 1999. Yersinia enterocolitica and Yersinia pseudotuberculosis. Clin. Lab. Med. 19:523-536. [PubMed] [Google Scholar]
- 42.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodriguez, and A. Juarez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 44.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prentice, M. B., and L. Rahalison. 2007. Plague. Lancet 369:1196-1207. [DOI] [PubMed] [Google Scholar]
- 46.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 47.Reverchon, S., C. Rouanet, D. Expert, and W. Nasser. 2002. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroder, O., and R. Wagner. 2002. The bacterial regulatory protein H-NS—a versatile modulator of nucleic acid structures. Biol. Chem. 383:945-960. [DOI] [PubMed] [Google Scholar]
- 49.Schulte, R., S. Kerneis, S. Klinke, H. Bartels, S. Preger, J. P. Kraehenbuhl, E. Pringault, and I. B. Autenrieth. 2000. Translocation of Yersinia enterocolitica across reconstituted intestinal epithelial monolayers is triggered by Yersinia invasin binding to beta1 integrins apically expressed on M-like cells. Cell. Microbiol. 2:173-185. [DOI] [PubMed] [Google Scholar]
- 50.Shin, M., M. Song, J. H. Rhee, Y. Hong, Y. J. Kim, Y. J. Seok, K. S. Ha, S. H. Jung, and H. E. Choy. 2005. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 19:2388-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 53.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. Stewart, P. Williams, and G. P. Salmond. 1997. The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 56.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran, H. J., A. K. Heroven, L. Winkler, T. Spreter, B. Beatrix, and P. Dersch. 2005. Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 280:42423-42432. [DOI] [PubMed] [Google Scholar]
- 58.Walker, K. A., C. L. Atkins, and R. Osuna. 1999. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J. Bacteriol. 181:1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker, K. A., and V. L. Miller. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weichart, D., R. Lange, N. Henneberg, and R. Hengge-Aronis. 1993. Identification and characterization of stationary phase-inducible genes in Escherichia coli. Mol. Microbiol. 10:407-420. [PubMed] [Google Scholar]