Abstract
In response to infection, Caenorhabditis elegans produces an array of antimicrobial proteins. To understand the C. elegans immune response, we have investigated the regulation of a large, representative sample of candidate antimicrobial genes. We found that all these putative antimicrobial genes are expressed in tissues exposed to the environment, a position from which they can ward off infection. Using RNA interference to inhibit the function of immune signaling pathways in C. elegans, we found that different immune response pathways regulate expression of distinct but overlapping sets of antimicrobial genes. We also show that different bacterial pathogens regulate distinct but overlapping sets of antimicrobial genes. The patterns of genes induced by pathogens do not coincide with any single immune signaling pathway. Thus, even in this simple model system for innate immunity, striking specificity and complexity exist in the immune response. The unique patterns of antimicrobial gene expression observed when C. elegans is exposed to different pathogens or when different immune signaling pathways are perturbed suggest that a large set of yet to be identified pathogen recognition receptors (PRRs) exist in the nematode. These PRRs must interact in a complicated fashion to induce a unique set of antimicrobial genes. We also propose the existence of an “antimicrobial fingerprint,” which will aid in assigning newly identified C. elegans innate immunity genes to known immune signaling pathways.
When infected by a pathogen, humans mount an immediate innate immune response as well as a slower but more specific adaptive immune response. The immediate response involves infiltration of phagocytic and cytotoxic cells at the site of infection and the release of antimicrobial compounds (25). The innate immune system also contributes to the activation of the adaptive immune response (20). Thus, innate immunity plays a vital role in pathogen defense. However, misregulation of innate immunity contributes to the pathogenesis of many human diseases, including sepsis, asthma, and atherosclerosis (6). While adaptive immunity is only present in vertebrates, many aspects of innate immunity are conserved throughout the animal kingdom.
The nematode C. elegans is susceptible to many of the pathogens that infect humans, including gram-positive Staphylococcus aureus and gram-negative Serratia marcescens and Pseudomonas aeruginosa (28, 31, 45, 49, 52). Like most pathogens that infect C. elegans, these three bacteria colonize the digestive tract and ultimately kill the nematode. In contrast, other bacteria such as Escherichia coli and Bacillus subtilis are usually not toxic to C. elegans (13). Many of the host genes involved in pathogen defense in mammals also function in host defense of C. elegans (3, 10, 35, 46). Similarly, many virulence genes used by pathogens to infect C. elegans are also required for mammalian infection (3, 8, 31, 39, 48, 50, 53-55, 57). Thus, C. elegans has proven to be a useful and relatively simple model with which to study innate immunity.
The C. elegans innate immune response consists of the production of numerous antimicrobial proteins, many of which are conserved in higher organisms. Many candidate antimicrobial genes have been identified in the C. elegans genome (17, 40, 46). Previous data have demonstrated that the expression of some of these genes is induced upon pathogen infection (7, 32, 41, 47, 56). Moreover, some of these putative antimicrobial genes are regulated by signaling pathways involved in pathogen defense in nematodes and mammals (7, 21, 36, 38, 56). To better understand how these genes function in host defense, we have undertaken a study of a large, representative sample of candidate antimicrobial genes encoded in the C. elegans genome (Table 1).
TABLE 1.
C. elegans candidate antimicrobial genes characterized
| Gene name | Gene ID | Gene description | Expression pattern |
|---|---|---|---|
| lys-1 | Y22F5A.4 | Lysozyme | Intestine, head neurons including one in amphid, two phasmid neurons |
| lys-7 | C02A12.4 | Lysozyme | Intestine, rectal gland cells, head neurons |
| lys-8 | C17G10.5 | Lysozyme | Intestine, head and tail neurons |
| spp-1 | T07C4.4 | Saposin-like | Intestine |
| spp-7 | ZK616.9 | Saposin-like | Pharyngeal muscles (pm7), intestine (posterior > anterior), head neurons |
| abf-1 | C50F2.9 | Homolog of the antibacterial factor ASABF from Ascaris suum | Intestine, weak pharyngeal lumen, head neurons including one in amphid |
| abf-3 | F54B8.5 | Homolog of the antibacterial factor ASABF from Ascaris suum | Intestine, rectal gland cells |
| clec-85 | Y54G2A.6 | C-type lectin | Intestine |
| nlp-29 | B0213.4 | Originally classified as neuropeptide-like protein | Epidermal syncytia but not seam cells |
| F55G11.4 | F55G11.4 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | |
| F08G5.6 | F08G5.6 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | Intestine, head neurons |
| dod-22 | F55G11.5 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | Intestine, rectal gland cells, head neurons |
| F10A3.4 | F10A3.4 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | Intestine, tail neurons |
| F54B11.11 | F54B11.11 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | |
| F55G11.7 | F55G11.7 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | Intestine, rectal gland cells, head neurons including one in amphid, a phasmid neuron |
| K08D8.5 | K08D8.5 | Contains similarity to Pfam domain PF02408 (CUB-like domain) | Intestine, rectal gland cells, head neurons |
To determine how these candidate antimicrobial genes are regulated, we examined the effects of three known immune signaling pathways on expression of these genes: the tir-1-nsy-1/SARM-mitogen-activated protein kinase (MAPK) signaling pathway, the dbl-1 transforming growth factor β (TGF-β) signaling pathway, and the daf-2-daf-16 insulin-like signaling pathway (10). Here we demonstrate that 14 different C. elegans candidate antimicrobial genes are expressed in tissues exposed to the environment, locations where they are well situated to fight off infection. Each of the three known C. elegans immune response pathways regulates a distinct but overlapping set of these genes. We suggest that this unique “antimicrobial fingerprint” of each signaling pathway will be useful for characterizing novel C. elegans innate immunity genes in future studies. We also found that different bacterial pathogens induce expression of unique subsets of antimicrobial genes to fight off infection. The pattern of pathogen-induced antimicrobial production depends on multiple immune signaling pathways. This implies that there are numerous, highly specific pathogen receptor mechanisms present in C. elegans that have yet to be identified.
MATERIALS AND METHODS
Construction of candidate antimicrobial::gfp fusion strains.
Promoter::gfp fusions were engineered using the PCR fusion technique described by Hobert (19) using primers listed in Table S1 in the supplemental material. Briefly, primer 1 and a primer that was the reverse complement of primer 3 were used to amplify the promoter for the individual antimicrobial genes from genomic DNA. This generated promoter DNA fused to a small piece of gfp. Likewise, primer 3 and a primer unique to gfp (5′-CCACTGAGCCTCAAACCCAAACCTTCTTCCG-3′) were used to amplify gfp from plasmid pPD95.67 (Addgene). This product contained gfp with a portion of the unique promoter at the 5′ end. These DNA products were pooled, and the complete promoter::gfp fusions were amplified by PCR using primer 2 and a second gfp primer (5′-CTTTCTTGCATCGTGCTCATCAATACTTGTG-3′). The identity of each promoter fusion was verified by DNA sequencing. In general, gfp was fused in frame directly downstream of the second or third codon of each gene, except for the abf-1 fusion. Unlike the other candidate antimicrobial genes, abf-1 has a large intron that we hypothesized could contain a regulatory sequence, so gfp was fused in frame to the exon shortly after the large intron. All 14 gfp fusions were injected into pha-1(e2123) animals at a concentration of 50 ng/μl using PBX (pha-1+) as a coinjection marker (16). Stable transgenic lines were selectively maintained at 23°C. A minimum of two independent lines were examined for each fusion.
GFP expression patterns were visualized using differential interference contrast optics and fluorescence microscopy. To quantitate total nematode fluorescence, the COPAS Biosort (Union Biometrica) was used. All 14 gfp strains expressed green fluorescent protein (GFP) that was visible by microscopy; however, only nine strains were bright enough to score reliably using the COPAS Biosort. Thus, GFP localization data are presented for all 14 GFP fusions, but COPAS Biosort data are only presented for the nine brightest gfp fusions. As a control for the RNA interference (RNAi) experiments, a gfp fusion under the control of the non-immune-regulated scm promoter was used (S. Alper and C. Kenyon, unpublished data). To determine whether the gfp fusions were expressed in chemosensory neurons, the animals harboring the gfp fusions were stained by dye filling using DiI, which labels some chemosensory neurons (18). In cases where previous expression data were available (either as a gfp fusion or in situ data), our expression data were consistent with those data (7, 24, 32; http://www.wormbase.org).
RNAi of antimicrobial::gfp nematodes.
RNAi was performed in liquid culture in a 96-well format, largely as described previously (2). Briefly, frozen stocks of E. coli RNAi bacteria were inoculated into LB medium supplemented with 80 μg/ml ampicillin plus 10 μg/ml tetracycline. After incubation overnight at 37°C, double-stranded RNA (dsRNA) production was induced by the addition of 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration). After four hours, the bacteria were recovered by centrifugation and suspended in 1/4 of the volume of nematode growth (NG) medium supplemented with 80 μg/ml ampicillin and 2 mM IPTG. This medium (50 μl) was added to the wells in a 96-well plate. Nematode eggs (10 μl) isolated by bleaching (100 to 200 eggs) (59) were then added to the wells. Plates were covered with Breathe Easy film (USA Scientific) and assayed three days later on the COPAS Biosort. The control RNAi strain and the daf-2(RNAi) strain were from reference 9; all other RNAi strains were from MRC Geneservice (23). The identities of the bacterial RNAi constructs were verified by DNA sequencing.
Fluorescence was assayed using the COPAS Biosort (Union Biometrica), which reports time of flight (nematode length) and green fluorescence (GFP-induced fluorescence) for each individual animal. Three types of data analysis were performed after an initial filtering of the data to remove obvious outliers. First, overlay plots of fluorescence versus time of flight were generated to visualize expression of test RNAi-treated animals compared with control RNAi-treated animals (as depicted in Fig. 2A). Second, using boxplots as depicted in Fig. 2C, fluorescence was compared between animals of similar sizes (typically measuring 200 to 400 in arbitrary COPAS Biosort time-of-flight units). Third, mean expression values as a percent of control mean RNAi bacteria treatment were calculated for each sample, as depicted in Fig. 3. The scripts used for data analysis are available upon request.
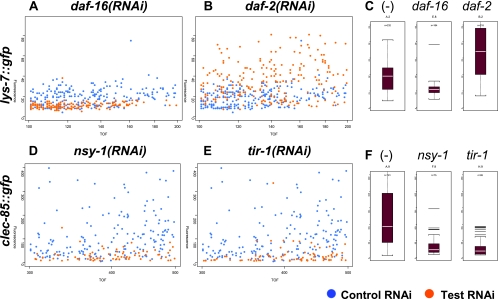
FIG. 2.
Use of the COPAS Biosort to assay changes in antimicrobial::gfp expression. Nematodes harboring either the lys-7::gfp fusion (panels A to C) or the clec-85::gfp fusion (panels D to F) were treated with the indicated dsRNA as described in Materials and Methods. Panels A, B, D, and E are overlay graphs comparing the indicated RNAi test treatment (orange dots) to control-treated animals (blue dots). Each dot represents a single animal. The x axis represents the time of flight (TOF), or length of each animal in arbitrary units, and the y axis represents total fluorescence in arbitrary units. Panels C and F are boxplots, displaying the median fluorescence (white horizontal bar) and the 25th and 75th percentiles of fluorescence (lower and upper limits of each boxplot, respectively). (-) indicates a control RNAi treatment.
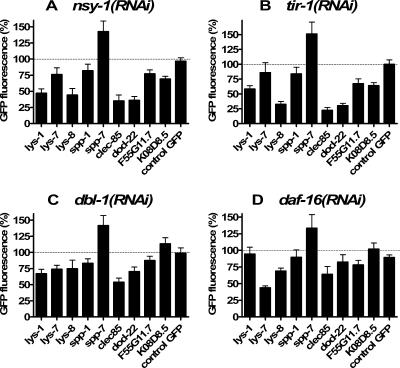
FIG. 3.
Use of RNAi to investigate the role of known immune signaling pathways in the regulation of antimicrobial gene expression. Nine different antimicrobial::gfp strains and one control::gfp strain were treated with one of the four indicated RNAi bacteria, and fluorescence was measured using the COPAS Biosort. In parallel, these gfp strains were treated with a control bacterial strain that was expected to have no effect on gfp expression. Mean fluorescence for each strain and each treatment was then calculated and normalized relative to the control RNAi bacteria treatment. GFP fluorescence is plotted as the percentage of this control treatment. Each assay was performed a minimum of four times.
Real-time RT-PCR to measure antimicrobial RNA production in mutant nematodes.
Nematodes were maintained as described previously (59). Strains used in real-time reverse transcriptase (RT)-PCR experiments were N2 (wild type), CF1038 daf-16(mu86) I, VC390 nsy-1(ok593) II, AU3 nsy-1(ag3) II, RB1085 tir-1(ok1052) III, LT121 dbl-1(wk70) V, and NU3 dbl-1(nk3) V (27, 30, 37, 51). The strains were grown on high-growth plates, and eggs were isolated by bleaching (59). Eggs were then transferred to T-175 flasks containing 10 ml NG medium supplemented with 2 mM IPTG, 80 μg/ml ampicillin, and the control E. coli RNAi strain as a food source. After growing for two days at 23°C, nematodes were harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C. RNA was isolated from the nematodes as described previously (42). Real-time RT-PCR was performed on an ABI 7900 instrument using a QIAGEN Quantitect real-time RT-PCR kit according to the manufacturer's instructions. Primers used for the PCR are listed in Table S1 in the supplemental material. The total RNA concentration was normalized using mlc-1 (myosin light chain) expression (primers shown in Table S1 in the supplemental material). Similar data were observed when act-1 (actin) was used for RNA normalization (data not shown). Antimicrobial gene expression in the mutant nematodes was compared to wild-type N2 nematodes as a reference using the ddCt method. Each strain was analyzed in triplicate.
Real-time RT-PCR to measure antimicrobial RNA production in nematodes exposed to pathogens.
CF512 [fer-15(b26) II; fem-1(hc17) IV] (12) eggs were isolated by bleaching and then plated on NG medium plates seeded with E. coli strain OP50. Eggs were allowed to hatch and were subsequently grown for two days at 25°C, a temperature at which these nematodes are sterile. Nematodes were then collected in M9 medium, washed with M9 supplemented with 10 μg/ml tetracycline, washed several times with M9 without supplement, and then plated. In the gram-negative experiments, synchronized CF512 animals were plated onto NG plates containing either E. coli strain OP50, S. marcescens strain IGX2 (11, 28), or P. aeruginosa strain PA14 (31, 44, 52). For the gram-positive experiments, the synchronized CF512 animals were plated onto brain heart infusion plates containing either the B. subtilis strain PY79 (60) or the S. aureus strain NCTC 8325 (22, 49) (except for the control experiment shown in Fig. 5C, in which S. aureus was grown on NG medium plates). After the indicated incubation (24 h in Fig. 5 or 12, 24, 36, or 48 h in Fig. S1 in the supplemental material), nematodes were processed and RNA production was measured by real-time RT-PCR as described above for the mutants. In these experiments, two additional duf-141 family members were examined by real-time RT-PCR that were not investigated in our GFP experiments.
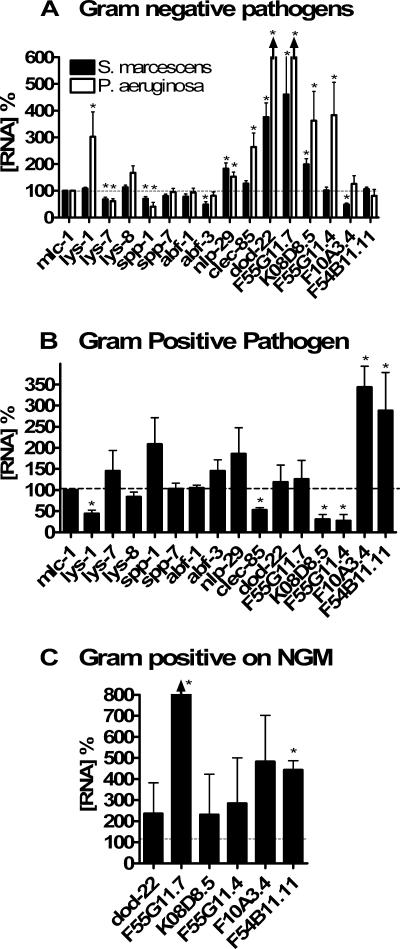
FIG. 5.
Different pathogens induce the expression of different antimicrobial genes in C. elegans. Nematodes were incubated in the presence of different bacteria as described in Materials and Methods. RNA was prepared, and antimicrobial gene expression was assayed by real-time RT-PCR using mlc-1 to normalize RNA concentration. Expression of the antimicrobial genes on gram-negative pathogens was normalized relative to E. coli. Expression of antimicrobials on the gram-positive pathogen was normalized relative to B. subtilis (B) or E. coli (C). Expression levels that were significantly different from control (P < 0.05) are indicated with an asterisk (P values were calculated using one-sample t tests). lys-1, clec-85, and F55G11.7 were the only genes whose expression was significantly different between the two gram-negative treatments (P < 0.05; t test). As indicated by the arrowheads in panel A, expression of dod-22 (775%) and F55G11.7 (>10,000%) in the presence of P. aeruginosa was off scale in this figure.
For the experiment in which mutant nematodes were exposed to pathogenic bacteria (see Fig. 6), exposures were carried out for 24 h on solid media as described in this section and not in liquid media as described in the previous section.
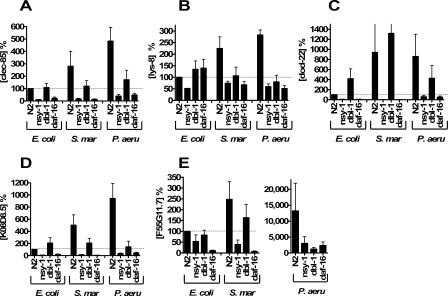
FIG. 6.
Role of immune pathways in regulation of pathogen-induced antimicrobial gene expression. The four indicated nematode strains [N2 (the wild type), nsy-1(ok593), dbl-1(nk3), daf-16(mu86)] were exposed to either E. coli, S. marcescens, or P. aeruginosa, RNA was isolated, and antimicrobial gene expression was monitored using mlc-1 to normalize for RNA concentration. Expression was measured relative to the wild-type N2 strain grown on E. coli. Note the change of scale for P. aeruginosa in panel E. Depicted are the results of three independent experiments.
RESULTS
Choice of candidate antimicrobials for investigation.
To investigate the C. elegans antimicrobial response, we fused the promoters of fourteen putative antimicrobial genes to gfp and generated transgenic lines with these fusions (see Materials and Methods and Table S1 in the supplemental material). The C. elegans genome contains numerous candidate antimicrobial genes, including homologs with established roles in host defense. These include 10 homologs of lysozyme (lys-1 to lys-10), 20 genes encoding saposin-like domains that are also present in NK-lysins and granulysins (spp-1 to spp-20), six genes somewhat similar to vertebrate defensins (abf-1 to abf-6), and a recently identified family of genes similar to nlp-29 (17, 40). We engineered gfp fusions to three lysozymes (lys-1, lys-7, and lys-8) that were previously implicated in innate immunity in C. elegans, because they were either induced by S. marcescens infection (all three) or regulated by the insulin signaling pathway (lys-7 and lys-8) (32, 38). Likewise, we fused gfp to the promoter for spp-1, which is also reported to be regulated by the insulin signaling pathway (38). spp-7, abf-1, and abf-3 were chosen randomly to represent each antimicrobial gene family. We also fused gfp to nlp-29, which was identified as a gene induced by S. marcescens and the fungal pathogen Drechmeria coniospora (7, 32).
NLP-29 and SPP-1 (4, 7) have demonstrated antimicrobial activity. The three lysozyme genes, spp-7, and the two abf genes are homologs of genes with demonstrated antimicrobial function. In contrast, other candidate antimicrobial genes that we have chosen to examine are in families induced by pathogen infection, but their precise antimicrobial function is poorly understood. These include clec-85, a member of the C-type lectin family (32). C. elegans C-type lectins are hypothesized to play a role in pathogen recognition or clearance (40). Another family of genes whose precise function is unknown but which is induced by infection encodes a protein family containing Pfam domain PF02408, formerly known as the domain of unknown function number 141 (we refer to this family as the duf-141 family). Several members of this family are also reported to be regulated by the insulin signaling pathway (32, 38, 41). We fused gfp to the promoter for five of these genes. For simplicity, we refer to all 14 gfp fusions as antimicrobial::gfp fusions, although the precise functions of members of the duf-141 family are still uncertain.
C. elegans antimicrobial genes are expressed in tissues exposed to the environment.
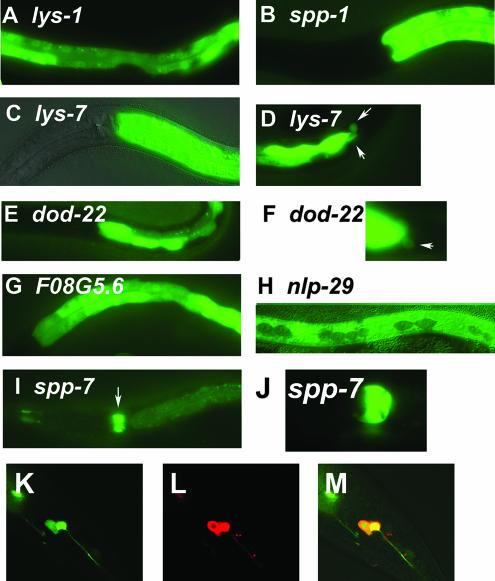
In the presence of the standard laboratory E. coli strain OP50 (59), thirteen of these fourteen antimicrobial::gfp fusions were expressed in the C. elegans intestine, a position where they would come into contact with ingested pathogens (Fig. 1A to G and I and Table 1). The pattern of this expression was largely uniform throughout the intestine, except in the case of spp-7, in which gfp expression was much stronger in the posterior intestine than in the anterior. spp-7 was also strongly expressed in several cells in the pharynx (primarily in the pm7 pharyngeal muscle cells), another organ in the digestive tract (Fig. 1I and J and Table 1). Although the intensity of gfp expression in the digestive tract was by far the strongest expression in all these antimicrobial::gfp strains, expression was also observed in other cells. This includes some head and tail neurons, including chemosensory neurons that are exposed to the environment, and the rectal gland cells, which are thought to secrete substances into the digestive tract (http://www.wormatlas.org) (Fig. 1D, F, and K to M and Table 1).
FIG. 1.
C. elegans candidate antimicrobial genes are expressed in tissues exposed to the environment. Panels A to J depict representative fluorescence micrographs of the indicated GFP fusion-bearing strains (panels C and H are overlays of fluorescence and Nomarski micrographs). Strong intestinal gfp expression is observed in panels A (mid-body view), B, C, E, and G (anterior to mid-body view), and D and F (posterior view). Weak intestinal gfp expression and strong pharyngeal gfp expression are observed in panel I. Panel J is a close-up view of the pharyngeal expression in panel I. White arrows point to rectal gland gfp expression in panels D and F or to pharyngeal expression in panel I. Panels K, L, and M are confocal microscopy images of the posterior part of the same nematode expressing lys-1::gfp. Panel K depicts gfp expression (green), panel L depicts animals filled with the dye DiI, which labels some chemosensory neurons (red), and panel M depicts the overlap (yellow), demonstrating that this gfp fusion is expressed in two chemosensory phasmid neurons. Anterior is to the left and ventral is down in all images. A complete summary of the gfp expression data for all 14 antimicrobial::gfp fusions is presented in Table 1.
nlp-29 was the only gene that was not visibly expressed in the intestine. Instead, as reported previously (7), it was expressed in the epidermal syncytia (but not the epidermal seam cells) (Fig. 1H and Table 1), the outermost cell layer in the nematode.
C. elegans antimicrobial genes are regulated by known immune signaling pathways.
Interestingly, all the antimicrobial::gfp fusions that we generated were expressed in the presence of the nonpathogenic strain of E. coli, OP50, often quite strongly. This raised the possibility that although OP50 is normally not toxic to C. elegans, it might be inducing an immune response. To test this hypothesis and to investigate the regulation of antimicrobial production in the nematode, we inhibited known immune response pathways using RNAi and examined the effect on antimicrobial production. To do this, we used E. coli that expressed dsRNA corresponding to genes in three known C. elegans immune signaling pathways: tir-1 and nsy-1 in the SARM-MAPKKK pathway, dbl-1 in the TGF-β pathway, and daf-2 and daf-16 in the insulin signaling pathway. tir-1, nsy-1, dbl-1, and daf-16 mutant animals are all more susceptible to infection and are predicted to have a weaker antimicrobial response. daf-2 mutants, on the other hand, are more resistant to pathogens and are expected to have a stronger antimicrobial response. E. coli expressing these dsRNAs were fed to nematodes that carried the antimicrobial::gfp fusions, and fluorescence was measured to determine the effect on antimicrobial::gfp production.
As a control, E. coli expressing daf-16 dsRNA were fed to animals carrying the lys-7::gfp fusion. Previous microarray data indicated that daf-16+ is required for expression of lys-7 (38). In keeping with this result, we found that inhibition of daf-16 by RNAi inhibited lys-7::gfp expression (Fig. 2A and C). In daf-2 mutant animals, daf-16 is constitutively active and lys-7 is overexpressed (38). When daf-2 was inhibited by RNAi, lys-7::gfp expression increased (Fig. 2B and C). Thus, as expected, we can observe opposing effects of daf-2 and daf-16 on lys-7 expression.
Inhibition of tir-1 by RNAi inhibits the expression of two antimicrobial genes, nlp-29 and nlp-31 (7). When we used RNAi to inhibit tir-1 or nsy-1 in animals bearing the clec-85::gfp fusion, we found that expression of clec-85::gfp was strongly inhibited (Fig. 2D to F). Thus, wild-type tir-1 and nsy-1 are required for proper expression of this candidate antimicrobial gene, in keeping with their proposed role in host defense.
We next examined the effect of inhibition of each of the three different immune signaling pathways on nine of our antimicrobial gene fusions and on a control gfp fused to a promoter not involved in immunity. The other five antimicrobial::gfp fusions, while bright enough to assay by fluorescence microscopy, were too weak to assay on the COPAS Biosort used to quantitate nematode fluorescence and were therefore not used in this assay. Inhibition of each immune pathway affected a different subset of antimicrobial genes (Fig. 3). For example, inhibition of nsy-1 by RNAi led to a strong reduction in expression of lys-1, lys-8, clec-85, and dod-22, as assayed by reduced GFP fluorescence, but had a much more moderate or no effect on expression of the other gfp fusions. Interestingly, inhibition of tir-1, which is thought to act upstream of nsy-1 (5, 29), had an almost identical effect (compare Fig. 3A to B), causing strong inhibition of lys-1, lys-8, clec-85, and dod-22 expression and a much more moderate effect on the other genes. We also found that nlp-29::gfp fluorescence exhibited a moderate decrease when either nsy-1 or tir-1 were inhibited (data not shown), consistent with previous work (7). However, the level of this fluorescence was very close to background on the COPAS Biosort.
Inhibition of dbl-1 by RNAi had a different effect on antimicrobial gene expression. clec-85 expression was strongly decreased when dbl-1 was inhibited by RNAi (Fig. 3C). Inhibition of dbl-1 by RNAi also caused a much more moderate decrease in expression of the three lysozyme genes and dod-22 (Fig. 3C). Thus, the pattern of genes regulated by dlb-1 overlaps with but is distinct from those regulated by nsy-1 and tir-1.
Inhibition of daf-16 by RNAi had a very different effect, causing the strongest inhibition of lys-7::gfp expression and weaker inhibition of several other genes including lys-8 and clec-85 (Fig. 3D).
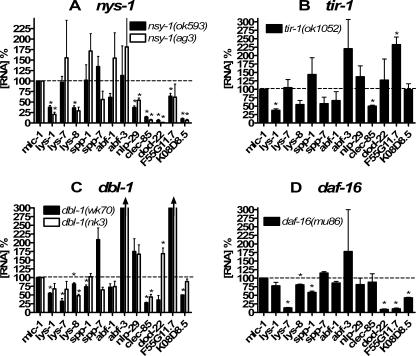
To verify the RNAi data generated with our antimicrobial::gfp fusions, we examined RNA production in available mutant animals using real-time RT-PCR. Wild-type and mutant animals were grown in liquid culture in the presence of E. coli, and RNA was isolated. Real-time RT-PCR was then used to examine antimicrobial RNA levels in different wild-type or mutant animals. In general, antimicrobial RNA production was decreased in mutant nematodes in a pattern analogous to that seen in the RNAi experiments using the antimicrobial::gfp animals (Fig. 4). For lys-1, lys-8, clec-85, nlp-29, and the duf-141 genes dod-22 and K08D8.5, RNA levels were decreased in animals harboring mutations in either of two alleles of nsy-1. Expression of the two defensin-like genes abf-1 and abf-3 and the two saposin-containing genes spp-1and spp-7 was not decreased. The pattern of genes affected by the tir-1 mutation was also similar to the pattern of those regulated by tir-1(RNAi), with lys-1, lys-8, and clec-85 expression all strongly reduced in the tir-1 mutant background.
FIG. 4.
Use of real-time RT-PCR to measure antimicrobial RNA in immune pathway mutant nematodes. The indicated nematode strains were prepared and collected. RNA was purified from each strain, and antimicrobial gene expression was assayed by real-time RT-PCR using mlc-1 to normalize RNA concentration. Expression was measured relative to the wild-type strain N2, which was grown in parallel. Depicted on the graph are the means of three independent experiments. Panel A depicts two different mutant variants of nsy-1 animals compared to the wild type. Panel B depicts tir-1, panel C dbl-1, and panel D daf-16. Expression levels that were significantly different from the wild type (P < 0.05) are indicated with an asterisk (P values were calculated using one-sample t tests). As indicated by the arrowheads in panel C, expression of abf-3 (432% and 329% in wk70 and nk3 alleles, respectively) and F55G11.7 (311% and 565% in wk70 and nk3 alleles, respectively) was off scale in this figure.
Consistent with the RNAi data, clec-85 RNA levels were strongly decreased in nematodes harboring either of two mutations in the TGF-β homolog dbl-1 (Fig. 4C). All three lysozyme RNAs also exhibited a very moderate decrease in dbl-1 mutant animals, similar to that observed with the dlb-1(RNAi) data; the other antimicrobials tested did not exhibit a consistent decrease in the two different alleles of dbl-1 mutant animals tested (Fig. 4C).
The daf-16 mutant animals exhibited a strong decrease in lys-7 expression, which was consistent with the RNAi data, as well as decreased expression of several other genes, particularly the three duf-141 family genes (Fig. 4D). lys-8 and spp-1 RNA levels exhibited much more moderate decreases in daf-16 mutant animals. Interestingly, several duf-141 family members that were positively regulated by daf-16+ in both assays were reported to be negatively regulated in previous microarray experiments (33, 38, 56). This is likely due to the fact that our experiments were carried out in an otherwise wild-type background where DAF-16 is minimally active, whereas the prior experiments were performed in a daf-2 mutant background in which DAF-16 is hyperactive.
Different pathogens induce expression of different antimicrobial genes.
To explore the effects of different pathogenic bacteria on C. elegans antimicrobial gene expression, we exposed animals to the gram-negative pathogens S. marcescens or P. aeruginosa (using E. coli as a control) and the gram-positive pathogen S. aureus (using B. subtilis as a control) and then examined antimicrobial RNA levels using real-time RT-PCR. Exposure to either S. marcescens or P. aeruginosa resulted in similar but not identical patterns of antimicrobial gene induction (Fig. 5A). Some of the strongest genes induced by both gram-negative bacteria were members of the duf-141 gene family, in particular dod-22, F55G11.7, and K08D8.5. Expression of nlp-29 and clec-85 was also induced by both pathogens, although more weakly. In contrast, the expression of several genes, including lys-7 and spp-1, was actually stronger on E. coli than on the pathogens. The two gram-negative pathogens did not have identical effects, since several other genes such as lys-1 and the duf-141 gene F55G11.4 were induced by P. aeruginosa but not S. marcescens. In fact, expression of the duf-141 gene F10A3.4 was moderately induced on P. aeruginosa but repressed on S. marcescens. Experiments in which gene inductions by pathogen were monitored over time suggest that the differences in gene induction by P. aeruginosa and S. marcescens are largely due to a difference in magnitude of the response, although different temporal patterns of induction were observed as well (see Fig. S1 in the supplemental material).
In contrast, the gram-positive pathogen S. aureus induced expression of a different set of genes (Fig. 5B). Again, the strongest effects were on the duf-141 gene family, although the duf-141 family members induced by S. aureus (F10A3.4 and F54B11.11) were distinct from those induced by the gram-negative pathogens. Several other genes exhibited weak induction by S. aureus (spp-1, lys-7, abf-3, and nlp-29) and quite a few were down-regulated on S. aureus, including several genes that were induced by gram-negative bacteria.
Because gram-positive infection experiments of C. elegans are typically performed on richer media than are gram-negative infection experiments, we also examined the expression of the duf-141 gene family in nematodes exposed to S. aureus grown on NG medium (the less-rich medium used to grow gram-negative pathogens) and used E. coli as a control. The pattern of duf-141 genes induced under these nonstandard gram-positive infection conditions was similar but not identical to that observed when S. aureus was compared to B. subtilis on rich medium but was distinct from the pattern of induction caused by gram-negative pathogens (Fig. 5C).
The pathogen-mediated induction of C. elegans antimicrobial genes is regulated by known immune signaling pathways.
To determine the effect of known immune signaling pathways on pathogen-mediated antimicrobial gene expression, we examined the effects of three different gram-negative bacteria (E. coli, S. marcescens, and P. aeruginosa) on nematodes harboring mutations in either nsy-1, dbl-1, or daf-16. For this analysis, we used real-time RT-PCR to monitor expression of the three duf-141 genes that were most strongly induced by gram-negative pathogens; we also examined pathogen-induced expression of the two genes that were regulated by all three immune signaling pathways in the presence of E. coli: clec-85 and lys-8. Interestingly, the effects of daf-16 and dbl-1 were more moderate in the presence of E. coli when the experiment was carried out on solid media (Fig. 6A and B) compared to liquid media (Fig. 4C and D), but all three pathways were required for full pathogen-mediated induction of clec-85 and lys-8 expression (Fig. 6A and B). Thus, all three pathways modulate clec-85 and lys-8 expression, regardless of the bacterial inducer. The duf-141 gene dod-22 was strongly regulated by nsy-1 and daf-16 but not dbl-1 when exposed to any of the three gram-negative bacteria (Fig. 6C). Similarly, all three pathways exerted differing effects on the regulation of F55G11.7 and K08D8.5 expression in the presence of different bacteria (Fig. 6D and E). Thus, these three signaling pathways affect not only the “basal” antimicrobial expression in the presence of nonpathogenic E. coli but also the “induced” antimicrobial expression when the nematodes are exposed to either S. marcescens or P. aeruginosa.
DISCUSSION
To more fully understand the C. elegans immune response, we have undertaken a study of a representative set of candidate antimicrobial genes to identify patterns in their regulation. Our results indicate that all the candidate antimicrobial genes examined are expressed in tissues that are potentially exposed to environmental pathogens, in agreement with their proposed function in host defense. The primary route of infection for most C. elegans pathogens is through the intestine (3, 48), and not surprisingly, all but one of the antimicrobials that we examined exhibited expression in the digestive tract (intestine and/or pharynx). The exception was nlp-29, which was expressed in the epidermis. nlp-29 was originally identified because of its role in defense against the fungal pathogen D. coniospora (7). D. coniospora hyphae invade through the C. elegans epidermis; thus, nlp-29 is expressed in a position where it can fight off that infection. Other sites where some of our antimicrobial genes are expressed include cells near the anus and chemosensory neurons, both of which are sites exposed to the environment, and are therefore locations where a defense against pathogens is necessary. Because homologous antimicrobial genes were sometimes expressed in different sets of cells, it is possible that they may serve distinct functions in host defense.
Interestingly, all of the antimicrobial::gfp fusions were expressed in the presence of E. coli, which is usually not pathogenic to C. elegans (although E. coli can be toxic to C. elegans under certain special conditions or late in life) (12, 13, 15). Previous C. elegans studies used DNA microarrays to investigate gene regulation in response to infection and compared nematodes exposed to pathogens with nematodes exposed to nonpathogenic E. coli (7, 32, 41, 47, 56). However, because C. elegans does not usually encounter E. coli, its food source in the laboratory, in its native soil environment, it is possible that E. coli exposure itself could be inducing immune response genes. Consistent with this hypothesis, inhibition of the immune response pathways affected the expression of numerous antimicrobial genes. We also note that inhibition of tir-1 inhibits not only nlp-29 expression in the presence of pathogens but also the “basal” level of expression in the presence of E. coli (7). Moreover, we found that although some antimicrobial genes were induced when exposed to pathogens, some actually exhibited stronger expression in the presence of the nonpathogenic E. coli or B. subtilis control strains. Therefore, it is reasonable to think of E. coli as just one more bacteria to which C. elegans can sense and respond.
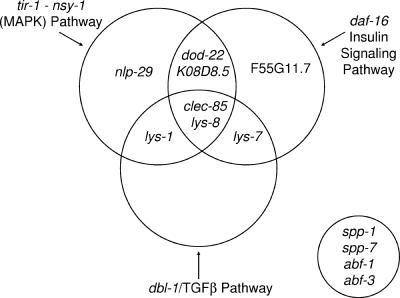
Several immune response signaling pathways have been identified in C. elegans. The best characterized pathway is the MAPK signaling pathway, which affects survival when nematodes are exposed to any of numerous pathogens. Previous data suggested that tir-1 functions through the nsy-1 MAPK pathway (5, 29). Moreover, tir-1 was shown to regulate two antimicrobial peptides, including nlp-29 (7). Here we show that many candidate antimicrobial genes are regulated by tir-1 and nsy-1 (Fig. 7). Notably, this pathway does not regulate all antimicrobial genes. Instead, the two genes in this pathway regulate an identical subset of the antimicrobial genes tested. The unique set of antimicrobial genes regulated by this pathway, or the tir-1 nsy-1 pathway “antimicrobial fingerprint,” is distinct from that regulated by the other immune response pathways tested and may be useful in characterizing novel innate immunity genes and assigning them to known pathways. Consistent with this hypothesis, we have identified several novel genes in a genomic screen for antimicrobial gene regulators that have an “antimicrobial fingerprint” identical to tir-1 and nsy-1 (S. Alper, J. H. Freedman, and D. A. Schwartz, unpublished data).
FIG. 7.
A model for the regulation of antimicrobial gene expression in C. elegans. Depicted in the Venn diagram are the genes regulated by each of the three immune signaling pathways in C. elegans. The data are a summary of the RNAi and real-time RT-PCR data. Genes that lie within two or three circles are regulated by multiple pathways. The four genes in the circle at the lower right were constitutively expressed and were not strongly regulated by any of the immune signaling pathways tested.
The RNAi data using our gfp fusions and the real-time RT-PCR data using mutants showed comparable results. There were a few cases where the antimicrobial expression was decreased more strongly by mutations than by RNAi, such as for the duf-141 gene K08D8.5 when nsy-1 was inhibited. RNAi reduces gene function but may not eliminate it entirely. Perhaps K08D8.5 is less sensitive than other antimicrobial genes to the decrease in nsy-1 activity caused by RNAi. Only when nsy-1 activity is completely absent in mutant animals is K08D8.5 expression reduced. Nevertheless, the agreement between the two assays is remarkably strong.
dbl-1 mutant animals are more susceptible to some pathogens, and several antimicrobial genes are reported to have altered expression in dbl-1 mutant animals, as determined using microarrays (32, 36). The set of genes regulated by dbl-1 in our study overlaps with but is distinct from those regulated by the tir-1 nsy-1 pathway (Fig. 7).
The daf-2 pathway regulates the rate of aging in C. elegans, with daf-2 mutant animals living longer than wild-type animals (26). The long life span of these daf-2 mutant animals depends on the activity of the downstream transcription factor daf-16 (26). daf-2 mutant animals are resistant to a wide variety of stresses, including pathogens, and this stress/pathogen resistance is also dependent on daf-16 (14). We identified several antimicrobial genes that were regulated by daf-16 (Fig. 7). Again, although there is overlap in the genes regulated by this signaling pathway and the two other pathways, the pattern of antimicrobial genes regulated by the insulin signaling pathway is still unique.
We find that a distinct set of genes is also induced by different pathogenic bacteria. The two gram-negative pathogens induced a similar but not identical set of antimicrobial genes. In contrast, the gram-positive pathogen tested induced a completely different set of genes. This pathogen-mediated gene induction required the same signaling pathways that were required for the “basal” antimicrobial gene expression present when nematodes were grown on E. coli. Not only were different genes induced by all three pathogens, but the genes induced cannot be assigned to any one genetic signaling pathway. Because no single pathway is activated by any given pathogen, this suggests that C. elegans can distinguish between different pathogens and activate multiple pathways that interact at some level to turn on the appropriate set of antimicrobial genes. In higher organisms, pathogen recognition receptors (PRRs) are thought to recognize a unique set of pathogenic compounds (termed pathogen-associated molecular patterns, or PAMPs) (34). The identities of the specific PAMPs recognized by C. elegans remain unclear, although Salmonella lipopolysaccharide has been identified as one potential PAMP (1). The identity of the C. elegans PRRs is still unclear, although our data as well as previously published microarray data (7, 21, 32, 41, 47, 56) suggest the existence of numerous PRRs that can uniquely identify different pathogens to induce a differential response. The most obvious candidate, tol-1, the only Toll-like receptor in C. elegans, plays a role in a pathogen avoidance response, but it does not affect pathogen susceptibility (43). Although the identity of the PRRs in C. elegans remains elusive, the complicated, highly specific response that we observed suggests that several exist. It remains to be seen whether these PRRs are similar to those used in higher organisms. However, the downstream immune signaling components identified in C. elegans so far are homologous to immune genes that function in higher organisms. It therefore seems likely that novel genes identified in C. elegans will play a role in immunity in higher organisms as well.
Although we have only examined several members of selected antimicrobial gene classes, we can draw some general conclusions based on our results and others reported in the literature. Different antimicrobial gene families exhibit distinct patterns of regulation in response to different pathogens. For example, we showed that different members of the duf-141 gene family are induced by each of the three pathogens and that this pattern is distinct from those family members induced by Microbacterium nematophilum (7, 32, 41, 47, 56). Likewise, different pathogens induce different members of the lysozyme family: lys-7 is induced by S. aureus, lys-1 and lys-8 are very weakly induced by S. marcescens (32) and more strongly induced by P. aeruginosa (as is lys-2) (47), and lys-3, lys-7, and lys-8 are induced by M. nematophilum (41). Different members of the C-type lectin family and the nlp-29 family are also induced by different pathogens (7, 32, 41, 47, 56). In contrast, the two defensin-like genes and the two saposin genes that we studied exhibited less change in response to pathogens and were not strongly regulated by any of the three immune signaling pathways, suggesting that their expression could be constitutive. As observed in higher organisms, we see some antimicrobial genes with inducible expression and some without.
We have identified many novel targets in each of the three signaling pathways tested. We note that two genes, clec-85 and lys-8, are regulated by all three signaling pathways. Therefore, clec-85::gfp and lys-8::gfp will be useful screening tools to identify novel innate immunity signaling genes or PRRs in C. elegans.
Innate immunity has traditionally been described as a relatively nonspecific response (in contrast to the highly specific adaptive immune response). More recently, a large body of work has demonstrated that far more specificity exists in the innate response than originally believed, and this response is critical to many aspects of host defense (58). Strikingly, even in the “more modest” C. elegans innate immunity model system, which lacks many of the cellular mechanisms of defense present in higher organisms, we can observe this complexity and specificity in response at the level of antimicrobial gene regulation.
Supplementary Material
Acknowledgments
This research was supported in part by the intramural research program of the NIH, National Institute of Environmental Health Sciences, National Heart, Lung, and Blood Institute, and the National Toxicology Program.
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
We thank Joy Alcedo, Cynthia Kenyon, Adam Driks, Emily Troemel, Fred Ausubel, and the NARSA repository for nematode and bacterial strains. Thanks to Julie Rice, Dan Snyder, and Windy Boyd for assistance with the COPAS Biosort, Brooke Baker for assistance with the confocal microscope, Dave Sherwood for use of his microscope, and Javier Apfeld for critical reading of the manuscript.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aballay, A., E. Drenkard, L. R. Hilbun, and F. M. Ausubel. 2003. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Ahringer, J. (ed.). 6 April 2006, posting date. Reverse genetics. In The C. elegans Research Community (ed.), WormBook. http://www.wormbook.org.
- 3.Alegado, R. A., M. C. Campbell, W. C. Chen, S. S. Slutz, and M. W. Tan. 2003. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell. Microbiol. 5:435-444. [DOI] [PubMed] [Google Scholar]
- 4.Banyai, L., and L. Patthy. 1998. Amoebapore homologs of Caenorhabditis elegans. Biochim. Biophys. Acta 1429:259-264. [DOI] [PubMed] [Google Scholar]
- 5.Chuang, C. F., and C. I. Bargmann. 2005. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 19:270-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook, D. N., D. S. Pisetsky, and D. A. Schwartz. 2004. Toll-like receptors in the pathogenesis of human disease. Nat. Immunol. 5:975-979. [DOI] [PubMed] [Google Scholar]
- 7.Couillault, C., N. Pujol, J. Reboul, L. Sabatier, J. F. Guichou, Y. Kohara, and J. J. Ewbank. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5:488-494. [DOI] [PubMed] [Google Scholar]
- 8.Dhakal, B. K., W. Lee, Y. R. Kim, H. E. Choy, J. Ahnn, and J. H. Rhee. 2006. Caenorhabditis elegans as a simple model host for Vibrio vulnificus infection. Biochem. Biophys. Res. Commun. 10:54. [DOI] [PubMed] [Google Scholar]
- 9.Dillin, A., D. K. Crawford, and C. Kenyon. 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298:830-834. [DOI] [PubMed] [Google Scholar]
- 10.Ewbank, J. J. 23 January 2006, posting date. Signaling in the immune response. In The C. elegans Research Community (ed.), WormBook. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 11.Flyg, C., K. Kenne, and H. G. Boman. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J. Gen. Microbiol. 120:173-181. [DOI] [PubMed] [Google Scholar]
- 12.Garigan, D., A. L. Hsu, A. G. Fraser, R. S. Kamath, J. Ahringer, and C. Kenyon. 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161:1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garsin, D. A., J. M. Villanueva, J. Begun, D. H. Kim, C. D. Sifri, S. B. Calderwood, G. Ruvkun, and F. M. Ausubel. 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300:1921. [DOI] [PubMed] [Google Scholar]
- 15.Gems, D., and D. L. Riddle. 2000. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics 154:1597-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granato, M., H. Schnabel, and R. Schnabel. 1994. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22:1762-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravato-Nobre, M. J., and J. Hodgkin. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7:741-751. [DOI] [PubMed] [Google Scholar]
- 18.Hedgecock, E. M., J. G. Culotti, J. N. Thomson, and L. A. Perkins. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111:158-170. [DOI] [PubMed] [Google Scholar]
- 19.Hobert, O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32:728-730. [DOI] [PubMed] [Google Scholar]
- 20.Hoebe, K., E. Janssen, and B. Beutler. 2004. The interface between innate and adaptive immunity. Nat. Immunol. 5:971-974. [DOI] [PubMed] [Google Scholar]
- 21.Huffman, D. L., L. Abrami, R. Sasik, J. Corbeil, F. G. van der Goot, and R. V. Aroian. 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl. Acad. Sci. USA 101:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iandolo, J. J. 2000. Genetic and physical map of the chromosome of Staphylococus aureus 8325. Academic Press, Washington, DC.
- 23.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 24.Kato, Y., T. Aizawa, H. Hoshino, K. Kawano, K. Nitta, and H. Zhang. 2002. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann, S. H. E., R. Medzhitov, and S. Gordon (ed.). 2004. The innate immune response to infection. ASM Press, Washington, DC.
- 26.Kenyon, C., J. Chang, E. Gensch, A. Rudner, and R. Tabtiang. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366:461-464. [DOI] [PubMed] [Google Scholar]
- 27.Kim, D. H., R. Feinbaum, G. Alloing, F. E. Emerson, D. A. Garsin, H. Inoue, M. Tanaka-Hino, N. Hisamoto, K. Matsumoto, M. W. Tan, and F. M. Ausubel. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623-626. [DOI] [PubMed] [Google Scholar]
- 28.Kurz, C. L., S. Chauvet, E. Andres, M. Aurouze, I. Vallet, G. P. Michel, M. Uh, J. Celli, A. Filloux, S. De Bentzmann, I. Steinmetz, J. A. Hoffmann, B. B. Finlay, J. P. Gorvel, D. Ferrandon, and J. J. Ewbank. 2003. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberati, N. T., K. A. Fitzgerald, D. H. Kim, R. Feinbaum, D. T. Golenbock, and F. M. Ausubel. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 101:6593-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, K., J. B. Dorman, A. Rodan, and C. Kenyon. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319-1322. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 32.Mallo, G. V., C. L. Kurz, C. Couillault, N. Pujol, S. Granjeaud, Y. Kohara, and J. J. Ewbank. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12:1209-1214. [DOI] [PubMed] [Google Scholar]
- 33.McElwee, J., K. Bubb, and J. H. Thomas. 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2:111-121. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov, R., and C. Janeway, Jr. 2000. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173:89-97. [DOI] [PubMed] [Google Scholar]
- 35.Millet, A. C., and J. J. Ewbank. 2004. Immunity in Caenorhabditis elegans. Curr. Opin. Immunol. 16:4-9. [DOI] [PubMed] [Google Scholar]
- 36.Mochii, M., S. Yoshida, K. Morita, Y. Kohara, and N. Ueno. 1999. Identification of transforming growth factor-beta-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl. Acad. Sci. USA 96:15020-15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, K., K. L. Chow, and N. Ueno. 1999. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126:1337-1347. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath, J. Ahringer, H. Li, and C. Kenyon. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277-283. [DOI] [PubMed] [Google Scholar]
- 39.Mylonakis, E., A. Idnurm, R. Moreno, J. El Khoury, J. B. Rottman, F. M. Ausubel, J. Heitman, and S. B. Calderwood. 2004. Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals. Mol. Microbiol. 54:407-419. [DOI] [PubMed] [Google Scholar]
- 40.Nicholas, H. R., and J. Hodgkin. 2004. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 41:479-493. [DOI] [PubMed] [Google Scholar]
- 41.O'Rourke, D., D. Baban, M. Demidova, R. Mott, and J. Hodgkin. 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16:1005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portman, D. S. 20 January 2006, posting date. Profiling C. elegans gene expression with DNA microarrays. In The C. elegans Research Community (ed.), WormBook. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 43.Pujol, N., E. M. Link, L. X. Liu, C. L. Kurz, G. Alloing, M. W. Tan, K. P. Ray, R. Solari, C. D. Johnson, and J. J. Ewbank. 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11:809-821. [DOI] [PubMed] [Google Scholar]
- 44.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 45.Schulenburg, H., and J. J. Ewbank. 2004. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulenburg, H., C. L. Kurz, and J. J. Ewbank. 2004. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198:36-58. [DOI] [PubMed] [Google Scholar]
- 47.Shapira, M., B. J. Hamlin, J. Rong, K. Chen, M. Ronen, and M. W. Tan. 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103:14086-14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119-127. [DOI] [PubMed] [Google Scholar]
- 49.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, Y., M. D. Yandell, P. J. Roy, S. Krishna, C. Savage-Dunn, R. M. Ross, R. W. Padgett, and W. B. Wood. 1999. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126:241-250. [DOI] [PubMed] [Google Scholar]
- 52.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, R. J., J. Breger, A. Idnurm, K. J. Gerik, J. K. Lodge, J. Heitman, S. B. Calderwood, and E. Mylonakis. 2005. Cryptococcus neoformans gene involved in mammalian pathogenesis identified by a Caenorhabditis elegans progeny-based approach. Infect. Immun. 73:8219-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 56.Troemel, E. R., S. W. Chu, V. Reinke, S. S. Lee, F. M. Ausubel, and D. H. Kim. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaitkevicius, K., B. Lindmark, G. Ou, T. Song, C. Toma, M. Iwanaga, J. Zhu, A. Andersson, M. L. Hammarstrom, S. Tuck, and S. N. Wai. 2006. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. USA 103:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vivier, E., and B. Malissen. 2005. Innate and adaptive immunity: specificities and signaling hierarchies revisited. Nat. Immunol. 6:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood, W. B. 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.