Abstract
Single-strand breaks are the commonest lesions arising in cells, and defects in their repair are implicated in neurodegenerative disease. One of the earliest events during single-strand break repair (SSBR) is the rapid synthesis of poly(ADP-ribose) (PAR) by poly(ADP-ribose) polymerase (PARP), followed by its rapid degradation by poly(ADP-ribose) glycohydrolase (PARG). While the synthesis of poly(ADP-ribose) is important for rapid rates of chromosomal SSBR, the relative importance of poly(ADP-ribose) polymerase 1 (PARP-1) and PARP-2 and of the subsequent degradation of PAR by PARG is unclear. Here we have quantified SSBR rates in human A549 cells depleted of PARP-1, PARP-2, and PARG, both separately and in combination. We report that whereas PARP-1 is critical for rapid global rates of SSBR in human A549 cells, depletion of PARP-2 has only a minor impact, even in the presence of depleted levels of PARP-1. Moreover, we identify PARG as a novel and critical component of SSBR that accelerates this process in concert with PARP-1.
Single-strand breaks (SSBs) are the commonest type of lesion arising in cells and can arise from direct attack of deoxyribose, as abortive intermediates of topoisomerase 1 activity, or as normal intermediates of base excision repair. One of the earliest responses to DNA strand breakage is the induction of poly(ADP-ribose) (PAR) synthesis (reviewed in references 17 and 35). Poly(ADP-ribose) polymerase 1 (PARP-1) is an abundant and stable component of chromatin and is the major source of PAR synthesis following DNA strand breakage (27, 40). PARP-1 rapidly binds to and is activated by DNA single- and double-strand breaks, resulting in covalent modification of itself and to a lesser extent other target proteins with long chains of PAR (4, 5, 15, 41, 42). The binding and activity of PARP-1 at DNA breaks are very transient because the ribosylated enzyme dissociates from DNA through charge repulsion (24, 60). Subsequently, a second DNA damage-activated PARP was identified in human cells and was called PARP-2 (1, 30). PARP-2 has 18-fold lower activity than PARP-1 but can support up to 25% of normal levels of DNA damage-induced PAR synthesis in the absence of PARP-1 (1, 49). While PARP-1 is the primary source of global PAR synthesis following DNA strand breakage, it is possible that PARP-2 fulfils an overlapping or backup role. In support of this, mice lacking either PARP-1 or PARP-2 are viable, but mice lacking both enzymes are not (38). The presence of high levels of PAR in cells following DNA strand breakage is very transient because the polymer is rapidly degraded by poly(ADP-ribose) glycohydrolase (PARG). Consequently, proteins that become ribosylated following DNA strand breakage are rapidly converted back to their unmodified form (16, 32, 56, 60). PARG is composed of a 110-kDa nuclear form and at least two cytoplasmic isoforms of 99 kDa and 103 kDa, each of which most likely arises from the same primary transcript (34, 39).
Despite their central roles in PAR metabolism, the relative importance of PARP-1, PARP-2, and PARG for chromosomal single-strand break repair (SSBR) is unclear. For example, both PARP-1 and PARP-2 interact with XRCC1, a scaffold protein that interacts with and recruits, stabilizes, or stimulates multiple enzymatic components of SSBR (13, 14, 54), and the loss of either PARP-1 or PARP-2 has been reported to slow SSBR (3, 19, 55). However, a separate study failed to observe an SSBR defect in Parp-1−/− mouse embryonic fibroblasts (MEFs) (59). Moreover, while numerous studies have shown that global PARP inhibition using small molecule inhibitors dramatically slows DNA strand break repair (2, 7, 8, 20-22, 46, 47, 50, 57, 58), such studies do not discriminate between PARP-1 and PARP-2 or between a positive role for these proteins and a dominant-negative effect of the catalytically inactivated enzyme binding to and occluding SSBs. The importance of PARG for SSBR and cell survival is also conflicting and unclear. For example, it has been reported that PARG interacts with XRCC1 (31) and that mice lacking PARG exhibit sensitivity to alkylating agents and gamma irradiation (16, 32). However, it has also been reported that depletion of PARG protects MEFs from H2O2-induced cell death and that PARG is dispensable for SSBR after oxidative DNA damage (6).
A major factor that confounds our understanding of the importance of PARP-1, PARP-2, and PARG for SSBR is the absence of any study in which these proteins have been examined together in a genetically defined cell type. To address this question, we have quantified SSBR rates in cells lacking or depleted of PARP-1, PARP-2, and PARG, both separately and in combination. These experiments reveal that while PARP-1 is critical for rapid rates of SSBR in both chicken DT40 and human A549 cells, depletion of PARP-2 has only a minor impact, even in the absence of PARP-1. We also identify PARG as a novel component of SSBR and show that this protein accelerates SSBR in concert with PARP-1.
MATERIALS AND METHODS
Cells and cell culture.
DT40 cells were cultured in suspension in RPMI 1640 medium (Gibco, Invitrogen) supplemented with 10% fetal calf serum, 1% chicken serum, and 1% glutamine. A549 cells were cultured in Dulbecco's minimal essential medium (Gibco, Invitrogen) supplemented with 15% fetal calf serum and 1% glutamine.
RNA interference-mediated depletion of PARP-1, PARP-2 and PARG.
A549 lung carcinoma cells were cotransfected using Genejuice (Novagen) with 2 μg of pCD2E vector encoding G418 resistance and 1 μg of either pSuper, pSuper-PARP-1, pSuper-PARP-2, or pSuper-PARG or 1 μg each of pSuper-PARP-1 and pSuper-PARP-2 or pSuper-PARP-1 and pSuper-PARG. After selection in 1.5 mg/ml G418 (Gibco, Invitrogen) for 6 days, cells were either harvested for clonogenic survival assays or incubated for a further 12 to 24 h in G418-free medium and then harvested for alkaline comet assays.
pSuper constructs and oligonucleotides.
High-performance liquid chromatography-purified oligonucleotides (MWG Biotech) containing appropriate regions of homology to PARP-1, PARP-2, or PARG were annealed, phosphorylated with T4 polynucleotide kinase (PNK), and subcloned into the BglII and HindIII restriction sites of pSuper (OligoEngine) (12). The identification of pSuper constructs harboring the correct oligonucleotide sequence was confirmed by sequencing. The sequences of the oligonucleotides cloned into pSuper were as follows (the 19-bp regions of gene specific homology are underlined): PARP-1 (forward, 5′-GATCCCCGGGCAAGCACAGTGTCAAATTCAAGAGATTTGACACTGTGCTTGCCCTTTTTGGAAA-3′; and reverse, 5′-AGCTTTTCCAAAAAGGGCAAGCACAGTGTCAAATCTCTTGAATTTGACACTGTGCTTGCCCGGG-3′), PARP-2 (forward, 5′-GATCCCCCAAGGCCAAGGAAATCTTTTTCAAGAGAAAAGATTTCCTTGGCCTTGTTTTTGGAAA-3′; and reverse, 5′-AGCTTTTCCAAAAACAAGGCCAAGGAAATCTTTTCTCTTGAAAAAGATT TCCTTGGCCTTGGGG-3′), and PARG (forward, 5′-GATCCCCGGAAACCGGAGAAACTTAATTCAAGAGATTAAGTTTCTCCGGTTTCCTTTTTGGAAA-3′; and reverse, 5′-AGCTTTTCCAAAAAGGAAACCGGAGAAACTTAATCTCTTGAATTAAGTTTCTCCGGTTTCCGGG-3′).
Antibodies and immunoblotting.
Cells (5,000/μl) were lysed in hot sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and incubated at 90°C for 5 min. Whole-cell extracts were fractionated by SDS-PAGE and transferred to nitrocellulose. Membranes were blocked for 1 h in Tris-buffered saline-Tween 20 (TBST) containing 5% nonfat dried milk (NFDM) and then incubated with appropriate primary antibodies. PARP-1 monoclonal antibody (MAb) A6.4.7 (kindly provided by Said Aoufouchi, now available from Serotec as clone A6.4.12) was employed at a 1/500 dilution in TBST for 1 h at room temperature (RT). PARP-2 polyclonal antibody (Yuc; Alexis) was employed overnight at 4°C at a 1/3,000 dilution in TBST containing 5% NFDM. PARG polyclonal antibody ab16060 (Abcam) was employed at a 1/200 dilution in TBST containing 1% NFDM overnight at 4°C. DNA ligase III polyclonal antibody TL25 (kindly provided by Thomas Lindahl) was employed at a 1/1,500 dilution in TBST for 1 h at room temperature. Affinity-purified XRCC1 polyclonal antibody SK3188, raised by Eurogentec against full-length recombinant human XRCC1 raised in baculovirus, was used at a 1/200 dilution in TBST for 1 h at room temperature. α-Actin MAb clone AC-40 (Sigma) was employed at a 1/2,000 dilution in TBST for 1 h at room temperature. Affinity-purified PNK polyclonal antibody, raised by Eurogentec against a recombinant fragment of human PNK (residues 1 to 130) expressed in Escherichia coli was employed at a 1/200 dilution in TBST containing 1% NFDM overnight at 4°C. Membranes were then washed in TBST and incubated in TBST containing horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG (DAKO), as appropriate, at a 1/5,000 dilution for 1 h at room temperature. Membranes were then washed with TBST, and antibody complexes were detected by enhanced chemiluminescence (Amersham).
Alkaline single-cell agarose gel electrophoresis (comet) assays.
DNA strand breaks were quantified using the alkaline comet assay as described previously (11). Average tail moments from 100 cells per sample were obtained using Comet Assay III software (Perceptive Instruments), and data are shown as means ± 1 standard error of the mean (SE) of this value from three or more independent experiments. Where indicated, representative scatter plots showing the distribution of individual tail moments within populations of 100 cells are also presented.
γ-H2AX assays.
Cells were grown on coverslips and mock treated or treated with H2O2 (100 μM) for 20 min on ice. Cells were then washed in phosphate-buffered saline (PBS) at RT and either harvested or incubated at 37°C for indicated repair periods. Cells were fixed with 4% paraformaldehyde for 5 min at RT, permeabilized with 0.2% Triton X-100 and, after two rinses in PBS, were blocked in PBS in 5% NFDM. The cells were then incubated in anti-γH2AX (Ser139) MAb (Upstate) at a 1/800 dilution in 1% NFDM in PBS for 1 h at RT. After three washes in PBS-TS (PBS, 0.1% Tween 20, 0.02% SDS), the cells were incubated in Alexa Fluor 488 goat anti-mouse IgG antibody (Invitrogen) at a 1/200 dilution in 1% NFDM for 1 h at RT, and after they were rinsed in PBS three times, cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vecta Laboratories). Cells were analyzed, and γH2AX foci were counted using a Nikon Eclipse 50i microscope at ×100 magnification. Twenty cells were scored per data point.
Direct fluorescence detection of monomeric red fluorescent protein-XRCC1.
HeLa cells were cotransfected using Genejuice (Novagen) with 1 μg of pCD2E vector encoding G418 resistance and 1 μg of either pSuper, pSuper-PARP-1, or pSuper-PARG. After selection in 0.8 mg/ml G418 (Gibco, Invitrogen) for 3 days, cells were seeded onto coverslips in G418 medium (0.8 mg/ml), and after a further 24-h incubation, cells were placed in complete medium lacking G418. The cells were transfected with 2 μg of monomeric red fluorescent protein (mRFP)-XRCC1 (29), using Genejuice transfection reagent (Novagen). Twenty-four hours posttransfection, cells were either treated or mock treated with H2O2 (100 μM) for 20 min at room temperature. Cells were then washed in PBS and incubated at 37°C for the indicated repair periods. Cells were fixed in 4% paraformaldehyde for 5 min at RT and permeabilized with 0.2% Triton X-100. Cells were counterstained with DAPI and mounted with Vectashield (Vecta Laboratories). Cells were analyzed and photographed with a Deltavision RT microscope with a 60× objective. Twenty mRFP-transfected cells were scored per data point.
Clonogenic survival assays.
A549 cells were plated into 10-cm dishes (4,000 per plate) in duplicate and incubated at 37°C for 4 h. Cells were treated with H2O2 in PBS at the indicated concentrations for 10 min at RT. Cells were then rinsed in PBS and incubated at 37°C in drug-free medium for 14 days. Colonies were fixed with 90% ethanol and stained with 1% methylene blue. Survival was calculated as a percentage, using the equation Nt/Nu × 100, where Nt is the number of colonies on treated plates and Nu is the number on untreated plates. Data are the means ± 1 SE of three independent experiments.
Indirect immunofluorescence detection of PAR.
Cells were grown on coverslips and mock treated or treated with H2O2 (10 mM) for 10 min on ice. Cells were then washed in PBS at RT and either harvested immediately or incubated at 37°C for the indicated repair period in a drug-free medium. The cells were then washed in PBS and fixed in 4% paraformaldehyde for 5 min at RT and ice cold methanol for 10 min at −20°C and, after two rinses in PBS, were blocked in PBS-TS (PBS, 0.1% Tween 20, 0.02% SDS) in 5% NFDM. The cells were then incubated in anti-PAR MAb 10H (Alexis) at a 1/200 dilution in 1% NFDM in PBS-TS for 2 h at RT. After cells were washed three times in PBS-TS, they were incubated in Alexa Fluor 488 goat anti-mouse IgG antibody (Invitrogen) at a 1/200 dilution in 1% NFDM for 1 h at RT, and after they were rinsed in PBS (three times), cells were counterstained with DAPI and mounted with Vectashield (Vecta Laboratories). Cells were analyzed and photographed with a Nikon Eclipse 50i microscope at ×100 magnification.
PARP and PARG activity assays.
Cell extracts were prepared from ∼6 × 106 normal, PARP-1-depleted, or PARG-depleted A549 cells from each of three independent experiments, and PARP and PARG activities were determined in triplicate. Colorimetric assay kits were employed as described by the manufacturer (Trevigen).
RESULTS
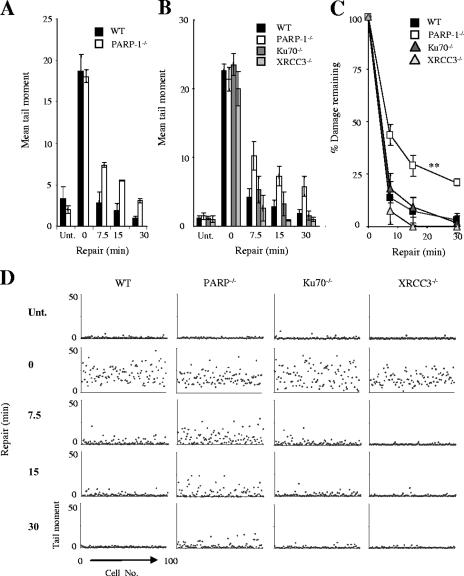
Because of conflicting reports in the literature (3, 19, 55, 59), we attempted to confirm that PARP-1 is required for rapid rates of DNA strand break repair. Initially, we employed PARP-1−/− chicken DT40 cells (28), since DT40 cells do not possess PARP-2 and so circumvent the issue of enzymatic redundancy. We treated PARP-1−/− DT40 cells with H2O2, a physiologically relevant agent that induces oxidative DNA damage, and measured the level of strand breakage using alkaline comet assays. Although H2O2 induced similar levels of DNA strand breakage in wild-type and PARP-1−/− DT40 cells, the two cell lines differed in the rates at which H2O2-induced breaks declined during a subsequent incubation in drug-free medium (Fig. 1A). It is likely that the defect observed for PARP-1−/− DT40 cells reflects a reduced ability to repair DNA SSBs, rather than DNA double-strand breaks (DSBs), because >99% of the DNA breaks induced by H2O2 are SSBs (9). However, to examine this further, we repeated these experiments with KU70−/− and XRCC3−/− DT40 cells, each of which lacks one of the two major pathways for DNA DSBR (52, 53). Ku70 and XRCC3 are required for the repair of DSBs by nonhomologous end joining (26, 45, 51) and homologous recombination, respectively (10, 36). We reasoned that if a significant proportion of the DNA strand breaks induced by 25 μM H2O2 are DSBs, then cells with established defects in DSBR should display reduced kinetics of DNA strand break repair in alkaline comet assays. However, only PARP-1−/− DT40 cells exhibited a delay in the rate of DNA strand break repair compared to that of wild-type cells (Fig. 1B-D). These data strongly suggest that the reduced repair rate observed for PARP-1−/− DT40 cells reflected a defect in SSBR (Fig. 1B to D).
FIG. 1.
Reduced rates of SSBR in PARP-1−/− chicken DT40 cells. (A) Total DNA strand breakage was quantified in wild-type (WT) and PARP-1−/− DT40 cells by alkaline comet assays with untreated cells (Unt.), with cells immediately after treatment with 25 μM H2O2 for 20 min on ice (0), and with H2O2-treated cells after the indicated repair period in H2O2-free medium. Data points are means (± 1 SE) of at least three independent experiments, with the average tail moment from 100 cells quantified in each experiment. (B) Total DNA strand breakage was quantified in wild-type (WT), PARP-1−/−, KU70−/−, and XRCC3−/− DT40 cells as described above. (C) The data from panel B was replotted as the fraction (%) of DNA strand breaks remaining at the indicated time points. **, PARP-1 and WT repair kinetics were significantly different (analysis of variance, P = 3 × 10−4). Ku70−/− and XRCC3−/− repair kinetics were not significantly different from those of the WT (P = 0.47 and P = 0.15, respectively). (D) A representative scatter plot of the raw data from one of the experiments used in panels B and C to show the level of variation in DNA strand breakage within single populations of cells. Each dot represents the tail moment of an individual cell, and 100 cells were scored per sample.
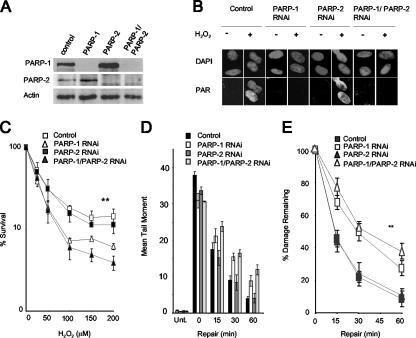
Mammalian cells possess a second DNA damage-dependent poly(ADP-ribose) polymerase, called PARP-2 (1, 30). To define the relative importance of PARP-1 and PARP-2 for SSBR in human cells, we transiently depleted these proteins from A549 lung carcinoma cells using RNA interference (RNAi) expression constructs, both separately and in combination (Fig. 2A). Depletion of PARP-1 greatly reduced cellular PAR synthesis following H2O2 treatment, as measured by indirect immunofluorescence (Fig. 2B). In addition, in vitro histone ribosylation assays revealed that the rate of PAR synthesis catalyzed by cell extracts from PARP-1-depleted cells was reduced by ∼95% compared to that of normal cell extracts (data not shown). In contrast, depletion of PARP-2 protein by >80% did not measurably affect global levels of cellular PAR synthesis (Fig. 2A and B), suggesting that PARP-1 was the predominant source of PAR synthesis following oxidative stress. We did note that PARP-2-depleted cells were slightly more sensitive to H2O2 than mock-depleted cells and that PARP-1/PARP-2-codepleted cells were slightly more sensitive than PARP-1-depleted cells, but these differences were not statistically significant (Fig. 2C).
FIG. 2.
Depletion of PARP-1 but not PARP-2 reduces rates of chromosomal SSBR and sensitizes human A549 cells to oxidative DNA damage. (A) Levels of PARP-1 and PARP-2 protein in total cells extracts from A549 cells transfected with pcD2E and either empty pSuper (Control), pSuper-PARP-1 (PARP-1), pSuper-PARP-2 (PARP-2), or both pSuper-PARP-1 and pSuper-PARP-2 (PARP-1/PARP-2), as measured by immunoblotting with appropriate antibodies. (B) Levels of PAR before and after (1-min repair) treatment with 10 mM H2O2 on ice with A549 cells depleted of the indicated proteins, as measured by indirect immunofluorescence microscopy. Cells were counterstained with DAPI to identify nuclear DNA. (C) Clonogenic survival of A549 cells depleted of the indicated proteins following exposure to the indicated concentrations of H2O2 in PBS for 10 min at RT. Cells were fixed after 14 days and stained with methylene blue, and the fraction (%) of surviving cells was calculated. Data are the means (± 1 SE) of three independent experiments. **, the survival curve for control cells was significantly different (by analysis of variance [ANOVA]) from those of PARP-1-depleted (P = 0.009) and PARP-1/PARP-2-depleted (P = 0.01) cells but not those of PARP-2-depleted cells (P = 0.57). The survival curves for PARP-1- and PARP-1/PARP-2-depleted cells were not significantly different (P = 0.76). (D) Total DNA strand breakage was quantified by comet assays with A549 cells depleted of the indicated proteins before (Unt.) and immediately after (0) treatment with 100 μM H2O2 for 20 min on ice and after the indicated repair periods in H2O2-free medium. Data points are the means (± 1 SE) of at least three independent experiments, with the average tail moment from 100 cells calculated in each experiment. (E) The data from panel D were replotted as the fraction (%) of DNA strand breaks remaining at the indicated DNA repair time points. **, the repair kinetics for control cells were statistically significantly (ANOVA) different from those of PARP-1-depleted (P = 7.4 × 10−7) and PARP-1/PARP-2-depleted (P = 0.68 × 10−8) cells but not those of PARP-2-depleted cells (P = 0.87). The repair kinetics for PARP-1- and PARP-1/PARP-2-depleted cells were not significantly different (P = 0.1).
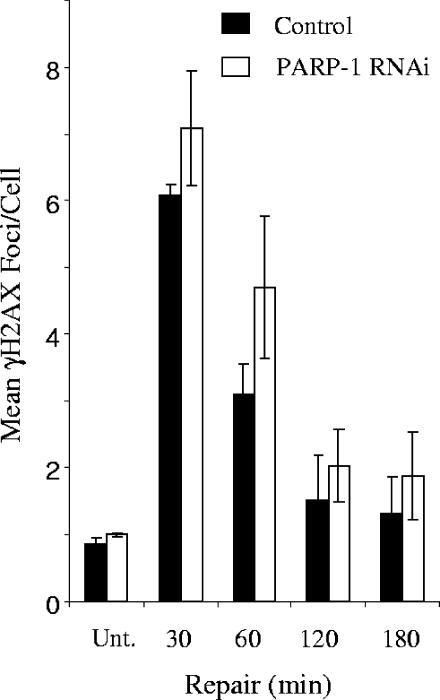
We next measured the rate of repair of H2O2-induced DNA strand breakage in normal cells and in cells depleted of PARP-1 and/or PARP-2. We noted in these experiments that H2O2 induced slightly more damage in control cells than in cells from which PARP-1 or PARP-2 was depleted (Fig. 2D). Despite this, while control and PARP-2-depleted cells repaired oxidative DNA strand breaks at similar rates, PARP-1-depleted cells exhibited significantly delayed rates of DNA strand break repair (Fig. 2D and E). When PARP-1 and PARP-2 were depleted simultaneously, only a small reduction in the initial rate of repair was observed compared to that for depletion of PARP-1 alone, and this was not statistically significant. γ-Ray calibration curves revealed that the mean tail moment observed for A549 cells (∼30) immediately after treatment with 100 μM H2O2 was equivalent to ∼30,000 total DNA strand breaks per cell (data not shown) and that the difference between normal and PARP-1-depleted cells at 30 to 60 min after H2O2 (∼5 mean tail moment units) was equivalent to ∼5,000 total DNA strand breaks per cell. To confirm that the majority of these breaks were SSBs, we quantified the level of H2O2-induced γ-H2AX immunofoci, a marker for DSBs, under the same experimental conditions. Approximately 6 γ-H2AX foci per cell were observed for both normal and PARP-1-depleted cells 30 min after H2O2 treatment, confirming that the vast majority of unrepaired DNA strand breaks detected in PARP-1-depleted cells following H2O2 are SSBs. The low level of DSBs observed for these experiments compared to the total number of DNA strand breaks observed under the same experimental conditions (10 to 15,000) is consistent with the low ratio (1:2,000) of DSBs:SSBs induced by H2O2 (9) (Fig. 3). We noted that the decline of γ-H2AX foci occurred more slowly with PARP-1-depleted cells, though this difference was not statistically significant. In summary, we conclude that PARP-1 is the primary poly(ADP-ribose) polymerase responsible for accelerating the global rate of SSBR in human cells following oxidative stress and that PARP-2 has only minor, if any, influence on this rate, even in cells in which PARP-1 is greatly depleted.
FIG. 3.
H2O2-induced formation and removal of γ-H2AX from normal and PARP-1-depleted A549 cells. γ-H2AX foci were quantified in normal and PARP-1-depleted A549 cells before (Unt.) treatment with 100 μM H2O2 for 20 min on ice and after the indicated repair periods in H2O2-free medium. Data points are the means (± 1 SE) of three independent experiments. Note that the kinetics at which γ-H2AX foci declined in normal and PARP-1-depleted cells were not significantly different (analysis of variance, P = 0.057).
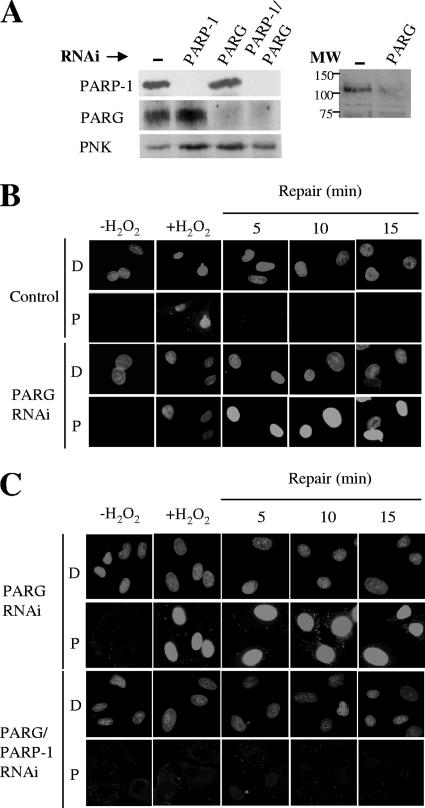
Following its activation and automodification, PARP-1 dissociates from DNA strand breaks through charge repulsion. The PAR chains are then rapidly degraded by PARG, restoring PARP-1 to its preactivated state. Despite the major impact of PARG on PAR metabolism, the influence of this enzyme on SSBR rates is unknown. To evaluate the importance of PARG for this process, we transiently depleted this protein from A549 cells (Fig. 4A). In vitro PARG assays employing ribosylated histones as a substrate suggested that the basal level of PARG activity in cell extracts from PARG-depleted cells was reduced by ∼50% (data not shown). However, the impact of PARG depletion on the cellular PAR levels following treatment with H2O2 was much more pronounced, with high levels of PAR persisting for much longer periods in PARG-depleted cells than in normal cells (Fig. 4B). These data indicate that PARG activity has a major influence on the level of PAR present in cells following oxidative DNA stress. Notably, the appearance of PAR in PARG-depleted cells at 5 to 15 min after H2O2 treatment was abolished by codepletion of PARP-1, further indicating that PARP-1 is the primary source of global PAR synthesis following oxidative stress (Fig. 4C).
FIG. 4.
Impact of PARP-1 and/or PARG depletion on levels of H2O2-induced PAR synthesis. (A) The left panel shows levels of PARP-1 and PARG proteins in total cell extracts from A549 cells transfected with pcD2E and either empty pSuper (Control), pSuper-PARP-1 (PARP-1), pSuper-PARG (PARG), or both pSuper-PARP-1 and pSuper-PARG (PARP-1/PARG), as measured by immunoblotting with anti-PARP-1 MAb, anti-PARG polyclonal antibody, and anti-PNK polyclonal antibody as a loading control. The right panel shows the immunoblot of a different set of cell extracts from normal (−) and PARG-depleted A549 cells with anti-PARG antibody, showing the specificity of the antibody in the region of the blot containing full-length (110-kDa) PARG. The positions of molecular weight (MW) standards are shown. (B) Levels of PAR (rows P) in normal (Control) or PARG-1-depleted (PARG RNAi) A549 cells before treatment with 10 mM H2O2 on ice (−H2O2), 1 min after H2O2 treatment (+H2O2), and after the indicated repair periods in drug-free medium, as measured by indirect immunofluorescence microscopy. Cells were counterstained with DAPI (rows D) to identify nuclear DNA. (C) Levels of PAR (rows P) in PARG-1-depleted (PARG RNAi) or PARG-1/PARP-1-depleted (PARG/PARP-1 RNAi) A549 cells treated as described in the legend to panel B. Cells were counterstained with DAPI (rows D) to identify nuclear DNA.
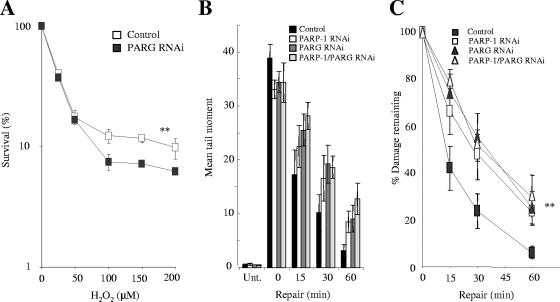
PARG-depleted cells were significantly more sensitive to H2O2 than normal cells at concentrations of H2O2 above 50 μM, supporting a role for this protein in protecting cells from elevated oxidative stress (Fig. 5A). More importantly, depletion of PARG reduced the rate of chromosomal SSBR to a level similar to that observed for cells depleted of PARP-1 (Fig. 5B and C), demonstrating that PARG is required for rapid global rates of SSBR in human cells. To address whether PARG might act separately or in concert with PARP-1, we compared chromosomal SSBR rates in cells depleted of either PARP-1 or PARG with those in cells depleted of both proteins together. Notably, the rate of SSBR in cells depleted of both proteins was no less than in cells depleted of either protein alone (Fig. 5B and C), suggesting that PARP-1 and PARG accelerate SSBR in concert with each other.
FIG. 5.
PARG accelerates SSBR in concert with PARP-1 in human A549 cells. (A) Clonogenic survival of A549 cells depleted or not of PARG following exposure to the indicated concentrations of H2O2 for 10 min at RT. Cells were fixed after 14 days and stained with methylene blue, and the fraction (%) of surviving cells was calculated. Data are the means (± 1 SE) of three independent experiments. **, the difference between the control and the PARG-depleted survival curves is statistically significantly (analysis of variance [ANOVA], P = 4.7 × 10−5). (B) Total DNA strand breakage was quantified by alkaline comet assays with control A549 cells (Control), PARP-1-depleted A549 cells (PARP-1 RNAi), PARG-depleted A549 cells (PARG RNAi), or PARP-1/PARG-depleted A549 cells (PARP-1/PARG RNAi) before (Unt.) and immediately after (0) treatment with 100 μM H2O2 for 20 min on ice and after the indicated repair periods in H2O2-free medium. Data points are the means (± 1 SE) of at least three independent experiments, with the average tail moment from 100 cells calculated in each experiment. (C) The data from panel B were replotted as the fraction (%) of DNA strand breaks remaining at the indicated DNA repair time points. **, statistically significant (ANOVA) differences were observed between the repair kinetics of control cells and those treated with either PARP-1 (P = 0.004), PARG (P = 0.001), or PARP-1/PARG (P = 0.001) RNAi. Repair kinetics of cells treated with PARP-1/PARG RNAi are not significantly different from those treated with PARP-1 or PARG RNAi alone.
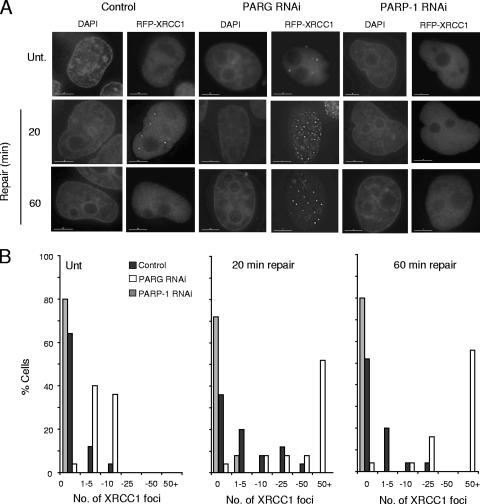
Why might PARG be required during SSBR? One function of PARP-1 during chromosomal SSBR following oxidative stress is to facilitate the accumulation of XRCC1 (23, 33, 43), a scaffold protein that can recruit, stimulate, and/or stabilize enzymatic components of the repair process (13, 54). The rapid accumulation of XRCC1 at sites of oxidative SSBs, as detected by the accumulation of this protein into discrete foci, requires PAR synthesis, most likely reflecting the observation that XRCC1 preferentially interacts with autoribosylated PARP-1 (18, 37, 44). Since PARG accelerates SSBR in concert with PARP-1, we considered the possibility that PARG might also regulate the accumulation of XRCC1 at oxidative DNA strand breaks. We reasoned that by de-ribosylating PARP-1 or some other ribosylated protein(s), PARG might negatively regulate XRCC1 accumulation. This might be important to prevent excessive XRCC1 accumulation, which could impede SSBR, or to disassemble XRCC1 protein complexes once repair is complete, thereby freeing this scaffold protein for reassembly at alternative sites. To address this question, we compared the impact of PARP-1 and PARG depletion on the appearance of RFP-XRCC1 foci following H2O2 treatment in transiently transfected HeLa cells. HeLa cells were employed in these experiments because of their superior transient transfection efficiency and because the achievable level of PARP-1 and PARG depletion was comparable to that of A549 cells (data not shown). Whereas RFP-XRCC1 assembled into discrete foci in normal HeLa cells, they largely failed to do so in PARP-1-depleted cells over the time course of the experiment (Fig. 6A and B). These data are in agreement with our previous report that XRCC1 fails to accumulate into subnuclear immunofoci in PARP-1−/− MEFs (23). More significantly, however, greatly increased levels and persistence of XRCC1 foci were observed for PARG-depleted cells following H2O2 treatment, with >50% of cells possessing more than 50 foci per cell. Moreover, XRCC1 foci were detected in the majority (∼80%) of PARG-depleted cells even in the absence of H2O2 treatment. This was in marked contrast to normal cells, of which less than 15% possessed XRCC1 foci. We conclude from these experiments that PARG negatively regulates the accumulation of XRCC1 protein scaffold during SSBR.
FIG. 6.
Accumulation of RFP-XRCC1 in normal and PARP-1- or PARG-depleted HeLa cells. (A) RFP-XRCC1 foci were detected with HeLa cells transiently transfected with an mRFP-XRCC1 expression construct and either an empty pSuper vector, a pSuper-PARP1 RNAi construct, or a pSuper-PARG RNAi construct by direct fluorescence microscopy. Cells were either mock treated (Unt.) or treated with 100 μM H2O2 for 20 min on ice followed by incubation in drug-free medium for the indicated repair periods. Nuclei were counterstained with DAPI. Bars represent 10 μm. Representative images are shown. (B) Quantification of the results from the experiment shown in panel A. Transfected (RFP-positive) cells were chosen at random, and the number of XRCC1 foci present was scored. Data are from a single experiment representative of multiple repeats.
DISCUSSION
Although there is evidence to support a positive role for PARP-1 in chromosomal SSBR, this hypothesis has been contentious. While there are many studies showing that poly(ADP-ribose) polymerase inhibitors block SSBR, it is difficult to exclude the possibility that this is a dominant-negative effect of the inactivated enzyme preventing access to the break by other proteins. Studies employing PARP-1−/− mouse cells or human cells in which PARP-1 was depleted by antisense expression have circumvented this issue but have resulted in conflicting conclusions (3, 19, 55, 59). A possible confounding factor in experiments addressing the role of mammalian PARP-1 is the existence of PARP-2, a second DNA damage-activated PARP that can contribute to levels of PAR synthesis following DNA damage (1, 30, 38).
To address this question, we first examined the importance of PARP-1 for SSBR in chicken DT40 cells because these cells have been reported to possess a single PARP (28). In addition, we defined the relative importance of PARP-1 and PARP-2 for SSBR in human A549 cells, using RNAi to deplete these proteins both separately and together. Our results confirm that PARP-1 is critical for high rates of chromosomal SSBR, both in chicken DT40 cells and in human A549 cells. It is noteworthy that we also observed similar results for HeLa cells (unpublished observations). In marked contrast, we failed to detect any significant impact of PARP-2 depletion on SSBR rates in A549 cells, even in cells that were simultaneously depleted of PARP-1. These data are surprising because PARP-2 has been reported to interact with XRCC1, a scaffold protein that accelerates the rate of SSBR, and reduced rates of SSBR were observed with PARP-2−/− MEFs following DNA alkylation (48). Although we cannot exclude the possibility that our data reflect the presence of residual levels of PARP-2 protein, we consider this unlikely. This is because the depletion of PARP-2 in our experiments (by >80%) failed to reduce global rates of SSBR, even in the absence of PARP-1, when the residual level of PAR synthesis (∼5%) was markedly limiting the rate of SSBR. While it is possible that PARP-2 plays a subtle role during SSBR, at a defined subset of breaks for example, our experiments strongly suggest that PARP-2 is not required for global rates of SSBR following oxidative stress, even in the presence of depleted levels of PARP-1.
It is noteworthy that removal or depletion of PARP-1 from DT40 and human cells, respectively, did not ablate SSBR but rather reduced the global rate of this process. This is also true of cells lacking XRCC1, the archetypal SSBR protein that plays a scaffold role. It is possible that the residual SSBR in cells lacking PARP-1 or XRCC1 reflects the presence of partially redundant factors. Alternatively, it is possible that XRCC1 and PARP-1 serve only to accelerate SSBR, by increasing the rate at which other DNA repair proteins access chromosomal SSBs, for example.
To date, the only protein shown to catabolize PAR following DNA damage is PARG (16, 32, 56). Despite this polypeptide's pivotal role in PAR metabolism, the importance and role of PARG following DNA damage is unclear. In the present study, we depleted PARG from human A549 cells and measured their sensitivity to H2O2 and their rate of chromosomal SSBR. Strikingly, we observed severely retarded rates of SSBR in PARG-depleted cells, to levels similar to those observed for PARP-1-depleted cells. These data demonstrate for the first time that PARG is critical for rapid rates of chromosomal SSBR. Significantly, codepletion of both PARP-1 and PARG did not slow the rate of SSBR any more effectively than did depletion of PARP-1 or PARG alone, suggesting that PARP-1 and PARG act in concert to accelerate SSBR.
What role might PARG play? One possibility is that the deribosylation of PARP-1 by PARG is required to restore PARP-1 to its preactivated state in preparation for subsequent SSBR events. In addition, our data suggest that deribosylation of PARP-1, or one or more other ribosylated proteins, by PARG might be required to regulate the accumulation of XRCC1 complexes at SSBs. This may be important to prevent excessive XRCC1 accumulation during SSBR, which may otherwise impede the repair reaction. Alternatively, PARG may disassemble XRCC1 complexes after a repair reaction has been completed, in readiness for reassembly of this scaffold protein at other sites. A role for PARG in disassembling or recycling XRCC1 and/or PARP-1 complexes for subsequent repair events might be particularly important at high levels of strand breakage, when the number or concentration of SSBs might exceed the localized availability XRCC1 and/or PARP-1 molecules. Our observation that PARG-depleted cells are hypersensitive to H2O2 only at higher concentrations is consistent with this possibility.
Recently, it was reported that a hypomorphic mutation that elevates PARG activity and reduces steady-state PAR levels results in decreased accumulation of XRCC1 foci and increased cellular sensitivity to DNA damage (25). Although that study addressed the cellular response to DNA alkylating agents, it is consistent with our findings. In contrast, another study failed to observe a defect in SSBR in PARG-depleted MEFs following H2O2 treatment (6). This may reflect differences in the efficiency of PARG depletion between this study and ours, since we employed short-term selection of plasmid-based RNAi constructs to eradicate nontransfected cells from our experiments, whereas Blenn et al. employed total populations of transfected cells.
In summary, we report that PARP-1 and PARG are critical for high global rates of chromosomal SSBR following oxidative stress and that these two proteins most likely act in concert. Our data suggest that one role of these proteins is to regulate the accumulation of an XRCC1 protein scaffold at sites of oxidative DNA damage, thereby maintaining optimal rates of SSBR.
Acknowledgments
This work was funded by an MRC Programme grant to K.W.C.
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Ame, J. C., V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia, and G. de Murcia. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860-17868. [DOI] [PubMed] [Google Scholar]
- 2.Atorino, L., S. Di Meglio, B. Farina, R. Jones, and P. Quesada. 2001. Rat germinal cells require PARP for repair of DNA damage induced by gamma-irradiation and H2O2 treatment. Eur. J. Cell Biol. 80:222-229. [DOI] [PubMed] [Google Scholar]
- 3.Beneke, R., C. Geisen, B. Zevnik, T. Bauch, W. U. Muller, J. H. Kupper, and T. Moroy. 2000. DNA excision repair and DNA damage-induced apoptosis are linked to poly(ADP-ribosyl)ation but have different requirements for p53. Mol. Cell. Biol. 20:6695-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin, R. C., and D. M. Gill. 1980. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J. Biol. Chem. 255:10493-10501. [PubMed] [Google Scholar]
- 5.Benjamin, R. C., and D. M. Gill. 1980. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 255:10502-10508. [PubMed] [Google Scholar]
- 6.Blenn, C., F. R. Althaus, and M. Malanga. 2006. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem. J. 396:419-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton, S., S. Kyle, and B. W. Durkacz. 1999. Interactive effects of inhibitors of poly(ADP-ribose) polymerase and DNA-dependent protein kinase on cellular responses to DNA damage. Carcinogenesis 20:199-203. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, K. J., D. R. Newell, A. H. Calvert, and N. J. Curtin. 2001. Differential effects of the poly(ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br. J. Cancer 84:106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley, M. O., and K. W. Kohn. 1979. X-ray induced DNA double-strand break production and repair in mammalian cells as measured by neutral filter elution. Nucleic Acids Res. 7:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenneman, M. A., A. E. Weiss, J. A. Nickoloff, and D. J. Chen. 2000. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat. Res. 459:89-97. [DOI] [PubMed] [Google Scholar]
- 11.Breslin, C., P. M. Clements, S. F. El-Khamisy, E. Petermann, N. Iles, and K. W. Caldecott. 2006. Measurement of chromosomal DNA single-strand breaks and replication fork progression rates. Methods Enzymol. 409:410-425. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 13.Caldecott, K. W. 2003. XRCC1 and DNA strand break repair. DNA Repair (Amsterdam) 2:955-969. [DOI] [PubMed] [Google Scholar]
- 14.Caldecott, K. W., S. Aoufouchi, P. Johnson, and S. Shall. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly(ADP-ribose) polymerase, and DNA ligase III is a novel molecular “nick-sensor” in vitro. Nucleic Acids Res. 24:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambon, P., J. D. Weill, and P. Mandel. 1963. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11:39-43. [DOI] [PubMed] [Google Scholar]
- 16.Cortes, U., W.-M. Tong, D. L. Coyle, M. L. Meyer-Ficca, R. G. Meyer, V. Petrilli, Z. Herceg, E. L. Jacobson, M. K. Jacobson, and Z. Q. Wang. 2004. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 24:7163-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 18.Dantzer, F., G. de La Rubia, J. Menissier-de Murcia, Z. Hostomsky, G. de Murcia, and V. Schreiber. 2000. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39:7559-7569. [DOI] [PubMed] [Google Scholar]
- 19.Ding, R., Y. Pommier, V. H. Kang, and M. Smulson. 1992. Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J. Biol. Chem. 267:12804-12812. [PubMed] [Google Scholar]
- 20.Durkacz, B. W., J. Irwin, and S. Shall. 1981. Inhibition of (ADP-ribose)n biosynthesis retards DNA repair but does not inhibit DNA repair synthesis. Biochem. Biophys. Res. Commun. 101:1433-1441. [DOI] [PubMed] [Google Scholar]
- 21.Durkacz, B. W., O. Omidiji, D. A. Gray, and S. Shall. 1980. (ADP-ribose)n participates in DNA excision repair. Nature 283:593-596. [DOI] [PubMed] [Google Scholar]
- 22.Durkacz, B. W., S. Shall, and J. Irwin. 1981. The effect of inhibition of (ADP-ribose)n biosynthesis on DNA repair assayed by the nucleoid technique. Eur. J. Biochem. 121:65-69. [DOI] [PubMed] [Google Scholar]
- 23.El-Khamisy, S. F., M. Masutani, H. Suzuki, and K. W. Caldecott. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31:5526-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferro, A. M., and B. M. Olivera. 1982. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J. Biol. Chem. 257:7808-7813. [PubMed] [Google Scholar]
- 25.Gao, H., D. L. Coyle, M. L. Meyer-Ficca, R. G. Meyer, E. L. Jacobson, Z. Q. Wang, and M. K. Jacobson. 2007. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp. Cell Res. 313:984-996. [DOI] [PubMed] [Google Scholar]
- 26.Getts, R. C., and T. D. Stamato. 1994. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J. Biol. Chem. 269:15981-15984. [PubMed] [Google Scholar]
- 27.Giri, C. P., M. H. West, M. L. Ramirez, and M. Smulson. 1978. Nuclear protein modification and chromatin substructure. 2. Internucleosomal localization of poly(adenosine diphosphate-ribose) polymerase. Biochemistry 17:3501-3504. [DOI] [PubMed] [Google Scholar]
- 28.Hochegger, H., D. Dejsuphong, T. Fukushima, C. Morrison, E. Sonoda, V. Schreiber, G. Y. Zhao, A. Saberi, M. Masutani, N. Adachi, H. Koyama, G. de Murcia, and S. Takeda. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25:1305-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iles, N., S. Rulten, S. F. El-Khamisy, and K. W. Caldecott. 2007. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27:3793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson, M. 1999. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics 57:442-445. [DOI] [PubMed] [Google Scholar]
- 31.Keil, C., T. Grobe, and S. L. Oei. 2006. MNNG-induced cell death is controlled by interactions between PARP-1, poly (ADP-ribose) glycohydrolase and XRCC1. J. Biol. Chem. 281:34394-34405. [DOI] [PubMed] [Google Scholar]
- 32.Koh, D. W., A. M. Lawler, M. F. Poitras, M. Sasaki, S. Wattler, M. C. Nehls, T. Stoger, G. G. Poirier, V. L. Dawson, and T. M. Dawson. 2004. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 101:17699-17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan, L., S. Nakajima, Y. Oohata, M. Takao, S. Okano, M. Masutani, S. H. Wilson, and A. Yasui. 2004. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. USA 101:13738-13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, W., J. C. Ame, N. Aboul-Ela, E. L. Jacobson, and M. K. Jacobson. 1997. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 272:11895-11901. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl, T., M. S. Satoh, G. G. Poirier, and A. Klungland. 1995. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 20:405-411. [DOI] [PubMed] [Google Scholar]
- 36.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 37.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menissier de Murcia, J., M. Ricoul, L. Tartier, C. Niedergang, A. Huber, F. Dantzer, V. Schreiber, J. C. Ame, A. Dierich, M. LeMeur, L. Sabatier, P. Chambon, and G. de Murcia. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22:2255-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer-Ficca, M. L., R. G. Meyer, D. L. Coyle, E. L. Jacobson, and M. K. Jacobson. 2004. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297:521-532. [DOI] [PubMed] [Google Scholar]
- 40.Mullins, D. W., Jr., C. P. Giri, and M. Smulson. 1977. Poly(adenosine diphosphate-ribose) polymerase: the distribution of a chromosome-associated enzyme within the chromatin substructure. Biochemistry 16:506-513. [DOI] [PubMed] [Google Scholar]
- 41.Ogata, N., K. Ueda, M. Kawaichi, and O. Hayaishi. 1981. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 256:4135-4137. [PubMed] [Google Scholar]
- 42.Ohgushi, H., K. Yoshihara, and T. Kamiya. 1980. Bovine thymus poly(adenosine diphosphate ribose) polymerase. Physical properties and binding to DNA. J. Biol. Chem. 255:6205-6211. [PubMed] [Google Scholar]
- 43.Okano, S., S. Kanno, S. Nakajima, and A. Yasui. 2000. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J. Biol. Chem. 275:32635-32641. [DOI] [PubMed] [Google Scholar]
- 44.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 45.Rathmell, W. K., and G. Chu. 1994. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 91:7623-7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satoh, M. S., and T. Lindahl. 1992. Role of poly(ADP-ribose) formation in DNA repair. Nature 356:356-358. [DOI] [PubMed] [Google Scholar]
- 47.Satoh, M. S., G. G. Poirier, and T. Lindahl. 1994. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry 33:7099-7106. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028-23036. [DOI] [PubMed] [Google Scholar]
- 49.Shieh, W. M., J. C. Ame, M. V. Wilson, Z. Q. Wang, D. W. Koh, M. K. Jacobson, and E. L. Jacobson. 1998. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 273:30069-30072. [DOI] [PubMed] [Google Scholar]
- 50.Smith, L. M., E. Willmore, C. A. Austin, and N. J. Curtin. 2005. The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin. Cancer Res. 11:8449-8457. [DOI] [PubMed] [Google Scholar]
- 51.Taccioli, G. E., T. M. Gottlieb, T. Blunt, A. Priestley, J. Demengeot, R. Mizuta, A. R. Lehmann, F. W. Alt, S. P. Jackson, and P. A. Jeggo. 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265:1442-1445. [DOI] [PubMed] [Google Scholar]
- 52.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and nonhomologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, L. H., and M. G. West. 2000. XRCC1 keeps DNA from getting stranded. Mutat. Res. 459:1-18. [DOI] [PubMed] [Google Scholar]
- 55.Trucco, C., F. J. Oliver, G. de Murcia, and J. Menissier-de Murcia. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ueda, K., J. Oka, S. Naruniya, N. Miyakawa, and O. Hayaishi. 1972. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 46:516-523. [DOI] [PubMed] [Google Scholar]
- 57.Veuger, S. J., N. J. Curtin, C. J. Richardson, G. C. Smith, and B. W. Durkacz. 2003. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 63:6008-6015. [PubMed] [Google Scholar]
- 58.Veuger, S. J., N. J. Curtin, G. C. Smith, and B. W. Durkacz. 2004. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene 23:7322-7329. [DOI] [PubMed] [Google Scholar]
- 59.Vodenicharov, M. D., F. R. Sallmann, M. S. Satoh, and G. G. Poirier. 2000. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 28:3887-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zahradka, P., and K. Ebisuzaki. 1982. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127:579-585. [PubMed] [Google Scholar]